Abstract
The type IIb class of plant Ca2+-ATPases contains a unique N-terminal extension that encompasses a calmodulin (CaM) binding domain and an auto-inhibitory domain. Binding of Ca2+-CaM to this region can release auto-inhibition and activates the calcium pump. Using multidimensional NMR spectroscopy, we have determined the solution structure of the complex of a plant CaM isoform with the CaM-binding domain of the well characterized Ca2+-ATPase BCA1 from cauliflower. The complex has a rather elongated structure in which the two lobes of CaM do not contact each other. The anchor residues Trp-23 and Ile-40 form a 1-8-18 interaction motif. Binding of Ca2+-CaM gives rise to the induction of two helical parts in this unique target peptide. The two helical portions are connected by a highly positively charged bend region, which represents a relatively fixed angle and positions the two lobes of CaM in an orientation that has not been seen before in any complex structure of calmodulin. The behavior of the complex was further characterized by heteronuclear NMR dynamics measurements of the isotope-labeled protein and peptide. These data suggest a unique calcium-driven activation mechanism for BCA1 and other plant Ca2+-ATPases that may also explain the action of calcium-CaM on some other target enzymes. Moreover, CaM activation of plant Ca2+-ATPases seems to occur in an organelle-specific manner.
Keywords: Calcium ATPase, Calmodulin, NMR, Plant, Protein Structure, Vacuolar ATPase, Calmodulin Complex, Soybean Calmodulin
Introduction
The calcium ion (Ca2+) is intimately involved as a secondary messenger in numerous signal transduction processes. Elevation of the cytoplasmic calcium levels leads to a variety of physiological responses in plants (1). Various stimuli such as abiotic stresses, hormones, wounds, and plant pathogens can trigger an influx of Ca2+ into the cytoplasm. Calcium signaling is well known to play a role in plant defense signaling pathways (2). The transient Ca2+ signals are translated into a physiological response by a variety of Ca2+-binding proteins including the ubiquitous Ca2+-sensor protein calmodulin (CaM)3 (3, 4). Unlike animals, which have only one form of the CaM protein, multiple CaM proteins occur in higher plant species (3, 5). For example, the model plant Arabidopsis thaliana, harbors nine CaM genes (AtCaM1–9) coding seven different CaMs (5, 6). Likewise, the plant Glycine max (soybean) contains not less than five CaM genes (SCaM1–5) coding for four distinct CaMs (7), whereas Oryza sativa (rice) also has at least five CaMs (8). Several studies have demonstrated that each CaM isoform is utilized to control different enzymes that are involved in specific physiological reactions (9, 10). In addition to multiple CaM isoforms, plants also contain up to 50 CaM-like proteins that have so far received limited attention (8, 11).
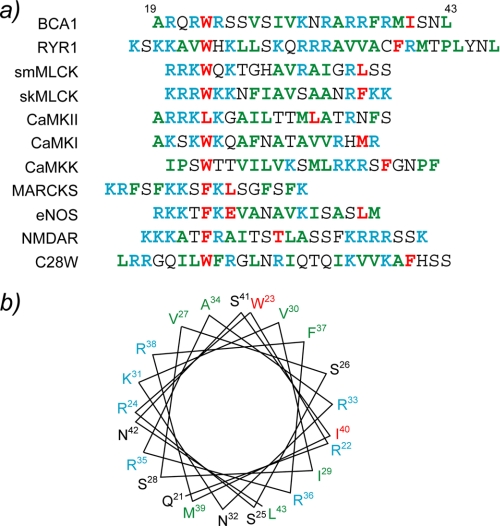
The cytosolic Ca2+ concentration is restored immediately after a stimulus to a low resting level by removing Ca2+ across the plasma membrane or by sequestering it into intercellular organelles, such as the vacuole, the endoplasmic reticulum, or the Golgi apparatus. Plant Ca2+-pumps, which are responsible for this ATP-driven transport process, belong to the superfamily of P-type Ca2+ pumping ATPases (12, 13). According to their ability to bind CaM, plant Ca2+-ATPases have been classified into two groups: type IIA and type IIB. Type IIB Ca2+-ATPases possess an N-terminal extension that harbors a CaM-binding domain (CaMBD). In contrast, their closest animal homologs, the plasma membrane Ca2+-ATPases, have an extension at the C-terminal end that encompasses their CaMBD. On the other hand, the activities of plant type IIA Ca2+-ATPases are CaM-independent. Although several type IIB Ca2+-ATPases have been identified in different plant species, the best studied organism is A. thaliana which contains 10 different type IIB Ca2+-ATPases (ACAs), which have different subcellular localizations (14). For example, ACA2 and ACA4 are located at the endoplasmic reticulum and the prevacuolar membrane, respectively, whereas ACA8 and ACA9 are located on the plasma membrane (15; reviewed in Refs. 16, 17). The BCA1 protein is a type IIB Ca2+-ATPase with a vacuolar localization that was discovered in the plant Brassica oleracea (cauliflower) (18). The CaMBD of BCA1 was found to be located in its N-terminal region between residues Ala-19 and Leu-43. This region is also part of the auto-inhibitory domain of the membrane pump, and it contains a novel CaM-binding motif (19 and Fig. 1a). Previously, we have characterized the interactions between different SCaM isoforms and a 25-residue synthetic peptide corresponding to the CaMBD of BCA1 (20). Isothermal titration calorimetry studies have revealed that both SCaM1 and SCaM4 can strongly bind to the BCA1 peptide with similar thermodynamic parameters. The results were also consistent with those obtained for well characterized CaM-target peptide interactions from mammalian proteins such as smooth muscle myosin light chain kinase (smMLCK) and CaM kinase I peptides. The CaMBD regions of these proteins bind with a fully α-helical structure to Ca2+-CaM. However, unexpectedly, the solution structure of the BCA1 peptide determined in 30% trifluoroethanol was not a complete α-helix, as under these conditions, the BCA1 peptide possessed a disordered region in the middle of the α-helical peptide (20).
FIGURE 1.
a, amino acid sequence alignment of CaMBDs from various CaM target proteins. The anchoring residues are highlighted in red. The basic and acidic residues are shown in cyan and magenta, respectively, whereas hydrophobic residues are displayed in green. b, a helical wheel representation of the CaMBD of BCA1. The residues are colored in the same manner as in a. NOS, endothelial nitric oxide synthase.
Here, we describe the solution structure of soybean CaM isoform 4 complexed with the CaMBD of BCA1. The bound BCA1 peptide adopts two separate α-helical regions that are connected by a bend rendering a unique angle between the two helical segments. Consequently, the relative orientation of the two lobes of SCaM4 is distinct from those seen in other CaM·peptide complexes. We have utilized backbone H-N residual dipolar coupling (RDC) NMR data and rotational diffusion dynamics obtained from 15N NMR relaxation data to confirm this novel orientation of the two lobes. We have also utilized the isotope-labeled peptide to study the motional and structural properties of the bound BCA1 peptide. This CaM binding mode drastically expands the variety of the possible sequence motifs, which can interact tightly with CaM. We also propose a unique mechanism of CaM target activation that can be elucidated on the basis of the SCaM4·BCA1 complex structure.
