Abstract
Friedreich ataxia (FRDA) is an autosomal recessive degenerative disease caused by insufficient expression of frataxin (FXN), a mitochondrial iron-binding protein required for Fe-S cluster assembly. The development of treatments to increase FXN levels in FRDA requires elucidation of the steps involved in the biogenesis of functional FXN. The FXN mRNA is translated to a precursor polypeptide that is transported to the mitochondrial matrix and processed to at least two forms, FXN42–210 and FXN81–210. Previous reports suggested that FXN42–210 is a transient processing intermediate, whereas FXN81–210 represents the mature protein. However, we find that both FXN42–210 and FXN81–210 are present in control cell lines and tissues at steady-state, and that FXN42–210 is consistently more depleted than FXN81–210 in samples from FRDA patients. Moreover, FXN42–210 and FXN81–210 have strikingly different biochemical properties. A shorter N terminus correlates with monomeric configuration, labile iron binding, and dynamic contacts with components of the Fe-S cluster biosynthetic machinery, i.e. the sulfur donor complex NFS1·ISD11 and the scaffold ISCU. Conversely, a longer N terminus correlates with the ability to oligomerize, store iron, and form stable contacts with NFS1·ISD11 and ISCU. Monomeric FXN81–210 donates Fe2+ for Fe-S cluster assembly on ISCU, whereas oligomeric FXN42–210 donates either Fe2+ or Fe3+. These functionally distinct FXN isoforms seem capable to ensure incremental rates of Fe-S cluster synthesis from different mitochondrial iron pools. We suggest that the levels of both isoforms are relevant to FRDA pathophysiology and that the FXN81–210/FXN42–210 molar ratio should provide a useful parameter to optimize FXN augmentation and replacement therapies.
Keywords: Iron-Sulfur Protein, Mitochondria, Neurodegeneration, Protein Assembly, Protein Processing, Frataxin, Friedreich Ataxia, Protein Augmentation
Introduction
Friedreich ataxia (FRDA)2 (OMIM number 229300) is an autosomal recessive disease with an estimated incidence of 1:40,000. Most FRDA patients are apparently healthy at birth and during the first 5–10 years of life; then their gait becomes increasingly unsteady and wide-based and their voluntary movements uncoordinated. Many patients develop hypertrophic cardiomyopathy as well as diabetes, muscle weakness, and skeletal deformities. Although cognitive functions remain largely intact during disease progression, patients develop significant communication difficulties due to dysarthria, which is often compounded by vision and hearing loss. The majority of patients eventually become wheelchair-bound and dependent on others for most daily activities. Cardiac failure is a frequent cause of death at a young age (1).
The FRDA locus encodes a mitochondrial protein designated frataxin (FXN), which is expressed at much lower levels in FRDA patients compared with normal individuals (2). In most patients, FXN deficiency results from the presence of an expanded GAA repeat in the first intron of the FRDA gene (2) that causes transcriptional silencing (reviewed in Ref. 3). Although FXN is ubiquitously expressed, certain cells (dorsal root ganglia neurons, cardiomyocytes, and pancreatic beta cells) are exquisitely sensitive to frataxin depletion, and the degenerative loss of these particular cells accounts for the major clinical aspects of FRDA (1).
Extensive biochemical studies have shown that frataxins across species are conserved iron-binding proteins that can either provide iron for Fe-S cluster assembly and heme synthesis or store iron as a stable mineral (reviewed in Ref. 4). The loss of these properties accounts for impaired iron utilization and increased iron toxicity linked to frataxin deficiency in the mitochondria of such diverse organisms as Saccharomyces cerevisiae, Drosophila, mouse, and humans (5–8). In humans, the mitochondrial alterations caused by FXN deficiency lead to tissue-specific changes in various cellular pathways involved in antioxidant, metabolic, and inflammatory responses, thereby amplifying the pathophysiology of FRDA and promoting disease progression (9–13).
The multifaceted pathophysiology of FRDA may explain why pharmacological treatments with mitochondrial-targeted antioxidants or iron chelators have thus far shown limited effects both in patients and animal models of the disease (14–17). On the other hand, FXN augmentation therapies (reviewed in Ref. 18) are predicted to be able to correct both the mitochondrial and other cellular alterations that drive FRDA onset and progression. The development of these therapies requires a clear understanding of the biogenesis of functional FXN, which involves mitochondrial import of a precursor polypeptide (FXN1–210) and multiple proteolytic events. At least four processing products of FXN1–210 have thus far been characterized in vitro and/or in vivo. These products, which include FXN42–210, FXN56–210, FXN78–210, and FXN81–210, are normally generated upon mitochondrial import (19–21), although FXN81–210 was also detected in the cytoplasmic fraction of cultured human cells (22, 23). The amino termini of FXN42–210 and FXN56–210 were initially defined by our group via processing of radiolabeled FXN1–210 with purified mitochondrial processing peptidase (MPP). We proposed that FXN42–210 and FXN56–210 represented a processing intermediate and the mature form, respectively (19). FXN78–210 and other ∼14-kDa FXN products were initially detected during expression of FXN56–210 in Escherichia coli (24, 25) and during import of FXN1–210 by isolated mitochondria (19). These products were later found to result from cleavages in the region between serine 56 and serine 81, at sites that do not match the canonical consensus sequence recognized by MPP (26–28). In vitro these cleavages could be catalyzed by the proteolytic activity of dihydrolipoamide dehydrogenase (27) and could also be induced by iron-mediated chemical cleavage (i.e. FXN79–210) (28). Formation of FXN78–210 was recently observed in human cells (20); however, the mechanism(s) responsible for formation of ∼14-kDa FXN products in vivo are not yet defined. Last, FXN81–210 is the shortest form of FXN identified thus far (20, 22). It encompasses the core structure of frataxin homologues across species (26) and results from the cleavage of FXN42–210 by MPP (20). We missed FXN81–210 in our earlier study (19) due to its abnormal electrophoretic mobility on 12% Tris glycine SDS-PAGE that makes it run more slowly than FXN78–210 and very close to FXN56–210 (20). In addition, the conditions we used for N-terminal radiosequencing of the putative mature form were suitable to identify only cleavage sites upstream of residue 62 (19).
FXN81–210 was the only form that could be detected in two independent studies in a variety of normal human cells and tissues under native conditions (20, 22). In contrast, FXN42–210, FXN56–210, and FXN78–210 were detected only after artificial overexpression and/or accumulation of FXN1–210 in cultured cells (20, 22). These findings have led to the conclusion that FXN81–210 represents the mature functional form, whereas the other forms are transient (i.e. FXN42–210) or are generated only when the production of FXN81–210 is impaired (i.e. FXN56–210 and FXN78–210) (20, 22). The seemingly exclusive presence of FXN81–210 in vivo was surprising to us because the N-terminal region upstream of serine 81 enabled recombinant FXN56–210 to oligomerize and store iron in a nontoxic form in vitro (25, 29). Moreover, broad distributions of FXN were observed in heart tissue extract, suggesting that oligomeric forms of the protein were also present in vivo (30). In addition, the ability to oligomerize enabled the E. coli and yeast frataxin homologues (CyaY and Yfh1) to detoxify iron and promote Fe-S cluster assembly (31–34). We reasoned that this property should be maintained in human cells, if not by FXN81–210, by another as yet undefined FXN isoform. The possibility that other functional isoforms might exist was also suggested by the fact that cells engineered to express primarily FXN42–210 and/or FXN56–210, without concomitant production of FXN81–210, were viable and exhibited aconitase activity, a sensitive marker of Fe-S cluster biosynthesis (20–22).
Here, by use of improved Western blotting we show that multiple isoforms of FXN are present in vivo, and that they are differentially depleted in FRDA cell lines and tissues. We characterize the longest and the shortest of these isoforms (i.e. FXN42–210 and FXN81–210) and demonstrate that they exist in oligomeric and monomeric configurations, respectively, which correlate with complementary roles in Fe-S cluster assembly. The data suggest that alternative processing of the FXN N terminus may respond to changes in metabolic requirements. Our findings highlight the importance of assessing how FXN augmentation and replacement therapies influence not only total FXN levels but also the biogenesis of specific FXN isoforms.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins Used in This Study
For expression of rec-FXN isoforms, E. coli strain BL21(DE3) (Novagen) was transformed with vector pETHF-42 (FXN cDNA coding for residues 42–210 in pET24a+ vector), pETHF-56 (FXN cDNA coding for residues 56–210 in pET24a+ vector) (25, 29), pETHF-78 (FXN cDNA coding for residues 78–210 in pET24a+ vector), or pETHF-81 (FXN cDNA coding for residues 81–210 in pET24a+ vector). Monomeric and oligomeric FXN species were expressed in E. coli and purified as described previously (25, 29). Detailed purification procedures for FXN42–210 and FXN81–210 are described under supplemental Methods.
For expression of human ISCUHis, a DNA fragment corresponding to the predicted mature form of ISCU (residues 35–168) (35) was cloned in the NdeI and XhoI sites of vector pCDFDuet (Novagen), which resulted in the addition of a C-terminal His6 tag. For production of untagged ISCU, a DNA fragment corresponding to residues 35–168 was cloned in the NdeI and XhoI sites of pET28b, which resulted in the addition of an N-terminal His6 tag followed by a thrombin cleavage site. Protein was expressed in E. coli strain Rosetta (DE3) pLysS (Novagen), and after nickel affinity chromatography, the histidine tag was removed by incubation with thrombin (Sigma) (0.1 mg of thrombin/100 mg of HisISCU) for ∼12 h at 4 °C. Uncleaved HisISCU was removed by nickel affinity chromatography, and fractions corresponding to untagged ISCU were purified by Superdex 75 gel filtration chromatography. Additional details are provided under supplemental Methods.
Two DNA fragments corresponding to residues 56–458 of NFS1 and 6–92 of ISD11 (32, 36) were cloned into the XhoI and BamHI sites of vector pET15b (Novagen), and the NcoI and EcoRI sites of vector pCDFDuet (Novagen), respectively. This resulted in the addition of an N-terminal His6 tag followed by a thrombin cleavage site on NFS1. HisNFS1 and ISD11 were co-expressed in E. coli and co-purified as a stable complex essentially as described for their yeast homologues (32). Detailed purification procedures are described under supplemental Methods.
