Abstract
Inward rectifier K+ (Kir) channels are activated by phosphatidylinositol-(4,5)-bisphosphate (PIP2), but G protein-gated Kir (KG) channels further require either G protein βγ subunits (Gβγ) or intracellular Na+ for their activation. To reveal the mechanism(s) underlying this regulation, we compared the crystal structures of the cytoplasmic domain of KG channel subunit Kir3.2 obtained in the presence and the absence of Na+. The Na+-free Kir3.2, but not the Na+-plus Kir3.2, possessed an ionic bond connecting the N terminus and the CD loop of the C terminus. Functional analyses revealed that the ionic bond between His-69 on the N terminus and Asp-228 on the CD loop, which are known to be critically involved in Gβγ- and Na+-dependent activation, lowered PIP2 sensitivity. The conservation of these residues within the KG channel family indicates that the ionic bond is a character that maintains the channels in a closed state by controlling the PIP2 sensitivity.
Keywords: G Proteins, Ion Channels, Membrane Proteins, Potassium Channels, X-ray Crystallography, Electrophysiology, Intracellular Na+, Inward Rectifier K+ Channel
Introduction
The regulation of gating and ion conduction in inward rectifier K+ (Kir) channels is fundamental to their physiological functions, such as the control of the deep resting membrane potential and cell excitability (1–3). The Kir channel is a tetramer with two structurally and functionally distinct transmembrane and cytoplasmic domains (4–6). The association of phosphatidylinositol-(4,5)-bisphosphate (PIP2)3 with the cytoplasmic domain of Kir channels causes channel activation (7–9). However, the G protein-gated Kir (KG) channels also require either G protein βγ subunits (Gβγ) (2, 10–12) or intracellular Na+ (13, 14) for their activation. The binding of either Gβγ or Na+ is thought to evoke conformational changes in the cytoplasmic domain of KG channels, which results in an increase in the channel sensitivity to PIP2, leading to channel opening (7, 15, 16). Thus, in the absence of either Gβγ or Na+ and in contrast with other Kir channels, KG channels are relatively insensitive to PIP2.
The crystal structures of whole Kir channels (4–6, 17) and parts of Kir channels (18–20) have provided valuable insights into the distribution of amino acids essential for the interaction with PIP2 (16, 21–23), Gβγ (24–26), and Na+ (16, 27–29). Furthermore, a series of crystal structures of the bacterial homologue Kir channel, KirBac3.1, reveals that the channel undergoes subunit rearrangement at the cytoplasmic domain accompanied by the alteration in the interaction between the N and C termini (17). However, the mechanism(s) underlying the specific regulation of KG channel activity and the ligand-induced conformational changes remained to be clarified.
To address these questions, we determined the crystal structure of the cytoplasmic domain of the homotetrameric KG channel subunit Kir3.2 in the absence of Na+ and performed the mutational analysis. The activator-free Kir3.2 possesses an ionic bond, which connects the N terminus (His-69) and the CD loop (Asp-228) of the C terminus. Because these residues are conserved within KG channel family subunits, this ionic bond seems to stabilize the closed state by preventing conformational changes in the cytoplasmic domain. The corresponding residues to His-69 and Asp-228 are known to be responsible for respective Gβγ-dependent (25) and Na+-dependent activation (16, 27, 28, 30). Furthermore, the CD loop of Kir channels has residues important for PIP2-dependent activation (16, 21–23). Therefore, the disruption of the bond is a common mechanism for the different activators to trigger channel opening by allowing the cytoplasmic domain to change its conformation and promoting the association with PIP2.