EXPERIMENTAL PROCEDURES
Sample Preparations
Uniformly 15N- and 15N,13C-labeled SCaM4 were prepared as described previously (21). The unlabeled BCA1 peptide corresponding to the amino acid sequence of residues 19–43 of BCA1, Ac-ARQRWRSSVSIVKNRARRFRMISNL-amide, was synthesized commercially by Anaspec, Inc. (San Jose, CA). Uniformly 15N and 2H, 15N,13C-labeled BCA1 peptides corresponding to the amino acid residues 19–47 of BCA1, PARQRWRSSVSIVKNRARRFRMISNLEKLA, with one Pro residue at the N terminus as a cloning artifact were prepared using the ketosteroid isomerase-fusion inclusion body approach as described previously (22). The peptides were further purified by reverse-phase HPLC, and their identity was confirmed by mass spectroscopy. All NMR samples contained ∼0.3 mm 15N- or 15N,13C-labeled SCaM4, 5 mm CaCl2, 100 mm KCl, 10 mm dl-1,4-dithiothreitol-d10, 0.03% NaN3, and 0.5 mm 2,2-dimethyl-2-silapentane-5-sulfonate in unbuffered 90% H2O/10% D2O or 99.99% D2O solution. The pH/pD values of samples are 7.0 without consideration of isotope effects. The molar ratio of SCaM4 and BCA1 peptide was 1:1.25 to ensure the complete saturation of SCaM4, whereas a ratio of 1:0.85 was used to ensure the absence of free BCA1 peptide in several NMR experiments where the SCaM4-bound BCA1 peptide was monitored. The samples used for RDC measurements also contained 20 mm bis-Tris (pH 7.0), 300 mm KCl, and ∼16 mg/ml filamentous phage Pf1 (Asla Biotech Ltd.).
NMR Measurements
All NMR experiments were performed at 30°C on Bruker Avance 500 or 700 MHz NMR spectrometers equipped with triple resonance inverse Cryoprobes with a single axis z-gradient. Sequential assignments of HN, N, CO, Cα, and Cβ resonances of BCA1 peptide-bound SCaM4 were achieved using several experiments including CBCANH, CBCA(CO)NH, HNCO, and HN(CA)CO (supplemental Fig. 4). The sequential backbone resonance assignments of the BCA1 peptide bound to SCaM4 were obtained by HNCACB, HN(CO)CACB, HNCO, HNCA, and HN(CO)CA. Side chain assignments of peptide-bound SCaM4 were obtained by C(CCO)NH-TOCSY, H(CCO)NH-TOCSY, HBHA(CBCACO)NH, and HCCH-TOCSY experiments. The assignments of the unlabeled SCaM4-bound BCA1 peptide was achieved based on NOE connectivity by analyzing 13C, 15N/F1, F2-filter NOESY, and 13C/F1, F2-filter correlation spectroscopy experiments. All NOESY experiments including 15N-edited NOESY-HSQC, 13C-edited NOESY-HSQC, and 13C/F3-filter 13C/F1-edited NOESY-HSQC were measured with a mixing time of 100 ms. H-N RDC measurements were performed using the in-phase/anti-phase (1H,15N)-HSQC experiment (23). All NMR experiments for the backbone dynamics studies of the unlabeled BCA1 peptide bound to 15N-labeled SCaM4 and the 15N-labeled BCA1 peptide bound to unlabeled SCaM4 were acquired at 50.68 MHz for the 15N frequency. The 15N T2 relaxation data were obtained using a Carr-Purcell-Meiboom-Gill-type T2 experiment, where the field strength of the 180° pulse was 5.6 kHz, and the 180° pulses were applied every 1 ms. The 15N T1ρ data were obtained by applying a 2.5 kHz spin-locking pulse during the relaxation delay time. Both T1 and T2 experiments were repeated twice to estimate the uncertainty in the peak intensities. {1H}-15N heteronuclear NOE data were obtained using a 5-s train of 120° proton pulses. All spectra were processed using NMRPipe/NMRDraw (24) and analyzed using NMRView software (25).
Structure Calculation
The initial structure of SCaM4·BCA1 was calculated with CYANA (version 2.0) (26) using distance restraints derived from the automated NOE assignment protocol implemented in CYANA, hydrogen bond restraints based on the secondary structure from the chemical shift index, and dihedral angle restraints predicted by TALOS (27). A total of 24 Ca2+-ligand restraints were introduced according to the well known Ca2+ coordination in EF-hand sites (28). Further structure refinement with the addition of backbone H-N RDC restraints were performed by XPLOR-NIH (version 2.19) (29). Initial estimation for the axial component of the molecular alignment tensor (Da) and the rhombicity (R) were obtained from the lowest energy structure calculated by CYANA using PALES (30). Finally, the 25 lowest energy structures from a total of 200 were selected and used for the analysis. All molecular graphics used in this manuscript were created using MolMol (31) or PyMOL software.
Rotational Diffusion Dynamics
The 15N T1 and T2 data were fitted with the program CurveFit (A. G. Palmer, Columbia University). The uncertainties for the peak intensity used for the fitting were estimated from duplicated data sets. The uncertainty for {1H}-15N NOE values were evaluated using the S.D. of the noise in empty spectral regions of the spectra. Those residues that show low NOE values (<0.65) were excluded from further analysis because these residues have a slow internal motion, which contributes to the T1 relaxation. Those residues, which are involved in chemical exchange process that affects the T2 relaxation time, were detected and removed as described by Tjandra et al. (32). For the remaining residues, the rotational correlation time for the global tumbling (τm) for each residue were estimated from the R2/R1 ratio using the program R2R1_τm (A. G. Palmer, Columbia University). The τm values were globally fit to the N- and C-terminal lobe of SCaM4 in SCaM4·BCA1 using the program quadric_diffusion (A. G. Palmer, Columbia University).