Analysis of Native FXN Isoforms in Human Lymphoblastoid Cells
Immortalized lymphoblast cell lines were obtained from the Coriell Cell Repository (Coriell Institute for Medical Research) and the Biospecimens Accessioning Processing Laboratory (Mayo Clinic). FRDA carrier and patient cell lines were from the Coriell Cell Repository, including GM16214 (15-year-old male with FRDA onset at 9 years of age; cardiomyopathy; areflexia; weakness; scoliosis; homozygous for the GAA expansion with alleles of ∼600 and 700 repeats), GM16197 (14-year-old male with FRDA onset at 8½ years of age; mild cardiomyopathy; areflexia; weakness; sensory loss; scoliosis with spinal fusion; homozygous for the GAA expansion with alleles of ∼760 and 830 repeats), GM16215 (41-year-old female, mother of GM16214 has an allele with GAA expansion of 830 repeats and one normal allele), GM16200 (34-year-old female, mother of GM16197 has an allele with GAA expansion of 830 repeats and a normal allele). Lymphoblastoid cell lines from healthy individuals were from the Mayo Biospecimens Accessioning Processing Laboratory (numbers 326365 and 287097) and the Coriell Cell Repository, including GM03798 (10-year-old male), GM05398 (44-year-old male), GM07521 (19-year-old female), GM14406 (41-year-old female), and GM14907 (28-year-old male). In all cases, cultures were initiated with frozen stocks, and cells were grown in 20 ml of RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum and 1% penicillin/streptomycin/glutamine for 2 weeks in T-75 tissue culture flasks. Every 3 days, 10 ml of culture medium were replaced with an equivalent volume of fresh medium. Fifteen ml of culture (cell density 1.6–2.5 × 105 cells/ml) were harvested by centrifugation, washed with saline buffer with protease inhibitors (0.85% NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 tablet/50 ml of complete protease inhibitor mixture without EDTA (Roche Applied Science)), and cell pellets were immediately frozen in dry ice and stored at −80 °C. Frozen cell pellets were suspended in 400 μl of ice-cold extraction buffer (1.5% lauryl maltoside, 100 mm NaCl, 25 mm HEPES-KOH, pH 7.3) with protease inhibitors as above, and incubated on ice for 30 min with periodical mixing. These samples were cleared of insoluble material by centrifugation at 20,000 × g for 15 min at 4 °C. Total protein concentration was measured by the BCA kit (ThermoFisher Scientific). Typically, this procedure yields ∼500 μl of total cell extract with a protein concentration of ∼3–4 mg/ml. For gel filtration chromatography, 250 μl of total cell extract was loaded on a Superdex 75 column. Eighteen fractions (0.5 ml each) covering the entire fractionation range of the column were collected and an aliquot from each fraction (40 μl) was analyzed by SDS-PAGE and Western blotting.
Western Blot Analysis of Native FXN Isoforms in Post-mortem FRDA Tissues
Tissues were provided by Dr. Arnulf H. Koeppen (Veterans Affairs Medical Center, Albany, NY). Patient 1 was a 38-year-old man with GAA expansions of 934 and 249 repeats; onset was in childhood but disability did not become complete until age 35; cardiac involvement was modest but at age 36 atrioventricular node ablation was required due to ectopic supraventricular tachycardia; much of the later care of the patient was for his dilated cardiomyopathy; patient 2 was a 10-year-old boy with GAA expansions of 1016 and 1016 repeats; diagnosis of cardiomyopathy was made at the age of 2; first neurological symptoms and signs occurred at the age of 6, and diabetes mellitus at the age of 8. Cerebellum was from nine non-FRDA males ranging from 49 to 82 years of age. Frozen tissue was minced with a razor blade in a Petri dish on dry ice, immediately transferred to a 1-ml tissue grinder, resuspended in ∼300–500 μl of buffer (1.5% lauryl maltoside, 100 mm NaCl, 25 mm HEPES-KOH, pH 7.3, 5 mm EDTA, 2 mm Pefabloc, and protease inhibitor mixture as above), and homogenized by hand on ice with a tightly fitting glass pestle. Tissue homogenates were incubated for 30 min on ice with periodic homogenization, transferred to Eppendorf tubes, and centrifuged at 20,000 × g for 10 min at 4 °C. Soluble supernatants were collected, aliquoted, and immediately frozen and stored at −80 °C. Aliquots were used for determination of protein concentration with the BCA kit (ThermoFisher Scientific) or Western blotting.
Gel Filtration Chromatography Coupled with Co-immunoprecipitation
Polyclonal antibodies were immobilized on Protein A magnetic beads (New England BioLabs) by cross-linking with dimethyl pimelidate dihydrochloride, per the manufacturer's protocol. High (from ∼190–100 kDa) and low (from ∼20.5–10 kDa) molecular mass fractions were pooled, 30 μl of beads with or without cross-linked antibody were added to each pool, and samples were incubated overnight at 4 °C on a Nutator mixer. Beads were collected by magnet, washed 4 times in 1 ml of 10 mm HEPES-KOH, pH 7.3, 100 mm NaCl, 1 mm EDTA, 0.01% Triton X-100, 2 mm phenylmethylsulphonyl fluoride and protease inhibitor mixture, and after the final wash, antibody-bound proteins were eluted by boiling in 40 μl of Laemmli buffer.
Fe-S Cluster Assembly Assay
To produce Fe3+-loaded FXN, FXN isoforms were aerobically incubated with Fe(NH4)2(SO4)2 at a Fe2+ atom/protein subunit ratio of 1:10 for ∼16 h in 20 mm HEPES-KOH, pH 7.3, 150 mm NaCl (32), in parallel with samples containing the same iron concentration but no FXN protein. For Fe-S cluster synthesis assays, all buffers and solutions were purged with argon gas (<0.2 ppm O2) in vials tightly sealed with rubber septa. Each reaction was assembled in a quartz cuvette that was purged with nitrogen gas before and after addition of each reaction component, and finally sealed with a tight fitting rubber septum. [2Fe-2S] synthesis was started with the addition of 50 μm iron (60 μl), provided as holo-FXN protein or directly, to anaerobic reactions containing 5 μm ISCUHis, 5 μm HisNFS1·ISD11, 5 mm DTT, and 2 mm l-cysteine in 20 mm HEPES-KOH, pH 7.3, 150 mm NaCl (600 μl final reaction volume). [2Fe-2S] synthesis was monitored spectrophotometrically at A426. Reactions containing Fe2+-loaded FXN protein were similarly set up except that rec-FXN isoforms were incubated with Fe(NH4)2(SO4)2 anaerobically for 20 min.
In Vitro Pulldown Assay
In vitro pulldown assays were performed essentially as described previously (32). See supplemental Methods for details.
SDS-PAGE Conditions
For analysis of different FXN isoforms we used homemade 14% separating gels (total length ∼12.5 cm) overlaid with 4% stacking gels prepared from a stock solution of acrylamide:bisacrylamide = 40:1.7. Electrophoresis was carried out at room temperature at 180 V for ∼3.5 h, and continued at 260 V until the bromphenol blue reached the bottom of the separating gel, and further continued for an additional 1.5 h.
RESULTS
Multiple FXN Isoforms Are Detected in Cultured Cells and Post-mortem Tissues
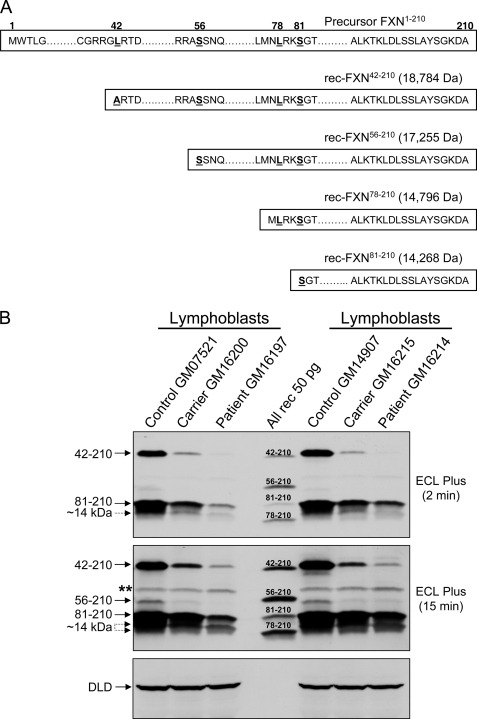
We produced recombinant (rec) FXN proteins corresponding to the FXN isoforms recently identified in human cells overexpressing FXN1–210 (20) (Fig. 1A), and generated two anti-FXN polyclonal antibodies in rabbits (PAC 2517 and PAC 2518). We then used these reagents as well as anti-FXN monoclonal antibody (MitoSciences, Inc.), coupled with high-resolution 14% SDS-PAGE, to define which form(s) of FXN are present in human cells and tissues at steady state. Lymphoblastoid cell lines from two different controls as well as two FRDA carriers and their affected children were cultured and processed in parallel, and total cell lysates were analyzed by Western blotting with PAC 2517 (Fig. 1B). The controls contained similar levels of two major protein bands that co-migrated with rec-FXN42–210 and rec-FXN81–210, respectively. They also contained at least one band migrating between rec-FXN81–210 and rec-FXN78–210, corresponding to a ∼14 kDa FXN product slightly larger than rec-FXN78–210, possibly FXN79–210 (28). Longer exposures of the blot revealed a protein band co-migrating with rec-FXN56–210, and an additional ∼14 kDa product co-migrating with rec-FXN78–210. All of these bands were depleted in cultured lymphoblastoid cells from two FRDA carriers, and were further depleted in cells from the carriers' affected children, as would be expected for authentic FXN isoforms (Fig. 1B). Another mitochondrial matrix protein (dihydrolipoamide dehydrogenase) was otherwise present in equivalent amounts in controls, carriers, and patients (Fig. 1B). Very similar FXN protein profiles were observed with PAC 2518 and the monoclonal antibody, although the latter antibody cross-reacted less strongly with all FXN isoforms (supplemental Fig. S1). Varying levels of the same isoforms were detected in cultured lymphoblastoid cells, fibroblasts, and bronchial epithelial cells from healthy males and females of varying ages, as well as a post-mortem heart tissue sample from an elderly male who had died of lung cancer, and in a yeast frataxin knock-out strain expressing the wild-type human frataxin precursor (yfh1Δ[FXN1–210]) (37) (supplemental Fig. S2, a and b).
FIGURE 1.