RESULTS
Structure of SCaM4·BCA1
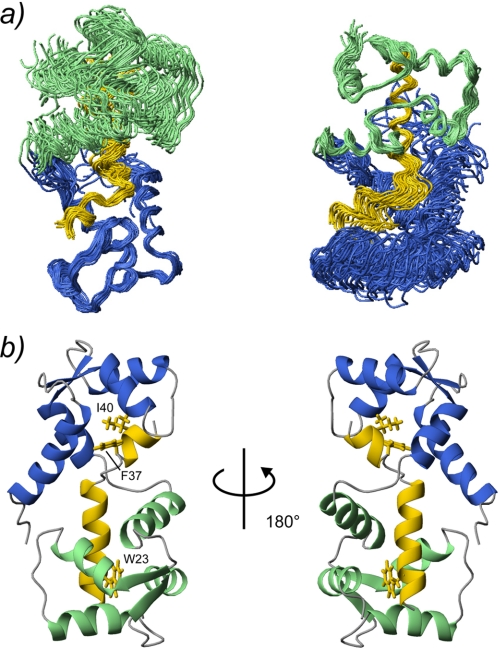
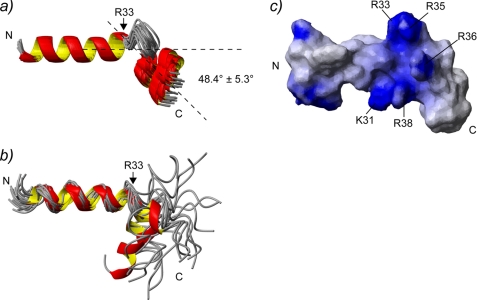
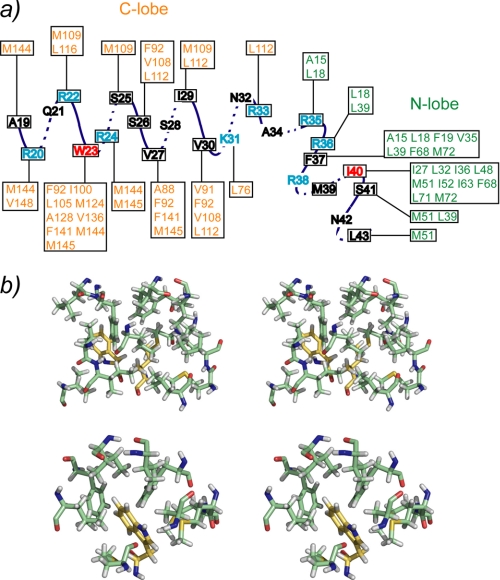
The SCaM4·BCA1 structure was calculated based on 1773 distance restraints derived from NOEs, including 188 intermolecular distances (supplemental Table 1). Because there were no interdomain NOEs detected between the N- and the C-terminal lobe of SCaM4, to determine the relative orientation of the two lobes, backbone H-N RDC data were also employed in the structure determination, as these provide information on long range order in multidomain proteins (33–35). Backbone H-N RDC values were available for 83% of the total residues. Fig. 2a shows the superposition of the 25 lowest energy structures. The backbone r.m.s.d. values in the folded regions are 0.58 ± 0.08 and 0.53 ± 0.10 Å when only the N-terminal unit (the N-terminal lobe of SCaM4 and the C-terminal part of BCA1) and the C-terminal unit (the C-terminal lobe of SCaM4 and the N-terminal part of BCA1) are superposed, respectively. However, the backbone r.m.s.d. value was 1.47 ± 0.59 Å when all of the folded regions of SCaM4·BCA1 were used for the analysis (supplemental Table 1). Fig. 2b displays the ribbon representation of the lowest energy structure of SCaM4·BCA1. The N-terminal lobe structure of SCaM4 in SCaM4·BCA1 is almost identical to that determined without peptide (21), where the backbone r.m.s.d. value in the folded region is 1.06 ± 0.06 Å (supplemental Fig. 1a). On the other hand, the C-terminal lobe structure of SCaM4 in SCaM4·BCA1 is somewhat different from that without the BCA1 peptide, in which the structure is altered to a more open conformation by the binding of the peptide (supplemental Fig. 1b). The backbone r.m.s.d. value in this folded region is 1.60 ± 0.06 Å. The structural alteration of SCaM4 upon BCA1 peptide binding could also be detected in the correlation between the experimentally derived RDC values of SCaM4·BCA1 and the best-fit RDC values calculated from the free Ca2+-SCaM4 structure. The obtained correlation factor (R) and the quality factor (Q) are 0.97/0.26 for the N-terminal lobe, and 0.82/0.56 for the C-terminal lobe. The SCaM4-bound BCA1 peptide adopts an α-helix in its N-terminal region (residues 20–33) followed by a bend and another turn of α-helix (residues 38–42). The bend changes the direction of the two separated α-helices by ∼48° (Fig. 3a). The bend region appears to be highly positively charged (Fig. 3c). The N- and C-terminal lobes of SCaM4 interact with the C- and N-terminal α-helices of the BCA1 peptide, respectively, to form the complex in an antiparallel orientation relative to BCA1 peptide as had been estimated previously from an analysis of near-UV CD spectra (20). The aromatic side chain of Trp-23 in the BCA1 peptide provides many intermolecular NOEs to the SCaM4 protein (Fig. 4a), and it is deeply buried in the hydrophobic target binding pocket of the C-terminal lobe of SCaM4 (Figs. 2b and 4b). This indicates that the side chain of Trp-23 provides the N-terminal anchor residue for the BCA1 peptide. The sidechain of residue Val-30 at position 8 also interacts with the hydrophobic patch between helices E and F in the C-terminal lobe of SCaM4. On the other hand, the side chains of both Phe-37 and Ile-40 in the BCA1 peptide show many hydrophobic contacts to the N-terminal lobe of SCaM4 (Fig. 4a). The side chain of Ile-40 is located almost at the center of the hydrophobic pocket of the N-terminal lobe, whereas that of Phe-37 interacts with the hydrophobic patch that is formed between helices A and B (Figs. 2b and 4b). This suggests that Ile-40 acts as the C-terminal anchor residue of the BCA1 peptide rather than the more bulky hydrophobic sidechain of Phe-37. Therefore, the two anchor residues of the BCA1 peptide are separated by 16 residues, and together with the Val residue, they represent a “1-8-18” motif.
FIGURE 2.
The 25 lowest energy structures of SCaM4·BCA1 are superimposed. The N- and C-terminal lobe of SCaM4 are shown in light green and purple, respectively, whereas the BCA1 peptide is shown in yellow. Only the folded regions of the N-terminal unit (SCaM4 residues 6–74 and BCA1 peptide residues 38–43) and the C-terminal unit (SCaM4 residues 81–145 and BCA1 residues 20–33) are superimposed in the right and left panel, respectively. b, ribbon structures of the lowest energy structure of SCaM4·BCA1 at different angles. The regions are colored in the same manner as in a. The side chains of the anchor residues of the BCA1 peptide, Trp-23 and Ile-40, are shown. The side chain of Phe-37, which also has several hydrophobic contacts to SCaM4, is indicated as well.
FIGURE 3.
a, the bound BCA1 peptide structures from the 25 lowest energy structures of SCaM4·BCA1. Only the first α-helical region (residues 20–33) is superimposed. The averaged angle between the first and the second α-helices for all the structures is 48.4° ± 5.3°. b, 20 BCA1 peptide structures determined in 30% trifluoroethanol are superimposed using the first α-helical region (residues 20–22). These structures were taken from Yamniuk and Vogel (20). c, the surface electrostatic properties of the BCA1 peptide. The residues that form a basic cluster in the middle of BCA1 peptide are labeled. C, C-terminal; N, N-terminal.
FIGURE 4.
Schematic showing the observed intermolecular NOEs between SCaM4 and the BCA1 peptide. The two anchor residues are depicted in red, whereas the basic residues are shown in cyan. b, stereo views of the local interactions between SCaM4 and BCA1 residues, Phe-37 and Ile-40 (top panel), and Trp-23 (bottom panel).