Recombinant FXN isoforms used in this study and native FXN isoform profiles in human lymphoblastoid cells. A, DNA fragments, including an ATG codon in-frame with codons 42–210, 56–210, 78–210, or 81–210 of human frataxin (FXN), were expressed in E. coli and purified as described under “Experimental Procedures.” The accurate molecular mass (shown in parentheses) of each rec-FXN species were determined by electrospray ionization mass spectrometry. According to the length parameter rule (51), the initiator methionine was cleaved from rec-FXN56–210 and rec-FXN81–210 during expression in E. coli, whereas cleavage of the initiator methionine from rec-FXN42–210 was achieved after substitution of alanine for leucine at position 42. This causes a slight increase in electrophoretic mobility relative to native FXN42–210 (see Fig. 1B). Leucine 78 in rec-FXN78–210 was not replaced and this protein retained the initiator methionine as expected without obvious effects on the electrophoretic mobility relative to native FXN78–210 (see Fig. 1B). B, lymphoblastoid cell lines from the Coriell Cell Repository were as follows: GM07521 (female, 19 years old, normal GAA repeats), GM16200 (FRDA carrier, female, 34 years old, mother of GM16197, 830/normal GAA repeats), GM16197 (FRDA male, 14 years old, 760 and 830 GAA repeats), GM14907 (male, 28 years old, normal GAA repeats), GM16215 (FRDA carrier, female, 41 years old, mother of GM16214, 830/normal GAA repeats), and GM16214 (FRDA male, 15 years old, 600 and 700 GAA repeats). Cell extracts were prepared from exponentially growing cell cultures, and each sample (50 μg of total protein) was analyzed by Western blotting with PAC 2517 anti-FXN polyclonal antibody, following the procedure described under “Experimental Procedures.” Rec-FXN proteins (All rec, 50 pg each) were mixed together and used as standards. The double asterisk denotes a nonspecific cross-reacting band that unlike FXN isoforms is present in equivalent amounts in the controls, carriers, and patients; this band was recognized weakly by PAC 2517 and more strongly by PAC 2518, whereas it was not recognized by a monoclonal antibody (see supplemental Fig. S1). DLD, dihydrolipoamide dehydrogenase. Two different exposures of the same blot are shown.
FXN Isoforms Are Differentially Reduced in Lymphoblastoid Cells and Disease-relevant Tissues from FRDA Patients
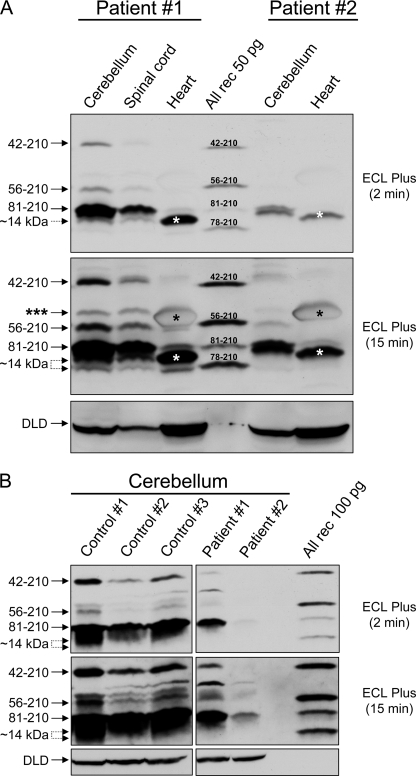
Next, we analyzed the FXN protein profile in some of the tissues most affected by frataxin deficiency (1). Cerebellum and heart were available from two FRDA patients (numbers 1 and 2) who had suffered from a more slowly and a more rapidly progressing disease, respectively; spinal cord was available from patient 1 only; cerebellum was available from nine non-FRDA individuals (see “Experimental Procedures” for clinical details).
As analyzed with PAC 2517, PAC 2518, or the monoclonal antibody, the FXN profiles in post-mortem FRDA tissues looked similar to those in FRDA lymphoblastoid cells, with all FXN isoforms being progressively depleted from patient 1 to patient 2 according to disease severity (Fig. 2A and supplemental Fig. S2c). The depletion of FXN isoforms in the two patients was especially apparent upon direct comparisons with non-FRDA samples of cerebellum (Fig. 2B) and heart (supplemental Fig. S2d). In all patient and control tissues analyzed, we detected similar levels of full-length dihydrolipoamide dehydrogenase, indicating that the FXN profiles described above probably did not result from generalized post-mortem autolysis of mitochondrial matrix proteins (Fig. 2, A and B, and supplemental Fig. S2d).
FIGURE 2.
FXN isoform profiles in FRDA post-mortem tissues. A, autoptic samples from the indicated tissues were obtained for FRDA patients 1 (male, 39 years old, 934 and 249 GAA repeats) and 2 (male, 10 years old, 1016 and 1016 GAA repeats). Tissue extracts were prepared as described under “Experimental Procedures.” One hundred μg of total protein was loaded in each lane, and analyzed by Western blotting with PAC 2517 as described above. The triple asterisk denotes a band in cerebellum and spinal cord, which does not co-migrate with any of the known FXN isoforms; this band was also detected with PAC 2518 and the monoclonal antibody (supplemental Fig. S2c) and its significance is undetermined. The single asterisk denotes an abundant protein present in heart, probably myoglobin, which migrates close to FXN56–210. In the hearts of both patients, the 14-kDa region of the blot was dominated by a strong band migrating between FXN81–210 and FXN78–210 (white asterisk). Further analyses using either PAC 2518 or the monoclonal antibody indicated that this band most likely consisted of low amounts of ∼14-kDa FXN that were detected by all antibodies, and larger amounts of a co-migrating protein that was only detected by PAC 2517 (supplemental Fig. S2c). DLD, dihydrolipoamide dehydrogenase. Two different exposures of the same blot are shown. B, comparison of autoptic samples of cerebellum from three non-FRDA individuals (males, 52 to 62 years old) and the two patients described above (100 μg of total protein in each case) as analyzed by Western blotting with PAC 2518.
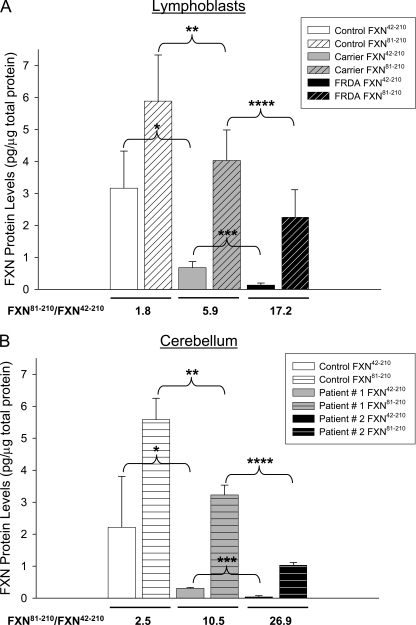
We focused on FXN42–210 and FXN81–210, the longest and shortest of the isoforms known to result from processing of FXN1–210 by MPP (19, 20, 22). To quantify these isoforms, known amounts of recombinant FXN42–210 and FXN81–210 were analyzed side by side with control, carrier, and patient samples (e.g. Figs. 1B and 2A), and used as internal standards. This demonstrated significantly different levels of both FXN81–210 and FXN42–210 between controls and carriers, and between carriers and patients (Fig. 3, A and B). In lymphoblastoid cells, the molar ratio of FXN81–210 to FXN42–210 increased in carriers versus controls (∼3-fold) and in patients versus carriers (∼3-fold) (Fig. 3A). In addition, cerebella from patients 1 and 2 showed molar ratios of FXN81–210 to FXN42–210 that were ∼4- and ∼10-fold higher, respectively, relative to controls, in a manner that appeared to correlate with disease severity (Fig. 3B).
FIGURE 3.
Quantification of FXN42–210 and FXN81–210 isoforms in lymphoblastoid cells and post-mortem tissues. A, densitometry data were obtained for 2 carriers and 2 patients (6 measurements per group from 4 different blots, of which two were probed with PAC 2517, one with PAC 2518, and one with the monoclonal antibody), and for 7 controls (13 measurements from 5 different blots, of which two were probed with PAC 2517, two with PAC 2518, and one with the monoclonal antibody). Shown are the mean ± S.D. p values were: *, p ≤ 2.3 × 10−6; **, p ≤ 0.0025; ***, p ≤ 2.7 × 10−5; ****, p ≤ 0.005. The molar ratio of FXN81–210 to FXN42–210 shown under each set of data were calculated from the mean values. B, densitometry data were obtained for patients 1 and 2 (3 measurements each from 3 different blots, of which two were probed with PAC 2517 and one with PAC 2518), and for 9 controls (18 measurements from 2 different blots, of which one was probed with PAC 2517 and one with PAC 2518). The 9 controls were males with ages ranging from 49 to 82 years. Shown are the mean ± S.D. p values were: *, p ≤ 2.9 × 10−5; **, p ≤ 4.5 × 10−5; ***, p ≤ 1.3 × 10−4; ****, p ≤ 1.5 × 10−4. The molar ratio of FXN81–210 to FXN42–210 shown under each set of data were calculated from the mean values. In both A and B, known amounts of rec-FXN42–210 and rec-FXN81–210 were analyzed side by side with unknown samples and served as internal standards, enabling conversion of densitometry values to pg of FXN protein per μg of total protein.
The Length of the FXN N Terminus Determines Ability to Oligomerize
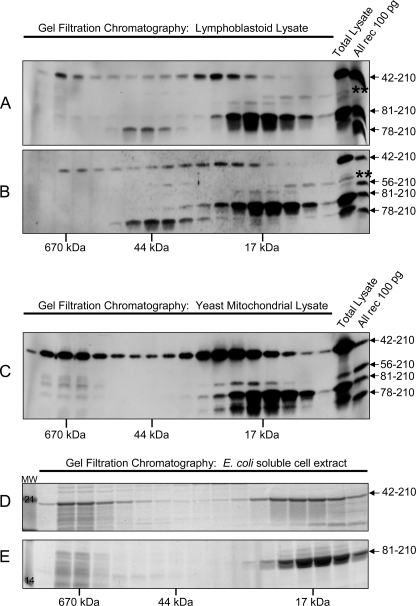
The consistent presence of FXN42–210 in cultured cells and tissues prompted us to investigate whether FXN42–210 might have an independent function, in addition to serving as a substrate for the production of FXN81–210 (20). We used gel filtration chromatography of soluble extracts prepared from lymphoblastoid cell lines from two normal controls to assess gross differences in the physical properties of native FXN42–210 and FXN81–210. This analysis consistently revealed a low molecular weight pool and a high molecular weight pool for native FXN42–210, in contrast to a well defined low molecular weight pool for native FXN81–210 (Fig. 4, A and B). Similar results were obtained upon gel filtration chromatography of soluble mitochondrial extracts from the yfh1Δ[FXN1–210] yeast strain (Fig. 4C). Similarly, upon overexpression of rec-FXN42–210 or rec-FXN81–210 in E. coli, gel filtration chromatography of soluble E. coli cell extracts revealed a low and a high molecular weight pool for rec-FXN42–210, and one low molecular weight pool for rec-FXN81–210 (Fig. 4, D and E). The low and high molecular weight pools of rec-FXN42–210 were independently purified and were found to represent stable monomeric and oligomeric forms, respectively, of rec-FXN42–210 (Fig. 5, A and B) (designated rec-FXN42–210 and oligomeric rec-FXN42–210, respectively). Purification of rec-FXN81–210 yielded a stable monomeric form (designated rec-FXN81–210) (Fig. 5C). In these analyses, native or recombinant FXN81–210 was eluted from the gel filtration column as a homogeneous peak of ∼17 kDa (Fig. 4, A–E). On the other hand, native FXN42–210 (i.e. the low molecular weight pool of the protein) was eluted with an apparent molecular mass slightly larger than observed for rec-FXN42–210 (Fig. 4, A–C versus D).
FIGURE 4.
Distributions of native and rec-FXN isoforms in cells. A and B, soluble extracts were prepared from two different lymphoblastoid cell lines from the Biospecimens Accessioning Processing Laboratory (Mayo Clinic) as described under “Experimental Procedures,” and ∼1 mg of total protein was analyzed by gel filtration chromatography on a Superdex 75 column. Fractions comprising the entire molecular mass fractionation range of the column were analyzed by Western blotting with PAC 2517. Rec-FXN proteins (All rec) were used as standards. The double asterisk denotes a nonspecific cross-reacting band as described in the legend to Fig. 1B. Native FXN78–210 was eluted with apparent molecular mass of 44 kDa in both A and B; the significance of this result remains to be established. C, a culture of the yfh1Δ[FXN1–210] strain was grown for ∼20 h at 30 °C in rich medium with galactose as the carbon source, after which mitochondria were isolated (32). The soluble mitochondrial fraction (∼5 mg of total protein) was analyzed by gel filtration chromatography and Western blotting as described above. D and E, the indicated recombinant FXN proteins were expressed in E. coli, and after sonication of bacterial cells and centrifugation, soluble cell extracts were analyzed by Superdex 75 gel filtration chromatography, SDS-PAGE, and staining with SYPRO Orange. MW, molecular weight markers.
FIGURE 5.
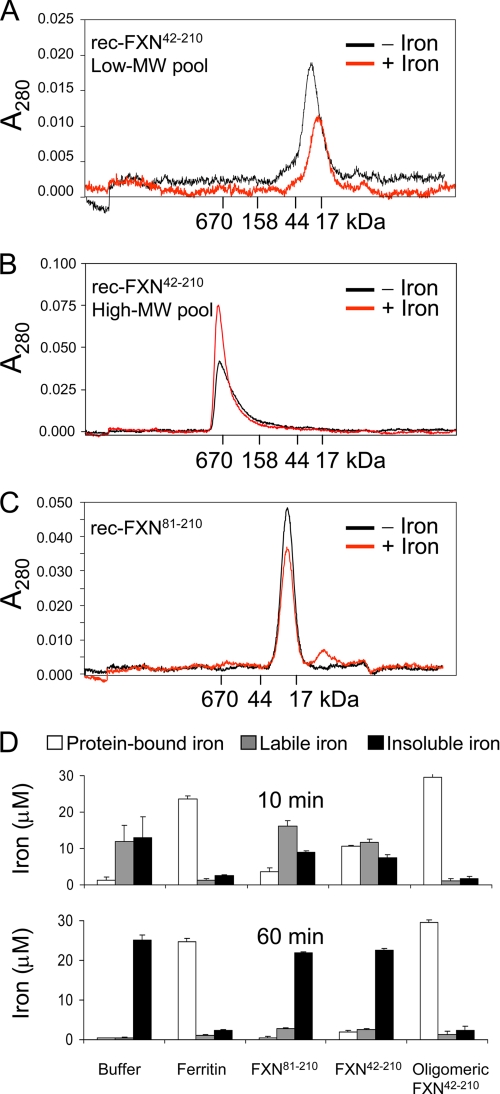
Oligomerization and Fe3+ chelation properties of rec-FXN42–210 and rec-FXN81–210. Purified low (A) and high (B) molecular weight (MW) pools of rec-FXN42–210 (20 μm each) or (C) the rec-FXN81–210 pool (40 μm), were incubated in the absence or presence of 10 molar eq of Fe2+, as described under “Experimental Procedures.” Each reaction was centrifuged for 5 min at 20,000 × g and the supernatant was directly analyzed by Superdex 200 (A and B) or Superdex 75 (C) gel filtration chromatography. In each chromatogram, −Iron and +Iron denote the iron-free and the iron-treated FXN protein. D, each purified rec-FXN protein or a commercial preparation of horse spleen ferritin (3 μm each) was incubated with 30 μm Fe2+ for 10 or 60 min under conditions that promote iron oxidation (10 mm HEPES-KOH, pH 7.3, at 30 °C); samples were then subjected to ultrafiltration in a Ultrafree-0.5 device (Millipore) at 10,000 × g for 10 min at 4 °C. Iron levels were measured in the concentrated (protein-bound iron), filterable (labile iron), and insoluble fractions with the chelator (α-α′-bipyridine) after addition of a strong reducing agent (dithionite) (29). Shown are the mean ± S.D. from 3 independent experiments.
Native FXN42–210 Is Not a Post-translationally Modified Version of FXN81–210
Prior to the present study, the N terminus of FXN42–210 had been defined by N-terminal radiosequencing in vitro (19); moreover, two independent groups had analyzed the identity of FXN81–210 and FXN42–210 in cultured human cells overexpressing FXN1–210 (20, 22). Both groups were able to directly identify the N terminus of FXN81–210. One group further showed that the assumed FXN42–210 isoform disappeared when residues 39–40 (i.e. two arginine residues upstream of the cleavage site used by MPP to generate FXN42–210 in vitro) were replaced with glycine residues. Given that MPP typically cleaves two residues downstream of an arginine residue (the arginine −2 rule) (38), this result demonstrated that the assumed FXN42–210 isoform was produced in vivo by cleavage between residues 40 and 41 or 41 and 42, the latter being the cleavage site identified with purified MPP in vitro (19). In addition, both groups showed that disruption of the cleavage site used by MPP to generate FXN81–210 caused the accumulation of FXN42–210, indicating that FXN42–210 was a processing intermediate in the two-step maturation of FXN81–210. Here, to further exclude the possibility that the assumed FXN42–210 isoform might be a post-translationally modified variant of FXN81–210, we analyzed it by peptide mass fingerprinting. Because the levels of native FXN isoforms present in lymphoblastoid cells were limited, we took advantage of the yfh1Δ[FXN1–210] strain overexpressing the human frataxin precursor. This approach was valid because yfh1Δ[FXN1–210] cells and human cells showed very similar patterns of FXN isoforms (supplemental Fig. S2a), and very similar distributions of these isoforms (Fig. 4, A and B versus C). Moreover, we will show below that in both cell types oligomeric FXN42–210 formed stable complexes with human NFS1 or yeast Nfs1, whereas FXN81–210 did not. Hence, each isoform maintained its unique properties in yfh1Δ[FXN1–210] cells, consistent with the fact that this strain was phenotypically normal (37).
As compared with the sequence of the human frataxin precursor (FXN1–210), peptide mass fingerprinting of the native FXN81–210 protein immunoprecipitated from yeast mitochondrial lysate showed (i) absence of the predicted tryptic peptides upstream of serine 81; (ii) presence of the most predicted tryptic peptides downstream of serine 81 (corresponding to ∼72% coverage of the 81–210 sequence); and (iii) identification of one tryptic peptide starting at serine 81, as reported previously by others (20) (supplemental Fig. S3). In contrast, in the case of the native FXN42–210 isoform, the data showed (i) absence of predicted tryptic peptides upstream of arginine 43; (ii) presence of the most predicted tryptic peptides downstream of arginine 43, including three peptides upstream of serine 81 (corresponding to ∼72% coverage of the 42–210 sequence); and (iii) identification of one tryptic peptide starting at residue 44 (supplemental Fig. S3). The corresponding recombinant isoforms yielded essentially identical peptide mass fingerprinting results, except for a more complete coverage in the C-terminal regions (supplemental Fig. S3). These data excluded the possibility that the assumed FXN42–210 isoform could be a post-translationally modified version of FXN81–210. The data also allowed us to map the N terminus of the assumed FXN42–210 isoform within three residues downstream of arginine 40, i.e. ±1 residue from leucine 42, the N terminus originally identified by N-terminal radiosequencing with purified MPP (19).
The Length of the FXN N Terminus Determines Stable Versus Labile Iron Binding
Next we analyzed if the monomeric rec-FXN42–210 or rec-FXN81–210 exhibited iron-dependent oligomerization when incubated aerobically with 10 or 40 molar eq of Fe2+, conditions that induce oligomerization of monomeric Yfh1p and stable accumulation of iron inside the protein (39, 40). As analyzed by gel filtration chromatography and SDS-PAGE, the distribution of each protein did not shift significantly in the absence or presence of iron (Fig. 5, A and C, and data not shown), consistent with the lack of obvious oligomerization. Moreover, during a centrifugation step performed before gel filtration chromatography, iron-containing samples exhibited iron precipitation, which correlated with a reduction in the intensity of the monomeric protein peak that was eluted from the column (Fig. 5, A and C), consistent with formation of insoluble iron-protein aggregates. In contrast, when oligomeric rec-FXN42–210 was incubated with 10 or 40 molar eq of Fe2+, there was no detectable iron precipitation upon centrifugation, and the intensity of the oligomeric protein peak was increased in iron-treated versus untreated samples (Fig. 5B), consistent with accumulation of Fe3+ inside oligomeric rec-FXN42–210 (30). In agreement with these results, iron-chelation assays revealed quantitative and stable chelation of iron by oligomeric rec-FXN42–210 but much lower and labile iron chelation by monomeric rec-FXN42–210 or rec-FXN81–210 (Fig. 5D).
The Length of the FXN N Terminus Determines the Stability of Interactions with Components of the Fe-S Cluster Biosynthetic Machinery in Vitro
Our recent study of highly conserved components of the yeast Fe-S cluster assembly machinery has shown that the iron-chaperone, Yfh1, and the sulfur-donor complex, Nfs1·Isd11, directly bind to each other, and that both Yfh1 and Nfs1·Isd11 can directly bind to the Fe-S cluster scaffold, Isu1 (32). Binding of Yfh1 to Nfs1·Isd11 or Isu1 requires oligomerization of Yfh1 and can occur in an iron-independent manner (32). Here, we produced recombinant forms of the human Fe-S cluster assembly machinery, the sulfur donor complex NFS1·ISD11 and the scaffold ISCU (homologous to yeast Nfs1·Isd11 and Isu1, respectively) (supplemental Fig. S4). The NFS1·ISD11 complex was purified as an apparent octamer ([NFS12-ISD112]2) and ISCU as an apparent dimer (data not shown) consistent with previous reports (36, 41). These proteins were tested for their ability to bind to rec-FXN81–210 and rec-FXN42–210 by use of pulldown assays using histidine-tagged versions of NFS1 or ISCU, a method used in previous studies of Yfh1/Isu1 and Yfh1/Nfs1 interactions (32, 42). For all rec-FXN isoforms and ISCU, the protein concentration was calculated per subunit; for the HisNFS1·ISD11 complex, it was calculated per NFS1·ISD11 dimer.