Structure Comparison to Other Complexes
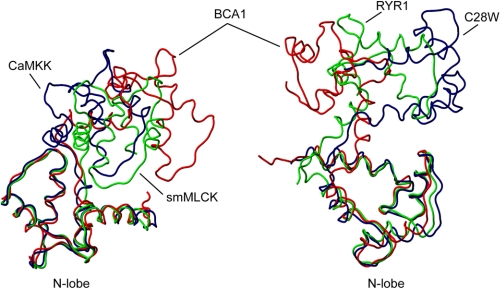
When only the N- or C-terminal lobe of the SCaM4·BCA1 complex are compared with those of CaM in previously reported CaM·peptide complexes, the structures closely resemble one another, where the backbone r.m.s.d. values in the well folded regions are only ∼1 Å for the majority of available complexes (data not shown). However, the relative orientations and the positions of the two lobes of SCaM4 in the SCaM4·BCA1 complex are distinct from any other CaM·peptide complexes. In Fig. 5, only the N-terminal lobe of SCaM4 is superposed on other CaM·peptide complexes, including CaM·smMLCK (36), CaM·CaM kinase kinase (37), the CaM·ryanodine receptor Ca2+ release channel (RYR1) (38), and the CaM·plasma membrane Ca2+-ATPases isoform 4b (C28W) (39). The difference in the relative orientations/positions of the C-terminal lobes relative to the N-terminal lobes is emphasized in Fig. 5. It is clear that the C-terminal lobe of SCaM4·BCA1 is positioned differently from the others. It is worthwhile to note that although the bound C28W CaMBD also possesses a 1-8-18 motif, the relative lobe orientation of CaM is very different from that of the SCaM4·BCA1 complex. The R/Q factors from the best-fit calculation of the NMR RDC data of SCaM4·BCA1 against the other CaM·peptide complexes are 0.82/0.56, 0.85/0.53, 0.84/0.54, and 0.74/0.69 for CaM·smMLCK, CaM·CaMKK, CaM·RYR1, and CaM·C28W, respectively (with some flexible regions (residues 1–5, 74–84, and 144–148/149) excluded). All the values for the other CaM complexes are summarized in supplemental Table 2. In this analysis, the structure with the closest relative orientation was determined as CaM·CaMKK. However, the C-terminal lobe of the CaM·CaMKK complex is located much closer to the N-terminal lobe and forms a more compact overall structure than SCaM4·BCA1 (Fig. 5). Because of the larger spacing between the two anchor residues (1–18 motif), the SCaM4·BCA1 structure seems more ellipsoidal and elongated than the other complexes where the overall dimension of the protein complex is ∼55 × 35 × 30 Å, whereas the reported dimension for CaM·smMLCK is ∼50 × 30 × 25 Å.
FIGURE 5.
The structure of SCaM4 in SCaM4·BCA1 (red) is compared with CaM in CaM·CaMKK (navy; Protein Data Bank code 1IQ5), CaM·smMLCK (green; Protein Data Bank code 1CDL) in the left panel and CaM·C28W (navy; Protein Data Bank code 2KNE) and CaM·RYR1 (green; Protein Data Bank code 2BCX) in the right panel. Only the well folded regions of the N-terminal lobe (residues 6–74) were overlaid in both panels.
Rotational Diffusion Dynamics
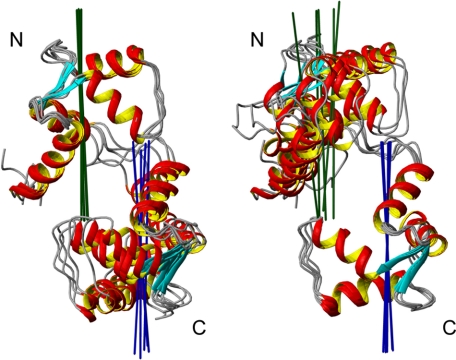
To confirm the unique relative orientation determined in the SCaM4·BCA1 structure, we have performed a rotational diffusion analysis using the available backbone 15N NMR relaxation data. When the two domains tumble or orient together in the protein structure, the experimentally derived diffusion tensors for each domain should be colinear, and therefore, it can be used to determine the relative domain orientation (40). Due to several overlapped resonances, 81.8% of the total residues (except for one Pro) were available, and τm values were estimated from the R2/R1 ratio for 66.9% of the total residues after filtration of the data (supplemental Fig. 2). The τm values ranged between 9.31 and 12.30 ns. After fitting the data to all three diffusion models including isotropic, axially symmetric, and fully anisotropic models, the F-static value for the entire SCaM4 protein was 26.21 (p < 1.0 × 10−5) in the axially symmetric model that represents a drastic improvement in the fitting over the isotropic model, whereas that in the fully anisotropic model against the axially symmetric model was only 0.51 (p = 0.60), which indicates that the axially symmetric model is the best describing the rotational diffusion properties of SCaM4·BCA1. The correlation time for the global molecular tumbling was obtained as 11.20 and 11.10 ns for the N- and C-terminal lobe of SCaM4, respectively. These values are almost identical to the correlation time of 11.12 ns for the entire SCaM4 protein (supplemental Table 3). The principle axes of the diffusion tensors for the N- and C-terminal lobe of SCaM4 are defined separately and visualized on the five lowest energy structures (Fig. 6). The two axes agree very well in all the structures analyzed and are reasonably aligned with the long axis of the SCaM4·BCA1 molecule, supporting the relative lobe orientation determined from the RDC data.
FIGURE 6.
The principle axis of the diffusion tensor is calculated separately for the N- and C-terminal lobe for SCaM4·BCA1 and displayed on the five lowest energy structures. Please note that the N- and C-terminal lobe were superimposed in the left and right panel, respectively.