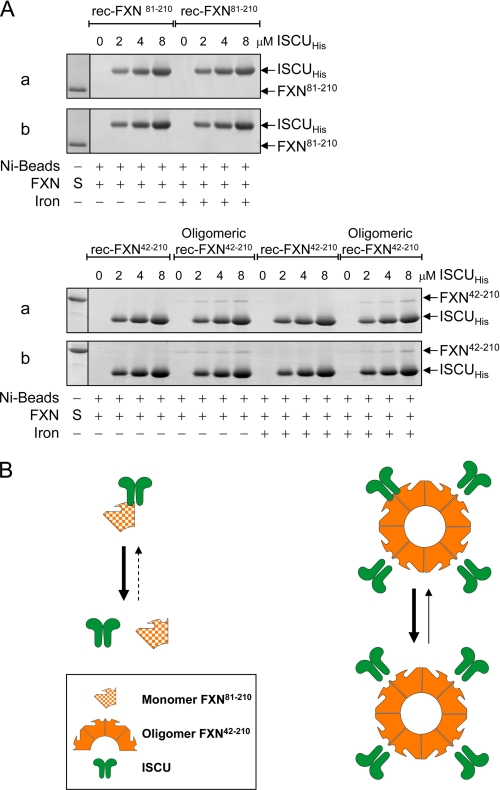
A fixed concentration of rec-FXN81–210 (4 μm) was not pulled-down with increasing concentrations of ISCUHis up to 8 μm, regardless of whether rec-FXN81–210 had been preincubated with iron (Fig. 6A). The same was true for rec-FXN42–210 (Fig. 6A). Weak but reproducible binding to ISCUHis was otherwise observed with oligomeric rec-FXN42–210, which was not significantly influenced by iron-loading of FXN (Fig. 6A) or by direct addition of iron to the binding reaction (data not shown). A possible interpretation of these results is that monomeric FXN (rec-FXN81–210 or rec-FXN42–210) forms limited and hence transient contacts with ISCUHis that cannot be detected by pulldown assay; on the other hand, oligomerization enables more extended contacts between ISCUHis and adjacent subunits of oligomeric rec-FXN42–210 such that a small fraction of the total rec-FXN42–210 present in the binding reaction can be pulled down with ISCUHis (Fig. 6B).
FIGURE 6.
Interactions of rec-FXN isoforms with purified ISCU. A, in vitro pulldown assays were performed with a fixed concentration (4 μm or 400 pmol of FXN subunit) of iron-free (−) or iron-loaded (+) preparations of the indicated rec-FXN isoforms and increasing concentrations of ISCUHis in 20 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 30 mm imidazole, 0.01% Triton X-100, 5 mm β-mercaptoethanol, as described in detail under supplemental Methods. After 60 min of incubation with Ni-NTA Superflow agarose beads (nickel beads), bound protein (100%) was separated on 14% BisTris SDS-PAGE (FXN81–210) or 14% Tris glycine SDS-PAGE (FXN42–210 or oligomeric FXN42–210), and detected by staining with SYPRO Orange. S denotes 100 pmol of each rec-FXN protein used as a reference. 0 denotes control reactions containing nickel beads and the indicated FXN isoform but not ISCUHis. The reproducible results of two independent assays (a and b) are shown. B, schematic representation of the pulldown results shown in A.
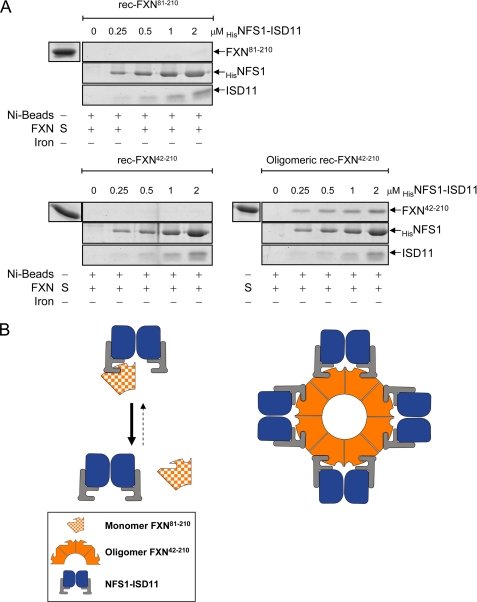
Binding of FXN proteins to the NFS1·ISD11 complex was similarly examined. Neither rec-FXN81–210 nor rec-FXN42–210 (2 or 4 μm) could be pulled down with increasing concentrations of HisNFS1·ISD11 up to 8 μm (Fig. 7A and data not shown). In contrast, oligomeric rec-FXN42–210 was able to bind tightly to HisNFS1·ISD11, with a binding affinity ∼10-fold higher than observed with ISCUHis (Figs. 6A versus 7A). Preloading the rec-FXN proteins with iron or adding iron directly to the binding reaction did not change their respective affinities for HisNFS1·ISD11 (data not shown). A possible interpretation of these results is that monomeric FXN (rec-FXN81–210 or rec-FXN42–210) can only form limited hence transient contacts with the HisNFS1·ISD11 complex, which cannot be detected by pulldown assay; however, oligomerization enables more extended and hence more stable contacts between the HisNFS1·ISD11 complex and adjacent subunits of oligomeric rec-FXN42–210 (Fig. 7B). Based on previous reports (32, 43), in Fig. 7B interactions between FXN isoforms and HisNFS1·ISD11 are hypothesized to be mediated by direct FXN-ISD11 contacts, although isolated ISD11 could not be generated to test this point directly.
FIGURE 7.
Interactions of rec-FXN isoforms with purified NFS1·ISD11. A, each FXN isoform (2 μm or 200 pmol of FXN subunit) was incubated with the indicated concentrations of HisNFS1·ISD11. Pulldown assays were carried out and bound proteins analyzed using the same conditions described in the legend to Fig. 6A. 0 denotes control reactions containing nickel beads and the indicated FXN isoform but not HisNFS1·ISD11. B, schematic representation of the pulldown results.
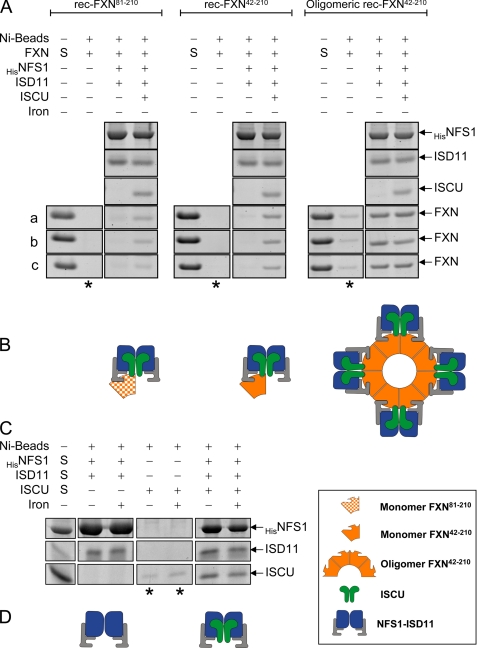
Next, rec-FXN proteins (2 μm) were incubated with an equivalent concentration of HisNFS1·ISD11 in the absence or presence of an equivalent concentration of untagged ISCU, and protein-protein interactions were again analyzed by pulldown assay. Interestingly, in the presence of ISCU, both rec-FXN81–210 and rec-FXN42–210 were pulled down with the HisNFS1·ISD11 complex (Fig. 8A). A possible interpretation of these results is that monomeric FXN (rec-FXN81–210 or rec-FXN42–210) binds more tightly to the HisNFS1·ISD11·ISCU complex than to isolated HisNFS1·ISD11 or ISCU, because of simultaneous ISD11-FXN and ISCU-FXN contacts (Fig. 8B). The possibility that monomeric FXN may stabilize HisNFS1·ISD11·ISCU contacts does seem less likely given that ISCU was efficiently pulled down with HisNFS1·ISD11 in the absence of any rec-FXN isoforms (Fig. 8C). Oligomeric rec-FXN42–210 once again exhibited strong binding to HisNFS1·ISD11, which was not apparently influenced by the absence or presence of ISCU (Fig. 8A). These data suggest the models proposed in Fig. 8, B and D.
FIGURE 8.
Interactions of rec-FXN isoforms with purified ISCU and NFS1·ISD11. A, all indicated proteins were used at a final concentration of 2 μm. Pulldown assays were carried out and analyzed using the same conditions described in the legend to Fig. 6A. Both HisNFS1·ISD11 complex and untagged ISCU were used in this experiment. The reproducible amounts of each rec-FXN isoform pulled down in three independent assays (a–c) are shown. In each set, the second lane (denoted by an asterisk under the lane) shows control reactions containing nickel beads and the indicated FXN isoform but not HisNFS1·ISD11. B, schematic representation of the pulldown results shown in A. C, HisNFS1·ISD11, untagged ISCU or all proteins together were incubated with nickel beads in binding buffer for 1 h at 4 °C, and the incubation was continued in the absence or presence of 20 μm Fe2+ at 37 °C for 30 min. The lanes denoted by an asterisk show control reactions containing nickel beads and ISCU but not HisNFS1·ISD11. D, schematic representation of the pulldown results shown in C.
The Length of the FXN N Terminus Determines Formation of Complexes with Native NFS1 and ISCU
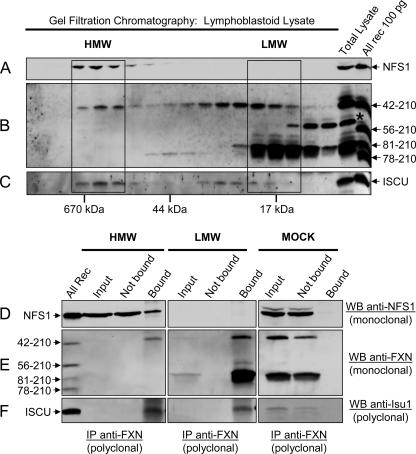
To assess interactions between native FXN isoforms and their Fe-S cluster assembly partners, a soluble extract prepared from a control human lymphoblastoid cell line was first fractionated by gel filtration chromatography as described above. The bulk of native NFS1 was eluted in one high molecular weight pool as expected (36), together with the high molecular weight pool of native FXN42–210 (Fig. 9, A and B). ISCU was eluted in two pools, a high and a low molecular weight pool, with a distribution closely paralleling that of native FXN42–210 (Fig. 9, B and C). The elution profile of native FXN81–210 did not appear to match that of NFS1 or ISCU (Fig. 9, A–C).
FIGURE 9.