Structural and Dynamic Properties of Isotope-labeled BCA1 Peptide in Complex
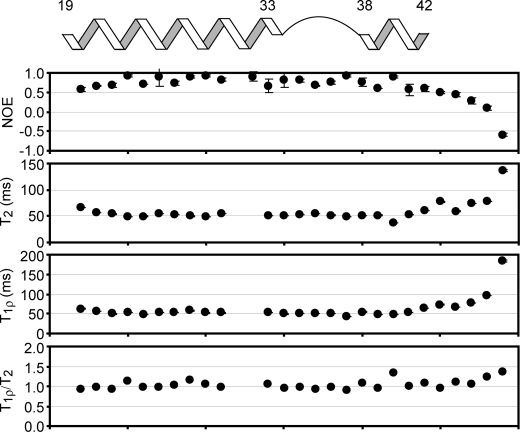
Because of the unique structure of the BCA1 peptide in the SCaM4·BCA1 complex, we sought to investigate the detailed structural and motional properties of the bound BCA1 peptide. Therefore, we have generated an isotope-labeded BCA1 peptide for multinuclear NMR studies. The isotope-labeled BCA1 peptide was bacterially synthesized and designed to be slightly longer than the chemically synthesized amidated BCA1 peptide to ensure that the C-terminal charge does not influence the SCaM4 binding. Supplemental Fig. 3a shows the 1H,15N-HSQC spectrum of the free BCA1 peptide. It is clear that the BCA1 peptide in aqueous solution does not possess any specific conformation, whereas the HSQC spectrum upon addition of Ca2+-SCaM4 represents characteristic folding of the BCA1 peptide (supplemental Fig. 3b). The backbone resonance assignments were obtained unambiguously except for some residues that experience severe broadening by a chemical exchange process. Subsequently, backbone dynamics experiments including {1H}-15N heteronuclear NOE, T2, and T1ρ measurements could be carried out for most residues (Fig. 7). These results showed that the BCA1 peptide retains a relatively rigid conformation for the entire SCaM4 bound region (residues 20–42). Also, the bend region does not appear to be flexible. We also analyzed the ratio between the T1ρ and T2 values to detect possible motions on a timescale of several hundred microseconds (41, Fig. 7). The T1ρ/T2 values except for Ile-40 and several C-terminal residues are very close to one, suggesting the absence of such motions in the bend region of BCA1. The isotope-labeled BCA1 peptide also allowed us to acquire H-N backbone RDC data for the bound BCA1 peptide. The RDC data were available for 27 residues out of 29 and used for structural validation of the bound BCA1 peptide. Although the RDC data were not included in the structure calculation, the R and Q factors for the best-fit calculation against the BCA1 peptide structure using the two helical regions (residues 20–32 and 39–42) are 0.93 and 0.15, respectively, supporting the presence of the unique fixed angle between the two helical portions of the bound BCA1 peptide. We note that the residues in the immediate bend region (residues 33–38) were excluded from the analysis as the structure of the bend region was not well determined in our structure calculation due to a paucity of structural restraints in this region. On the other hand, the best-fit calculation against the straight 1–18 C28W peptide using the same RDC data set produced rather poor correlations (R and Q factors are 0.77 and 0.26, respectively).
FIGURE 7.
15N-relaxation data for BCA1 peptide bound to SCaM4. The measured {1H}-15N NOE, T2, T1ρ, and T1ρ/T2 are plotted as a function of the residue number. A diagram showing the positions of the two helices and the bend in the BCA1 peptide structure is also displayed on top of the panels. The relaxation data show that the bending region of BCA1 peptide is not flexible in the complex.
DISCUSSION
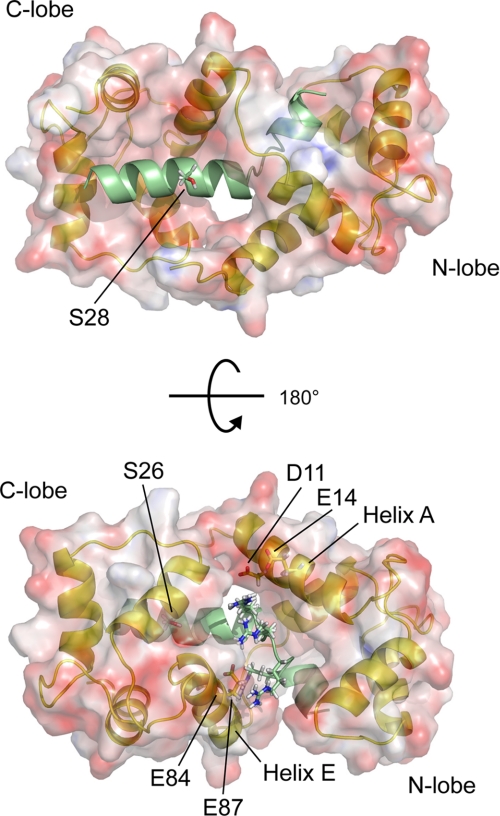
The structures of the CaMBDs in previously determined CaM·peptide complexes have often been classified into “1-5-10”, “1-8-14”, or “1-16” motifs, although there are some CaMBDs, such as the myristoylated alanine-rich C kinase substrate peptide, that do not conform to this pattern (42–45). The solution structure of the SCaM4·BCA1 complex revealed a novel 1-8-18 CaM-binding mode that was dictated by the unique structure of the bound BCA1 peptide. A 1-8-18 CaM-binding motif has been predicted previously for the CaMBD of the plasma membrane Ca2+-ATPase isoform 4b (46) and most recently, an NMR solution structure has been determined with the peptide corresponding to its CaMBD (C28W) (39). Unlike the CaM-bound C28W peptide, which forms a nearly perfect α-helix for the entire region, the structure of the bound BCA1 peptide in SCaM4·BCA1 resembles the reported structure of the BCA1 peptide determined in a 30% trifluoroethanol solution, where the BCA1 peptide consists of two short α-helices that are separated by a disordered region around the basic “RARR” cluster (Fig. 3b) (20). It is known that CaMBD peptides are unstructured in aqueous solution and form a helical structure in trifluoroethanol solution, which resembles that seen in the CaM·peptide complexes (47). This earlier result suggested that the basic cluster portion of the BCA1 peptide was intrinsically flexible. A remarkable feature of the bound BCA1 peptide structure in SCaM4·BCA1 is that the basic cluster forms a well defined relatively fitted bend and the angle between the two α-helices can be defined as 48° ± 5.3° (Fig. 3a). Consequently, the relative orientation of the two lobes of SCaM4 is distinct from any previously reported CaM·peptide complexes (Fig. 5). Although the bending region of the BCA1 peptide is relatively poorly converged in the calculated SCaM4·BCA1 structures (Figs. 2a and 3a), the correlation times obtained for the N- and C-terminal lobe of SCaM4 are almost identical to that of the entire SCaM4 protein (supplemental Table 3). This indicates that the bend region of the BCA1 peptide is not flexible and that it does not allow the N- and C-terminal units of SCaM4·BCA1 to behave independently as is seen for the free CaM protein (48). The two principle axes determined for the diffusion tensors for the N- and C-terminal lobe of SCaM4 are also consistent with the orientation of the two lobes in the structure of SCaM4·BCA1 (Fig. 6). However, it is possible that the bend region undergoes a slow internal motion, which could influence our analysis. It has been discussed that RDC data can be influenced by long time averaged information, whereas 15N relaxation experiments are only sensitive to nanosecond motions (49). The recently determined crystal structure of CaM·RYR1 also demonstrated a large “1-17” spacing of the two anchor residues without any interlobe interactions of the two domains of CaM, and the authors obtained a discrepancy between the resulting structure and the H-N RDC data (38). This might also be caused by a slow motion in the middle of the bound RYR1 peptide, and indeed, the presence of such a dynamic region in the CaM-bound RYR1 peptide was suggested by fluorescence resonance energy transfer measurements (50). Importantly, the CaMBD of RYR1 also contains a basic cluster in the middle of the sequence (Fig. 1a). To address these issues for the vacuolar ATPase complex, we have studied the dynamic properties of the bound isotope-labeled BCA1 peptide. The {1H}-15N NOE and T2 relaxation data and the T1ρ/T2 analysis clearly demonstrated that the bend region has no flexibility compared with the helical regions on the time scales of either nanoseconds or a few hundred microseconds (Fig. 7). We have therefore concluded that the relatively poorly determined structure around the bend is solely due to a lack of structural information rather than dynamic properties of the bound BCA1 peptide. As the hairpin-like loop that was seen in the CaM-bound CaMKK peptide also involved a highly basic region (Fig. 1a) (37, 51), it seems that a basic cluster in the middle of a CaMBD peptide can induce a bound peptide conformation that deviates from a regular continuous α-helix. Although there is no direct answer to the question why the BCA1 peptide has a bend at a particular angle, we can offer two explanations. First, should BCA1 form a continuous α-helical structure, the basic residues located around the bend region, including Arg-33, Arg-35, Arg-36, and Arg-38 will be equally distributed around the surface of the α-helix, which is obvious in Fig. 1b. This would cause direct contacts between these positive charges and the hydrophobic target binding patch of the N-terminal lobe of CaM. Second, and more importantly, in our structure, the basic cluster of the bound BCA1 peptide is positioned between helices A and E of SCaM4 and seemingly forms favorable electrostatic contacts with several negatively charged SCaM4 surface residues that include Asp-11, Glu-14, Glu-84, and Glu-87 (Fig. 3c and Fig. 8). These ion-pair interactions are probably essential to keep the two lobes of SCaM4 in a specific orientation leading to a relatively well defined angle between the two α-helices of the bound BCA1 peptide.