Interactions of native FXN isoforms with native NFS1 and ISCU. Lymphoblastoid cell lysate was analyzed by Superdex 75 size exclusion chromatography. Fractions comprising the entire molecular mass fractionation range of the column were analyzed by Western blotting with (A) anti-NFS1 monoclonal antibody MyBioSource, (B) anti-FXN, or (C) anti-Isu1 (32) polyclonal antibodies. The high and low molecular weight fractions were pooled (HMW and LMW box, respectively), and immunoprecipitation was performed with PAC 2517 anti-FXN antibody immobilized on Protein A Magnetic beads, as described under “Experimental Procedures.” Aliquots of each pool (HMW or LMW) before immunoprecipitation (Input, ∼5% of total volume), the flow-through fraction (Not bound; ∼5% of total volume), and the affinity-purified fraction (Bound; 100% of total volume) were analyzed by Western blotting (WB) with anti-NFS1 monoclonal antibody (D) or anti-FXN monoclonal antibody (E) or anti-Isu1 polyclonal antibody (F). FXN and ISCU were detected in individual HMW fractions (B and C) but not in the HMW fraction pool (E and F, input and not Bound) because ∼4 times less total protein was loaded on the gel in the latter analysis. For the control (MOCK), the Protein A Magnetic beads without antibody were incubated overnight with non-fractionated lymphoblastoid cell lysate, and Input, Not bound, and Bound proteins were analyzed as described above.
Fractions containing high molecular weight FXN42–210 were pooled. Fractions containing the bulk of FXN81–210, along with small amounts of low molecular weight FXN42–210, were also pooled. FXN42–210 was immunoprecipitated from the high molecular weight fractions together with comparable amounts of NFS1 and ISCU (Fig. 9, D–F, HMW, Bound), which was consistent with the results of our in vitro pulldown assays (see Fig. 8A). However, NFS1 was present in a large excess relative to FXN42–210 (Fig. 9, D versus E, HMW, Input), and most NFS1 was not associated in a stable manner with either FXN42–210 or ISCU (Fig. 9, D–F, HMW, Not bound). Interestingly, ISCU was co-immunoprecipitated with the FXN81–210 and/or FXN42–210 present in the low molecular weight fractions, which did not contain any detectable NFS1 (Fig. 9, D–F, LMW, Bound). This result was in contrast with the results of our in vitro pulldown assays where neither rec-FXN81–210 nor rec-FXN42–210 had exhibited stable contacts with ISCU in the absence of NFS1·ISD11 (see Fig. 6A).
Because FXN expression could complement the lack of the yeast homologue Yfh1 (37, 44), protein-protein interactions were also analyzed upon gel filtration chromatography of mitochondrial extract from the yfh1Δ[FXN1–210] strain. Similar to what we had observed for lymphoblastoid lysate, FXN42–210 was co-eluted with Nfs1 (yeast NFS1 homologue) and/or Isu1 (yeast ISCU homologue), whereas most FXN81–210 was eluted in fractions that did not contain any detectable Nfs1 or Isu1 (supplemental Fig. S5, a–c). The relatively high levels of FXN isoforms present in the fractions eluted from the column enabled us to carry out reciprocal co-immunoprecipitations using either an anti-FXN or an anti-Nfs1 polyclonal antibody. In the high molecular weight fractions, we observed co-immunoprecipitation of Nfs1 and Isu1 with FXN42–210, and of FXN42–210 and Isu1 with Nfs1 (supplemental Fig. S5, d–f), indicating that FXN42–210 and yeast Nfs1 formed a stable complex. In both co-immunoprecipitations, the levels of Isu1 that were pulled down were proportional to those of Nfs1, not FXN42–210 (supplemental Fig. S5, d–f), suggesting that the interaction of Isu1 with FXN42–210 was indirect. The low molecular weight fractions containing the bulk of FXN81–210 did not contain any detectable Isu1 or Nfs1, and accordingly neither protein was co-immunoprecipitated with FXN81–210 (supplemental Fig. S5, a–f).
The Length of the FXN N Terminus Controls Fe2+ or Fe3+ Availability for Fe-S Cluster Assembly on ISCU
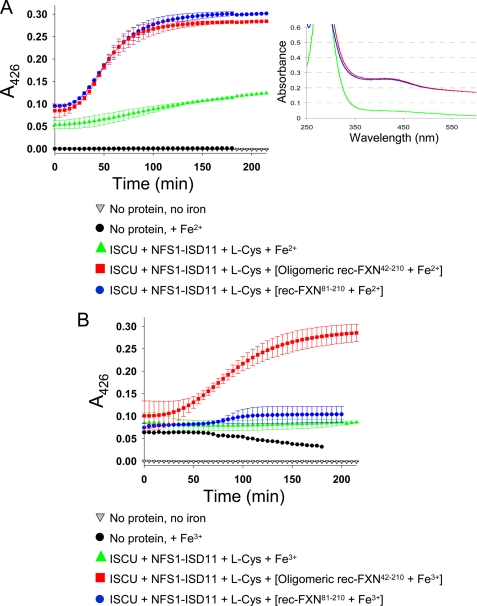
We tested whether the different rec-FXN isoforms might be able to provide iron for Fe-S cluster synthesis on the scaffold ISCU, with elemental sulfur being provided by the HisNFS1·ISD11 complex via cysteine desulfuration. Rec-FXN proteins were loaded with iron under anaerobic or aerobic conditions to prevent or favor, respectively, oxidation of Fe2+ to Fe3+ (32). We then used an anaerobic assay that monitors [2Fe-2S] cluster formation over time by continuous UV-visible absorption measurements at 426 nm (34). Reactions containing ISCUHis, HisNFS1·ISD11, and Fe2+ without rec-FXN showed a small increase in A426 relative to control reactions containing buffer with or without iron (Fig. 10A). In the presence of oligomeric rec-FXN42–210 or rec-FXN81–210, there was a much greater and faster increase in A426, which was almost indistinguishable between the two isoforms (Fig. 10A). A similar result was obtained with rec-FXN42–210 (not shown). In contrast, when rec-FXN isoforms were loaded with iron aerobically, we observed a significant increase in A426 in reactions containing oligomeric rec-FXN42–210, but only a slight increase in reactions containing rec-FXN81–210 or rec-FXN42–210 (Fig. 10B and not shown for FXN42–210). The rate of the reaction was slightly lower with Fe3+-loaded versus Fe2+-loaded oligomeric rec-FXN42–210 but overall this protein appeared to be able to efficiently donate Fe3+, as has been described for both the E. coli and yeast frataxin homologues (32, 34).
FIGURE 10.
FXN isoforms differentially use Fe2+ or Fe3+ to support [2Fe-2S] assembly on ISCU. Rec-FXN proteins were incubated with 10 eq of Fe2+ for 20 min anaerobically (A) or for ∼12 h aerobically (B) in 20 mm HEPES-KOH, pH 7.3, 150 mm NaCl. The [2Fe-2S] cluster synthesis reaction was started with the addition of 50 μm iron, provided as FXN-bound or free iron, to anaerobic reactions containing 5 μm ISCUHis, 5 μm HisNFS1·ISD11 complex, 5 mm dithiothreitol, and 2 mm l-cysteine in 20 mm HEPES-KOH, pH 7.3, 150 mm NaCl. Each plot represents the mean ± S.D. from 2 (oligomeric FXN42–210) and 3 (FXN81–210) independent experiments. Data fitting was performed using a sigmoidal equation in the SigmaPlot program. The inset shows the individual UV-visible absorption spectra for the completed anaerobic reactions containing ISCU + NFS1·ISD11 + l-Cys + Fe2+ (green), ISCU + NFS1·ISD11 + l-Cys + [oligomeric rec-FXN42–210 + Fe2+] (red), and ISCU + NFS1·ISD11 + l-Cys + [rec-FXN81–210 + Fe2+] (blue).
DISCUSSION
Different FXN Isoforms Are Normally Present in Human Cells and Tissues
FXN56–210 was thought to represent the native mature form of human frataxin until the recent identification of FXN81–210 (20, 22). In these studies, FXN81–210 was the only isoform that could be detected in a variety of normal human cells and tissues under native conditions (20, 22). In contrast, FXN42–210, FXN56–210, and FXN78–210 were detected only after overexpression of FXN1–210 and/or deletion of the 41–42 or 80–81 cleavage sites, which led to the conclusion that these forms of the protein are not produced in vivo under normal conditions (20, 22). However, by use of high resolution SDS-PAGE and Western blotting with three different antibodies we have detected all of the previously observed FXN isoforms in actively dividing cells, as well as post-mortem tissues from several controls and FRDA carriers and patients. These data clearly demonstrate that, like FXN81–210, FXN42–210, FXN56–210, and ∼14 kDa FXN are produced not only during the normal biogenesis of human frataxin but also when the expression of FXN1–210 is inherently reduced. Interestingly, relatively high levels of ∼14-kDa FXN products are present in FRDA cells and tissues. These products can originate from cleavage of FXN1–210, FXN42–210, or FXN56–210 via mechanisms that have been characterized in vitro but remain to be elucidated in vivo (20, 27, 28). The role of these mechanisms in FRDA should be addressed in future studies as it is possible that cleavage to ∼14-kDa products enhances depletion of the longest FXN isoforms and indirectly also contributes to FXN81–210 depletion.
The FXN81–210/FXN42–210 Molar Ratio Is Altered in FRDA Cells and Tissues
The apparent molar ratio of FXN81–210 to FXN42–210 was consistently greater in carriers versus controls and patients versus carriers, raising the interesting possibility that the rate of conversion of FXN42–210 to FXN81–210 becomes faster in FRDA cells. Feedback inhibition by the end product is a common regulatory mechanism for enzymes, and thus it is conceivable that the affinity of MPP for FXN42–210 increases when the levels of FXN81–210 decrease below certain levels. This suggests that it may be necessary to assess how FXN augmentation and replacement therapies influence not only total FXN levels but also the biogenesis of specific FXN isoforms. The FXN81–210/FXN42–210 molar ratio could be a useful parameter to optimize frataxin up-regulation strategies in a tissue-specific manner.
N-terminal Processing, Not Iron Binding, Controls Human Frataxin Oligomerization
To assess whether different isoforms have distinct functions, we have focused on FXN42–210 and FXN81–210. Through the analysis of both recombinant and native proteins we have shown that FXN81–210 exists as a stable monomer, whereas FXN42–210 exists in both monomeric and oligomeric configurations. The biological significance of frataxin oligomerization has been a matter of debate in the literature (for an overview of this subject, see Refs. 4, 18, and 32). There was an early report that oligomerization-deficient Yfh1 could interact with native Isu1 and sustain normal Fe-S cluster biogenesis, as determined from the lack of obvious growth defects (45). However, subsequent studies showed that similar Yfh1 mutant proteins formed weak contacts with Isu1 (32, 46) that correlated with slower kinetics of Fe-S cluster biosynthesis (46) and increased susceptibility to excess iron and other sources of oxidative stress (31, 46). Accordingly, from recent studies (32, 34) and our present work, oligomerization is emerging as a conserved property that enables frataxin across species to form stable contacts with the core components of the Fe-S cluster assembly machinery and to provide iron for Fe-S cluster assembly. In particular, recent studies have revealed that Yfh1p and CyaY can exist in monomeric or oligomeric configurations with drastically different effects on Fe-S cluster assembly (32, 34, 47). Under anaerobic conditions, CyaY and Yfh1 are stable monomers that bind stoichiometric concentrations of Fe2+ (34, 48). Monomeric CyaY-Fe2+ binds to the sulfur donor IscS and inhibits [2Fe-2S] cluster assembly on the scaffold IscU (47); similarly, in the presence of the sulfur donor complex Nfs1·Isd11, monomeric Yfh1-Fe2+ supports only very low rates of [2Fe-2S] cluster assembly on the scaffold Isu1 (32). On the other hand, under aerobic conditions, CyaY and Yfh1 oligomerize in an iron concentration-dependent manner, catalyze iron oxidation, and bind several atoms of Fe3+ per subunit (33, 34, 49). Fe3+-loaded CyaY or Yfh1 oligomers form stable complexes with their Fe-S cluster assembly partners, and promote assembly of [2Fe-2S] cluster on the scaffold IscU/Isu1 (32, 34). Thus, the oxidation state of the iron bound to CyaY and Yfh1 appears to define the ability of these proteins to inhibit (Fe2+-bound monomeric CyaY or Yfh1) or promote (Fe3+-bound oligomeric CyaY or Yfh1) Fe-S cluster assembly. This mechanism might enable the facultative anaerobes E. coli and S. cerevisiae to achieve different rates of Fe-S cluster synthesis in anaerobic versus aerobic conditions. Here we have shown that, unlike CyaY and Yfh1, FXN achieves oligomeric or monomeric configurations via retention or removal of the N-terminal region upstream of Ser-81, in a manner that appears independent of the presence of iron. This is consistent with our previous findings that rec-FXN56–210 was oligomerization-proficient, whereas rec-FXN78–210 was not (30). In addition, mutations in a conserved acidic patch implicated in iron binding (reviewed in Ref. 4) affected the iron-dependent oligomerization of Yfh1 (31, 45) but did not influence oligomerization of FXN56–210.3 These data together suggest that FXN has developed an iron-independent mechanism to initiate oligomerization involving its N-terminal region. Although some portions of the 1–81 amino acid sequence are poorly conserved even among closely related mammals, there is a high level of conservation among mammalian frataxins between residues 64 and 87 (supplemental Fig. S6). The conservation extends nearly 20 residues upstream of serine 81 and it is likely to have a functionally important role, in addition to forming the MPP cleavage site for production of FXN81–210. Interestingly, although Yfh1 and FXN use apparently different mechanisms to initiate oligomerization, both use trimer as the building block of larger oligomers (40),4 which ultimately enables both proteins to form stable contacts with components of the Fe-S cluster assembly machinery (Ref. 32 and this study).
Oligomeric Versus Monomeric Configuration Controls FXN Interactions with NFS1
In pulldown assays, oligomerization enabled rec-FXN42–210 to form stable complexes with NFS1·ISD11 in the absence of ISCU or other proteins or iron. Complexes comprising native oligomeric FXN42–210 and NFS1 and ISCU were also observed upon fractionation of cell lysate by gel filtration chromatography followed by co-immunoprecipitation, which underscores the stability of these complexes. Native oligomeric FXN42–210 could also form stable complexes with Nfs1 and Isu1 in yeast mitochondria, which is not surprising given that human frataxin can functionally replace yeast frataxin and maintain respiratory function and iron balance in S. cerevisiae (37, 44). Reciprocal co-immunoprecipitations showed that the amount of Isu1 present in oligomeric FXN42–210·Nfs1·Isu1 complexes was proportional to the amounts of Nfs1, not oligomeric FXN42–210, substantiating the view that unlike oligomeric Yfh1, oligomeric FXN42–210 does not directly bind to the scaffold (43). These observations are evidence of a physiological role of oligomeric FXN42–210 in Fe-S cluster synthesis.
Unlike oligomeric rec-FXN42–210, rec-FXN81–210 could bind to NFS1·ISD11 only in the presence of ISCU, which was once again independent of the presence of iron. In the presence of NFS1·ISD11·ISCU complex, simultaneous contacts of monomeric FXN81–210 with both ISD11 and ISCU may stabilize NFS1·SD11·ISCU·FXN81–210 complexes. This is in line with a previous report that interactions between FXN and ISCU could only be observed in the presence of ISD11 (43). Upon gel filtration chromatography of human cell lysates, native FXN81–210 was not recovered in high molecular weight fractions, indicating that this isoform does not form stable complexes with NFS1, or that complexes between native FXN81–210 and NFS1 are not stable enough to withstand gel filtration chromatography and co-immunoprecipitation procedures. Interestingly, native ISCU was co-immunoprecipitated with native FXN isoforms (mainly FXN81–210 and FXN42–210) in fractions that did not contain any detectable NFS1. Given that the distribution of native ISCU paralleled closely that of native FXN42–210 (Fig. 9, B and C), this particular result may reflect association of ISCU with native monomeric FXN42–210, which would explain why this protein was eluted from the gel filtration column with a larger molecular mass than expected (Fig. 4, A versus D). It is also possible that the fractions analyzed in this experiment contained free ISD11 or another adaptor molecule that mediated stable contacts between monomeric FXN isoforms (FXN42–210 and/or FXN81–210) and ISCU.
Complementary Roles of FXN42–210 and FXN81–210 in Fe-S Cluster Assembly
FXN81–210 was consistently more abundant than FXN42–210 in heart or cerebellum, however, both forms were almost equally abundant in actively dividing lymphoblastoid or yeast cells. A possible interpretation is that in non-dividing tissues at steady state most FXN42–210 is cleaved to FXN81–210, whereas during cell growth and conditions that induce global mitochondrial biogenesis, a significant proportion of FXN42–210 is not proteolytically processed and can be used to produce oligomeric FXN42–210 (Fig. 11). Inhibition of MPP by increasing levels of FXN81–210 (i.e. product inhibition) could be a simple mechanism to enhance the levels of oligomeric FXN42–210 during mitochondrial biogenesis. Given that oligomeric FXN42–210 has a unique ability to coordinate Fe3+, raising the levels of this isoform may enable the cell to increase mitochondrial iron uptake and achieve higher rates of Fe-S cluster synthesis without increasing the labile iron pool and its potential toxicity (Fig. 11). The recent identification of a mammalian siderophore involved in iron donation to mitochondria (50) supports the possibility that Fe3+ is an important ion in Fe-S cluster formation.
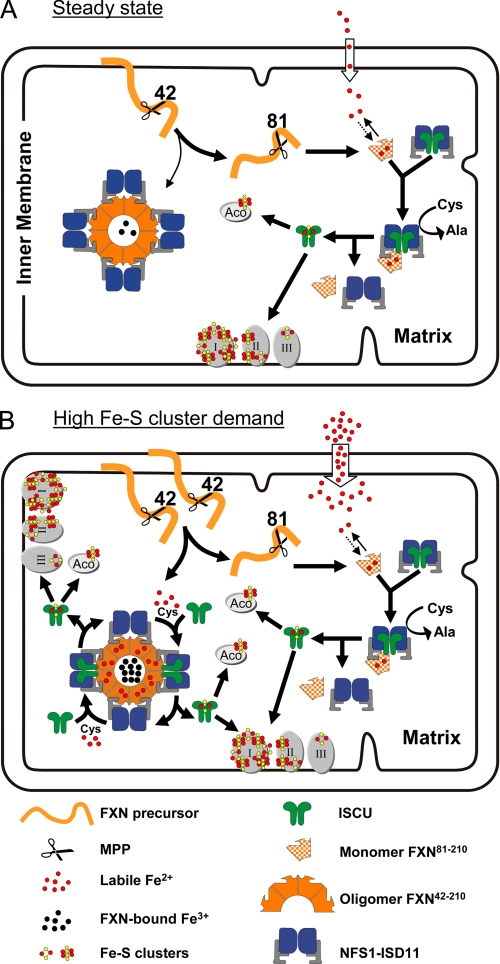
FIGURE 11.
Proposed functions of FXN isoforms in Fe-S cluster synthesis. The FXN precursor is processed by MPP to FXN42–210, which can undergo further processing to FXN81–210 or oligomerize. A, under basal conditions, most FXN42–210 is cleaved to FXN81–210, which controls the labile Fe2+ pool and supports the bulk of [2Fe-2S] cluster assembly through dynamic interactions with the NFS1·ISD11·ISCU complex. Stable complexes of oligomeric FXN and NFS1·ISD11 form in an iron-independent manner but may not significantly participate in Fe-S cluster assembly under basal conditions. B, under conditions of increased Fe-S cluster demand and increased mitochondrial iron uptake (e.g. increased mitochondrial biogenesis), a larger proportion of FXN42–210 can be converted to oligomeric FXN42–210, which can chelate any iron that exceeds the limited iron-binding capacity of FXN81–210. Stable complexes between iron-loaded oligomeric FXN42–210 and NFS1·ISD11 become an additional site of Fe-S cluster assembly on ISCU. I, II, and III, respiratory chain complexes I, II, and III; Aco, mitochondrial aconitase. Components involved in the transfer of [2Fe-2S] cluster from ISCU to Apo enzymes (52) are not shown.
Our model is based on important functional differences we have observed between FXN81–210 and oligomeric FXN42–210. FXN81–210 cannot chelate iron in vitro and therefore it is not expected to be able to store or transport iron in the mitochondrial matrix. Moreover, FXN81–210 forms stable contacts only with the complete NFS1·ISD11·ISCU complex in vitro, and it is recovered mostly as a free monomer in cell lysate. Thus, FXN81–210 may only transiently bind Fe2+ and support basal levels of Fe-S cluster assembly through dynamic interactions with the NFS1·ISD11·ISCU complex (Fig. 11A). On the other hand, oligomeric FXN42–210 chelates Fe3+ tightly in vitro and may be able to store Fe3+ in mitochondria whenever iron uptake is high and the labile Fe2+ pool exceeds the iron-binding capacity of FXN81–210. In addition, oligomeric FXN42–210 forms stable complexes with NFS1·ISD11 both in vitro and in cell lysate. Thus, stable complexes of oligomeric FXN42–210 and NFS1·ISD11 may provide a standby mechanism to increase the rate of Fe-S cluster synthesis when the demand exceeds the rate allowed by transient FXN81–210/NFS1·ISD11/ISCU interactions (Fig. 11B).