FIGURE 8.
The surface structure of SCaM4 is displayed together with a ribbon structure of SCaM4·BCA1. The side chains of the two Ser residues that have been reported to become phosphorylated by protein kinase C are shown (53). The acidic side chains of SCaM4 and the basic side chains of the BCA1 peptide that can form electrostatic interactions are also highlighted.
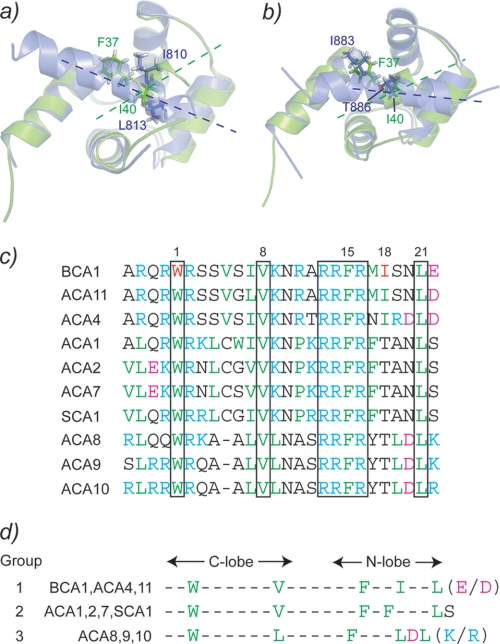
The first anchor residue of the BCA1 peptide Trp-32 shows almost identical hydrophobic contacts to the hydrophobic pocket of the C-terminal lobe of SCaM4 to those seen in many other CaM·target complexes (e.g. CaM·smMLCK, CaM·RYR1, and CaM·CaM kinase I) (Fig. 4). On the other hand, the second anchor residue Ile-40 is somewhat differently positioned in the hydrophobic pocket of the N-terminal lobe of SCaM4. In Fig. 9a, the interaction of the N-terminal lobe with the peptide is compared with that seen in the CaM·smMLCK complex. It appears that Ile-40 is located closer to the center of the hydrophobic pocket than the second anchor residue Leu-813 of smMLCK. The Phe-37 side chain of the BCA1 peptide also interacts with the hydrophobic patch of the N-terminal lobe of SCaM4, unlike the corresponding residue Ile-810 of smMLCK, which points to the other side of the peptide. Consequently, the orientation of the BCA1 peptide relative to the N-terminal lobe of SCaM4 is clearly different from that found in CaM·smMLCK (Fig. 9a). On the other hand, the recently reported CaM complex with the NMDA receptor peptide revealed a polar residue, Thr-886, as an anchor residue for the N-terminal lobe that is positioned almost the same as Ile-40 in SCaM4·BCA1 (Fig. 9b) (52). In addition, Ile-883 in the CaM·NMDA receptor complex also occupies a similar position as Phe-37 in SCaM4·BCA1.
FIGURE 9.
The interaction between the N-terminal lobe of SCaM4 and the BCA1 peptide are compared with those of CaM·smMLCK (a) and CaM·NMDA receptor (b). The N-terminal lobe of SCaM4 is colored green in both panels. c, the sequence alignment of the CaMBD of BCA1 with those of other type IIB Ca2+-ATPases. The conserved residues among all of the CaMBDs are boxed. d, the CaMBDs are classified into three groups according to the sequence alignment in b.
Because several different type IIB Ca2+-ATPases have been identified in different plant species, we have compared the amino acid sequence of the BCA1 peptide with those of the CaMBDs from the other type IIB Ca2+-ATPases (Fig. 9c). In A. thaliana and rice, these have been classified in four subfamilies based on genetic analysis (14). It should be noted that no CaMBD could be detected in the N-terminal region in one subfamily that includes ACA12 and ACA13 (19). Therefore, the known CaMBDs of type IIB Ca2+-ATPases can be classified into three different groups according to their amino acid sequences as shown in Fig. 9d. Group 1 includes BCA1, ACA4, and ACA11, and these calcium pumps are all localized on the prevacuolar or vacuolar membrane, and all the key hydrophobic residues are conserved. Consequently, all the members in this group are expected to have a CaM-binding mode that is identical to that determined here for the SCaM4·BCA1 complex. For the group2 ATPases, which include ACA1, ACA2, ACA7, and soybean type IIB Ca2+-ATPase (SCA1), the Ile residue at position 18 is replaced by a Thr, and instead, an extra Phe residue is located at position 17. This would correspond to the 1-17 CaMBD motif reported for the CaM·RYR1 complex. Group 3, which includes ACA8, ACA9, and ACA10, contains a typical 1-8-14 motif because of a deletion at position 5. The latter calcium pumps are therefore more likely to form a CaM complex that resembles CaM·smMLCK and CaM·CaM kinase I (53). Group 2 and 3 Ca2+-ATPases appear to be located on the chloroplast/endoplasmic reticulum and plasma membranes, respectively. It is interesting that the Ca2+-CaM induced structures for CaMBDs in Ca2+-ATPases located in physically distinct locations in the cell all have a different structure. Consequently, they must have a somewhat different activation mechanism.
Similar to many other CaM-target proteins, the CaMBDs of the type IIB Ca2+-ATPases can become phosphorylated in vivo. For BCA1, it has been shown that Ser-26 and/or -28 are potential target sites for protein kinase C and that the phosphorylation of Ser-28 had no effect on CaM binding (54). In the SCaM4·BCA1 structure, due to the unusual relative positioning of the two lobes of SCaM4, residue Ser-28 is not covered by the N-terminal lobe and is fully exposed to the solvent, which is consistent with the protein phosphorylation results (Fig. 8). On the other hand, Ser-26 is completely covered by the hydrophobic patch of the C-terminal lobe of SCaM4, and hence, phosphorylation of Ser-26 should markedly influence and probably prevent the binding of Ca2+-CaM to BCA1.