Previous studies have shown that FXN42–210 maintains aconitase activity and cell viability in cells depleted of FXN81–210 (20–22). Our present work further suggests that FXN42–210 and FXN81–210 have complementary roles in Fe-S cluster assembly (Fig. 11, A and B), consistent with our findings that both isoforms are normally present in vivo and that each isoform has unique biochemical properties. In future studies, cells engineered to harbor only FXN42–210 or FXN81–210 could help elucidating the specific roles of these proteins under different metabolic conditions, and in particular the importance of oligomeric FXN42–210 and its enhanced affinity for NFS1·ISD11.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Arnulf H. Koeppen, Veterans Affairs Medical Center, Albany, NY, for providing the autopsy tissue samples and clinical data of the patients with Friedreich ataxia and controls, and for critical reading of the manuscript. We thank Daniel L. Kraft, Mayo Clinic Biochemical Genetics Laboratory, and Jeremy D. Huston, Mayo Biospecimens Accessioning Processing Laboratory, for their generous help with lymphoblastoid cell cultures; Benjamin J. Madden, Mayo Proteomics Research Center, for help with peptide mass fingerprinting analysis; and Rachael Vaubel and Hongqiao Li, Isaya lab, for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AG15709, and grants from the Friedreich Ataxia Research Alliance, Muscular Dystrophy Association (to G. I.), and the Mayo Clinic Department of Pediatric & Adolescent Medicine (to T. B. and D. S. B.). Mayo Clinic has a financial interest associated with technology used in this research (rec-FXN56–210), which has been licensed to MitoSciences, Inc. Mayo Clinic, but not the investigators, has received royalties of less than the federal threshold for significant financial interest.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
H. A. O'Neill and G. Isaya, unpublished results.
S. Al-Karadaghi, personal communication.
- FRDA
- Friedreich ataxia
- FXN
- human frataxin
- rec-FXN
- recombinant FXN
- Yfh1
- yeast frataxin homolog
- CyaY
- E. coli frataxin homologue
- NFS1/Nfs1
- human/yeast cysteine desulfurase
- ISD11/Isd11
- human/yeast NFS1/Nfs1-binding protein
- ISCU/Isu1
- human/yeast Fe-S cluster scaffold protein
- MPP
- mitochondrial processing peptidase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Harding A. E. (1981) Brain 104, 589–620 [DOI] [PubMed] [Google Scholar]
- 2.Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 3.Wells R. D. (2008) FASEB J. 22, 1625–1634 [DOI] [PubMed] [Google Scholar]
- 4.Bencze K. Z., Kondapalli K. C., Cook J. D., McMahon S., Millán-Pacheco C., Pastor N., Stemmler T. L. (2006) Crit. Rev. Biochem. Mol. Biol. 41, 269–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karthikeyan G., Santos J. H., Graziewicz M. A., Copeland W. C., Isaya G., van Houten B., Resnick M. A. (2003) Hum. Mol. Genet. 12, 3331–3342 [DOI] [PubMed] [Google Scholar]
- 6.Anderson P. R., Kirby K., Hilliker A. J., Phillips J. P. (2005) Hum. Mol. Genet. 14, 3397–3405 [DOI] [PubMed] [Google Scholar]
- 7.Huang M. L., Becker E. M., Whitnall M., Rahmanto Y. S., Ponka P., Richardson D. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16381–16386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan G., Chen L. S., Lonnerdal B., Gellera C., Taroni F. A., Cortopassi G. A. (2001) Hum. Mol. Genet. 10, 2099–2107 [DOI] [PubMed] [Google Scholar]
- 9.Pianese L., Busino L., De Biase I., De Cristofaro T., Lo Casale M. S., Giuliano P., Monticelli A., Turano M., Criscuolo C., Filla A., Varrone S., Cocozza S. (2002) Hum. Mol. Genet. 11, 2989–2996 [DOI] [PubMed] [Google Scholar]
- 10.Sparaco M., Gaeta L. M., Santorelli F. M., Passarelli C., Tozzi G., Bertini E., Simonati A., Scaravilli F., Taroni F., Duyckaerts C., Feleppa M., Piemonte F. (2009) J. Neurol. Sci. 287, 111–118 [DOI] [PubMed] [Google Scholar]
- 11.Paupe V., Dassa E. P., Goncalves S., Auchère F., Lönn M., Holmgren A., Rustin P. (2009) PloS One 4, e4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppola G., Marmolino D., Lu D., Wang Q., Cnop M., Rai M., Acquaviva F., Cocozza S., Pandolfo M., Geschwind D. H. (2009) Hum. Mol. Genet. 18, 2452–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C., Schoenfeld R., Shan Y., Tsai H. J., Hammock B., Cortopassi G. (2009) Biochim. Biophys. Acta 1792, 1052–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kearney M., Orrell R. W., Fahey M., Pandolfo M. (2009) Cochrane Database of Systematic Reviews, CD007791. [DOI] [PubMed] [Google Scholar]
- 15.Seznec H., Simon D., Monassier L., Criqui-Filipe P., Gansmuller A., Rustin P., Koenig M., Puccio H. (2004) Hum. Mol. Genet. 13, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 16.Boddaert N., Le Quan Sang K. H., Rötig A., Leroy-Willig A., Gallet S., Brunelle F., Sidi D., Thalabard J. C., Munnich A., Cabantchik Z. I. (2007) Blood 110, 401–408 [DOI] [PubMed] [Google Scholar]
- 17.Whitnall M., Rahmanto Y. S., Sutak R., Xu X., Becker E. M., Mikhael M. R., Ponka P., Richardson D. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9757–9762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmucker S., Puccio H. (2010) Hum. Mol. Genet. 19, R103–110 [DOI] [PubMed] [Google Scholar]
- 19.Cavadini P., Adamec J., Taroni F., Gakh O., Isaya G. (2000) J. Biol. Chem. 275, 41469–41475 [DOI] [PubMed] [Google Scholar]
- 20.Schmucker S., Argentini M., Carelle-Calmels N., Martelli A., Puccio H. (2008) Hum. Mol. Genet. 17, 3521–3531 [DOI] [PubMed] [Google Scholar]
- 21.Long S., Jirku M., Ayala F. J., Lukes J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13468–13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Condò I., Ventura N., Malisan F., Rufini A., Tomassini B., Testi R. (2007) Hum. Mol. Genet. 16, 1534–1540 [DOI] [PubMed] [Google Scholar]
- 23.Acquaviva F., De Biase I., Nezi L., Ruggiero G., Tatangelo F., Pisano C., Monticelli A., Garbi C., Acquaviva A. M., Cocozza S. (2005) J. Cell Sci. 118, 3917–3924 [DOI] [PubMed] [Google Scholar]
- 24.Musco G., Stier G., Kolmerer B., Adinolfi S., Martin S., Frenkiel T., Gibson T., Pastore A. (2000) Structure Fold Des. 8, 695–707 [DOI] [PubMed] [Google Scholar]
- 25.Cavadini P., O'Neill H. A., Benada O., Isaya G. (2002) Hum. Mol. Genet. 11, 217–227 [DOI] [PubMed] [Google Scholar]
- 26.Adinolfi S., Trifuoggi M., Politou A. S., Martin S., Pastore A. (2002) Hum. Mol. Genet. 11, 1865–1877 [DOI] [PubMed] [Google Scholar]
- 27.Babady N. E., Pang Y. P., Elpeleg O., Isaya G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6158–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon T., Dizin E., Cowan J. A. (2007) J. Biol. Inorg. Chem. 12, 535–542 [DOI] [PubMed] [Google Scholar]
- 29.O'Neill H. A., Gakh O., Park S., Cui J., Mooney S. M., Sampson M., Ferreira G. C., Isaya G. (2005) Biochemistry 44, 537–545 [DOI] [PubMed] [Google Scholar]
- 30.O'Neill H. A., Gakh O., Isaya G. (2005) J. Mol. Biol. 345, 433–439 [DOI] [PubMed] [Google Scholar]
- 31.Gakh O., Park S., Liu G., Macomber L., Imlay J. A., Ferreira G. C., Isaya G. (2006) Hum. Mol. Genet. 15, 467–479 [DOI] [PubMed] [Google Scholar]
- 32.Li H., Gakh O., Smith D. Y., 4th, Isaya G. (2009) J. Biol. Chem. 284, 21971–21980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bou-Abdallah F., Adinolfi S., Pastore A., Laue T. M., Chasteen N. D. (2004) J. Mol. Biol. 341, 605–615 [DOI] [PubMed] [Google Scholar]
- 34.Layer G., Ollagnier-de Choudens S., Sanakis Y., Fontecave M. (2006) J. Biol. Chem. 281, 16256–16263 [DOI] [PubMed] [Google Scholar]
- 35.Tong W. H., Rouault T. (2000) EMBO J. 19, 5692–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marelja Z., Stöcklein W., Nimtz M., Leimkühler S. (2008) J. Biol. Chem. 283, 25178–25185 [DOI] [PubMed] [Google Scholar]
- 37.Cavadini P., Gellera C., Patel P. I., Isaya G. (2000) Hum. Mol. Genet. 9, 2523–2530 [DOI] [PubMed] [Google Scholar]
- 38.Hendrick J. P., Hodges P. E., Rosenberg L. E. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 4056–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamec J., Rusnak F., Owen W. G., Naylor S., Benson L. M., Gacy A. M., Isaya G. (2000) Am. J. Hum. Genet. 67, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlberg T., Schagerlöf U., Gakh O., Park S., Ryde U., Lindahl M., Leath K., Garman E., Isaya G., Al-Karadaghi S. (2006) Structure 14, 1535–1546 [DOI] [PubMed] [Google Scholar]
- 41.Gerber J., Mühlenhoff U., Lill R. (2003) EMBO Rep. 4, 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T., Craig E. A. (2008) J. Biol. Chem. 283, 12674–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan Y., Napoli E., Cortopassi G. (2007) Hum. Mol. Genet. 16, 929–941 [DOI] [PubMed] [Google Scholar]
- 44.Wilson R. B., Roof D. M. (1997) Nat. Genet. 16, 352–357 [DOI] [PubMed] [Google Scholar]
- 45.Aloria K., Schilke B., Andrew A., Craig E. A. (2004) EMBO Rep. 5, 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foury F., Pastore A., Trincal M. (2007) EMBO Rep. 8, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adinolfi S., Iannuzzi C., Prischi F., Pastore C., Iametti S., Martin S. R., Bonomi F., Pastore A. (2009) Nat. Struct. Mol. Biol. 16, 390–396 [DOI] [PubMed] [Google Scholar]
- 48.Cook J. D., Bencze K. Z., Jankovic A. D., Crater A. K., Busch C. N., Bradley P. B., Stemmler A. J., Spaller M. R., Stemmler T. L. (2006) Biochemistry 45, 7767–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park S., Gakh O., Mooney S. M., Isaya G. (2002) J. Biol. Chem. 277, 38589–38595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devireddy L. R., Hart D. O., Goetz D. H., Green M. R. (2010) Cell 141, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.