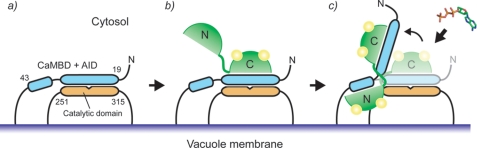
Here, we propose a possible mechanism in which the bend CaMBD structure that is caused by SCaM4 binding is important for the activation of the calcium pump. BCA1 consists of seven transmembrane domains, and the catalytic domain is located between the second and the third transmembrane domain (residues 251–315) (Fig. 10) (54). The N-terminal auto inhibitory domain partially overlaps with the CaMBD of BCA1. In resting cells with a low Ca2+ concentration, SCaM4 would be either free from BCA1 or it interacts with its CaMBD through the C-terminal lobe because it is well known that the Ca2+ affinity of the C-terminal lobe of CaM is very high in the presence of a target (Fig. 10b). This would facilitate an immediate response to a Ca2+ influx. However, Ca2+ binding to the N-terminal lobe of SCaM4 is required for full activation of BCA1. If the CaM-CaMBD·AID complex remains at the catalytic domain after their binding, there could still be steric hindrance to prevent substrate binding. Therefore, it has been discussed that additional mechanisms must be required to dislocate the AID from the catalytic domain. In the case of the activation of smMLCK and NAD kinase by CaM, the importance of additional interactions between the surface of the CaM-CaMBD·AID complex and other region of these enzymes have been described as a requirement for full activation (55–57). Such interactions are not required for BCA1, as removal of the AID from the catalytic domain can be directly achieved by lifting up the AID and the neighboring region by CaM binding (Fig. 10c). The unique bend complex structure would create enough room to allow access for a relatively small ATP molecule to the active site of BCA1.
FIGURE 10.
Model for a possible target activation mechanism from SCaM4·BCA1. a, only the N-terminal cytosolic domain, which contains the CaMBD and AID, and the second cytosolic domain, which contains the catalytic domain, are shown. b, at a low level of Ca2+, SCaM4 would be either free from the CaMBD or bound to the CaMBD only through its C-terminal lobe. c, at a high level of Ca2+, the N-terminal lobe binds Ca2+ and interacts with the CaMBD of BCA1 and induces a bend causing the release of the AID from the active site region leading to the full activation of BCA1.
As the vacuole is the main Ca2+ storage site in plant cells, vacuolar Ca2+-ATPases play a central role in Ca2+ homeostasis in plants. In this work, we have described the novel solution structure of SCaM4·BCA1, a plant CaM in complex with a target protein from a plant species, and proposed a novel CaM-target activation mechanism. Because of the highly conserved amino acid sequence and tertiary structure of CaM in various eukaryotes, a similar mechanism of action can be expected for CaM target protein activation in other organisms including mammals. Importantly, the observation that CaMBDs can have a break in the middle of an α-helical sequence drastically expands the variety of potential CaM-binding motifs. The standard helical wheel prediction that is commonly used to predict anchor residues for CaMBDs obviously does not apply in this case. In fact, a recent protein array study to screen for CaM-binding targets among 37,200 human brain proteins revealed 72 novel CaM-binding proteins, most of which did not contain a typical CaMBD motif (58). Finally, although the CaMBDs identified in the other isoforms of type IIB Ca2+-ATPases with a different subcellular localization appear to belong to well known motifs (1-14 or 1-17), they all possess the same cluster of basic residues that forces a bend in the middle of the BCA1 CaMBD sequence (Fig. 9c). Therefore, we cannot rule out the possibility that the bound CaMBDs from the other Ca2+-ATPase isoforms will also adopt a bend and have a similar activation mechanism for full enzyme activation. Further structural investigations of the other isoforms is required to fully understand the isoform- and organelle-specific CaM regulation of plant type IIB Ca2+-ATPases.
Supplementary Material
Acknowledgment
We thank Dr. Deane McIntyre for the maintenance of the NMR instrumentation in the Bio-NMR Centre.
This work was supported by an operating grant from the Natural Science and Engineering Research Council of Canada.
The atomic coordinates and structure factors (code 2L1W) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–4.
- CaMBD
- CaM-binding domain
- smMLCK
- smooth muscle myosin light chain kinase
- RDC
- residual dipolar coupling
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- NOE
- nuclear Overhauser effect
- r.m.s.d.
- root mean square deviation
- CaM
- calmodulin
- AID
- autoinhibitory domain
- CaMKK
- calcium/calmodulin-dependent protein kinase kinase
- TOCSY
- total correlation spectroscopy
- NOESY
- nuclear Overhauser effect spectroscopy
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1.Reddy A. S. (2001) Plant Sci. 160, 381–404 [DOI] [PubMed] [Google Scholar]
- 2.Lecourieux D., Ranjeva R., Pugin A. (2006) New Phytol. 171, 249–269 [DOI] [PubMed] [Google Scholar]
- 3.Snedden W. A., Fromm H. (2001) New Phytol. 151, 35–66 [DOI] [PubMed] [Google Scholar]
- 4.DeFalco T. A., Bender K. W., Snedden W. A. (2010) Biochem. J. 425, 27–40 [DOI] [PubMed] [Google Scholar]
- 5.Zielinski R. E. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697–725 [DOI] [PubMed] [Google Scholar]
- 6.Zielinski R. E. (2002) Planta. 214, 446–455 [DOI] [PubMed] [Google Scholar]
- 7.Lee S. H., Kim J. C., Lee M. S., Heo W. D., Seo H. Y., Yoon H. W., Hong J. C., Lee S. Y., Bahk J. D., Hwang I., Cho M. J. (1995) J. Biol. Chem. 270, 21806–21812 [DOI] [PubMed] [Google Scholar]
- 8.Boonburapong B., Buaboocha T. (2007) BMC Plant Biol. 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S. H., Kim M. C., Heo W. D., Kim J. C., Chung W. S., Park C. Y., Park H. C., Cheong Y. H., Kim C. Y., Lee K. J., Bahk J. D., Lee S. Y., Cho M. J. (1999) Biochim. Biophys. Acta 1433, 56–67 [DOI] [PubMed] [Google Scholar]
- 10.Lee S. H., Johnson J. D., Walsh M. P., Van Lierop J. E., Sutherland C., Xu A., Snedden W. A., Kosk-Kosicka D., Fromm H., Narayanan N., Cho M. J. (2000) Biochem. J. 350, 299–306 [PMC free article] [PubMed] [Google Scholar]
- 11.McCormack E., Braam J. (2003) New Phytol. 159, 585–598 [DOI] [PubMed] [Google Scholar]
- 12.Evans D. E., Williams L. E. (1998) Biochim. Biophys. Acta 1376, 1–25 [DOI] [PubMed] [Google Scholar]
- 13.Geisler M., Axelsen K. B., Harper J. F., Palmgren M. G. (2000) Biochim. Biophys. Acta 1465, 52–78 [DOI] [PubMed] [Google Scholar]
- 14.Baxter I., Tchieu J., Sussman M. R., Boutry M., Palmgren M. G., Gribskov M., Harper J. F., Axelsen K. B. (2003) Plant Physiol. 132, 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolte S., Brown S., Satiat-Jeunemaitre B. (2004) J. Cell Sci. 117, 943–954 [DOI] [PubMed] [Google Scholar]
- 16.Kabala K., Klobus G. Y. (2005) Acta Physiol. Plant 27, 559–574 [Google Scholar]
- 17.Boursiac Y., Harper J. F. (2007) J. Bioenerg. Biomembr. 39, 409–414 [DOI] [PubMed] [Google Scholar]
- 18.Malmström S., Askerlund P., Palmgren M. G. (1997) FEBS Lett. 400, 324–328 [DOI] [PubMed] [Google Scholar]
- 19.Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) J. Struct. Funct. Genomics 1, 8–14 [DOI] [PubMed] [Google Scholar]
- 20.Yamniuk A. P., Vogel H. J. (2004) J. Biol. Chem. 279, 7698–7707 [DOI] [PubMed] [Google Scholar]
- 21.Ishida H., Huang H., Yamniuk A. P., Takaya Y., Vogel H. J. (2008) J. Biol. Chem. 283, 14619–14628 [DOI] [PubMed] [Google Scholar]
- 22.Jaroniec C. P., Kaufman J. D., Stahl S. J., Viard M., Blumenthal R., Wingfield P. T., Bax A. (2005) Biochemistry 44, 16167–16180 [DOI] [PubMed] [Google Scholar]
- 23.Ottiger M., Delaglio F., Bax A. (1998) J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR. 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 25.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 26.Jee J., Güntert P. (2003) J. Struct. Funct. Genomics 4, 179–189 [DOI] [PubMed] [Google Scholar]
- 27.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR. 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 28.Gifford J. L., Walsh M. P., Vogel H. J. (2007) Biochem. J. 405, 199–221 [DOI] [PubMed] [Google Scholar]
- 29.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 30.Zweckstetter M., Bax A. (2000) J. Am. Chem. Soc. 122, 3791–3792 [Google Scholar]
- 31.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 29–32 [DOI] [PubMed] [Google Scholar]
- 32.Tjandra N., Wingfield P., Stahl S., Bax A. (1996) J. Biomol. NMR. 8, 273–284 [DOI] [PubMed] [Google Scholar]
- 33.Lipsitz R. S., Tjandra N. (2004) Annu. Rev. Biophys. Biomol. Struct. 33, 387–413 [DOI] [PubMed] [Google Scholar]
- 34.Prestegard J. H., Bougault C. M., Kishore A. I. (2004) Chem. Rev. 104, 3519–3540 [DOI] [PubMed] [Google Scholar]
- 35.Bax A., Grishaev A. (2005) Curr. Opin. Struct. Biol. 15, 563–570 [DOI] [PubMed] [Google Scholar]
- 36.Meador W. E., Means A. R., Quiocho F. A. (1992) Science 257, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 37.Kurokawa H., Osawa M., Kurihara H., Katayama N., Tokumitsu H., Swindells M. B., Kainosho M., Ikura M. (2001) J. Mol. Biol. 312, 59–68 [DOI] [PubMed] [Google Scholar]
- 38.Maximciuc A. A., Putkey J. A., Shamoo Y., Mackenzie K. R. (2006) Structure 14, 1547–1556 [DOI] [PubMed] [Google Scholar]
- 39.Juranic N., Atanasova E., Filoteo A. G., Macura S., Prendergast F. G., Penniston J. T., Strehler E. E. (2010) J. Biol. Chem. 285, 4015–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fushman D., Varadan R., Assfalg M., Walker O. (2004) Prog. Nucl. Mag. Res. Sp. 44, 189–214 [Google Scholar]
- 41.Tjandra N., Kuboniwa H., Ren H., Bax A. (1995) Eur. J. Biochem. 230, 1014–1024 [DOI] [PubMed] [Google Scholar]
- 42.Hoeflich K. P., Ikura M. (2002) Cell 108, 739–742 [DOI] [PubMed] [Google Scholar]
- 43.Vetter S. W., Leclerc E. (2003) Eur. J. Biochem. 270, 404–414 [DOI] [PubMed] [Google Scholar]
- 44.Yamniuk A. P., Vogel H. J. (2004) Mol. Biotechnol. 27, 33–57 [DOI] [PubMed] [Google Scholar]
- 45.Ishida H., Vogel H. J. (2006) Protein Pept. Lett. 13, 455–465 [DOI] [PubMed] [Google Scholar]
- 46.Penheiter A. R., Filoteo A. G., Penniston J. T., Caride A. J. (2005) Biochemistry 44, 2009–2020 [DOI] [PubMed] [Google Scholar]
- 47.Zhang M., Yuan T., Vogel H. J. (1993) Protein Sci. 2, 1931–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. (1992) Biochemistry 31, 5269–5278 [DOI] [PubMed] [Google Scholar]
- 49.Ryabov Y., Fushman D. (2006) Magn. Reson. Chem. 44, S143–151 [DOI] [PubMed] [Google Scholar]
- 50.Boschek C. B., Sun H., Bigelow D. J., Squier T. C. (2008) Biochemistry 47, 1640–1651 [DOI] [PubMed] [Google Scholar]
- 51.Osawa M., Tokumitsu H., Swindells M. B., Kurihara H., Orita M., Shibanuma T., Furuya T., Ikura M. (1999) Nat. Struct. Biol. 6, 819–824 [DOI] [PubMed] [Google Scholar]
- 52.Ataman Z. A., Gakhar L., Sorensen B. R., Hell J. W., Shea M. A. (2007) Structure 15, 1603–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baekgaard L., Luoni L., De Michelis M. I., Palmgren M. G. (2006) J. Biol. Chem. 281, 1058–1065 [DOI] [PubMed] [Google Scholar]
- 54.Malmström S., Akerlund H. E., Askerlund P. (2000) Plant Physiol. 122, 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S. H., Seo H. Y., Kim J. C., Heo W. D., Chung W. S., Lee K. J., Kim M. C., Cheong Y. H., Choi J. Y., Lim C. O., Cho M. J. (1997) J. Biol. Chem. 272, 9252–9259 [DOI] [PubMed] [Google Scholar]
- 56.Krueger J. K., Gallagher S. C., Zhi G., Geguchadze R., Persechini A., Stull J. T., Trewhella J. (2001) J. Biol. Chem. 276, 4535–4538 [DOI] [PubMed] [Google Scholar]
- 57.Van Lierop J. E., Wilson D. P., Davis J. P., Tikunova S., Sutherland C., Walsh M. P., Johnson J. D. (2002) J. Biol. Chem. 277, 6550–6558 [DOI] [PubMed] [Google Scholar]
- 58.O'Connell D. J., Bauer M. C., O'Brien J., Johnson W. M., Divizio C. A., O'Kane S. L., Berggård T., Merino A., Akerfeldt K. S., Linse S., Cahill D. J. (2010) Mol. Cell. Proteomics 9, 1118–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.