EXPERIMENTAL PROCEDURES
Crystallization, Data Collection, and Model Building of Cytoplasmic Domain of Kir3.2
The longest isoform of mouse Kir3.2 (Kir3.2c) is made up of 425 amino acids with two more amino acids on the N terminus compared with human Kir3.2. Therefore, His-69 and Asp-228 in mouse Kir3.2 correspond to His-67 and Asp-226 in human Kir3.2, respectively (27). The concatemer of cytoplasmic N and C termini of Kir3.2 (amino residues 53–74 and 200–381, respectively) was expressed in bacteria and purified without the addition of Na+ as described previously (20). Crystals were grown by vapor diffusion at 4 °C by mixing equal volumes of protein solution (5 mg/ml) and reservoir solution of 7.5–12.5% (v/v) ethanol, 0.1 m imidazole HCl (pH 8.0), 100 mm MgCl2, and 10–30 mm spermine. Crystals were immersed in reservoir solution supplemented with 25% (v/v) glycerol for cryoprotection. The data were collected at the BL41XU beam line at the SPring-8. Crystals belong to the space group I422 with cell dimensions a = b = 81.943 Å and c = 172.769 Å. The data were indexed and scaled using the HKL2000 suite (31). The initial phase was determined by molecular replacement using the coordinates of the cytoplasmic domain of Kir3.2 (Protein Data Bank code 2E4F) (20). Model refinement was carried out with programs in the CCP4i suite (32). The statistics of the crystallographic analyses are presented in Table 1. The illustrations of the structures were prepared using PyMOL (DeLano Scientific).
TABLE 1.
Summary of x-ray diffraction data for the cytoplasmic domain of Kir3.2 in the absence of Na+
| Data collection statistics | |
| Space group | I422 |
| Unit cell dimensions | a = b = 81.943 Å, c = 172.769 Å; α = β = γ = 90° |
| No. per a.u.a | 1 |
| Wavelength (Å) | 1.00 |
| Resolution (Å) | 2.2 |
| Total observation | 145,544 |
| Unique observation | 14,563 |
| Rmerge (%)b,c | 6.3 (62.9) |
| Completenessc | 99.4 (98.2) |
| I/σ(I)c | 61.0 (2.7) |
| Redundancyc | 9.5 (9.1) |
| Refinement statistics | |
| Resolution (Å) | 30-2.2 |
| Rwork (%)d | 19.8 |
| Rfree (%)e | 23.6 |
| No. of protein atoms | 1574 |
| No. of water molecules | 140 |
| Average B value | |
| Overall | 31.8 |
| Main | 29.7 |
| Side chain/water | 33.5 |
| r.m.s.d. | |
| Bonds (Å) | 0.012 |
| Angles | 1.27° |
| Ramachandran plotf | 88.6/11.4/0/0 |
a a.u. represents the number of protein molecules per asymmetric unit. r.m.s.d. represents root mean square deviation.
b Rmerge = ΣhΣi(|Ii(h)| − |〈I(h)〉|)/ΣhΣiIi(h), where Ii(h) is the observed intensity and 〈I(h)〉 is the mean intensity observed from multiple measurements.
c Values in parentheses define the resolution range at the highest shell of data.
d Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc denote observed and calculated structure factors, respectively.
e Five percent of the reflections were set aside for the calculation of the Rfree value.
f Shown is the percentage of amino acids that distribute in most favored/allowed/generous/disallowed regions of Ramachandran plot.
Electrophysiological Analyses
The cDNA of mouse Kir3.2d (33) were subcloned into the expression vector pcDNA3 (Invitrogen) and transfected with porcine m2-muscarinic receptor into HEK293T cells with Lipofectamine (Invitrogen). Point mutations were introduced using a QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. Mutations were confirmed with direct DNA sequencing. Inward Kir currents were recorded at a holding potential of −100 mV in excised inside-out patches using a patch clamp amplifier (Axopatch 200A, Axon Instruments). The current was low pass-filtered at 1 kHz and digitized at 10 kHz. Glass pipettes had resistances of 2.5–3.5 megohms when filled with pipette solution. The pipette solution contained 135 mm KCl, 1 mm CaCl2, 1.6 mm MgCl2, 5 mm HEPES, and 5 μm acetylcholine, and the bath solution was composed of 135 mm KCl, 5 mm EGTA, 2 mm MgCl2, and 5 mm HEPES. PIP2 (Sigma-Aldrich) dissolved in chloroform was stored at −20 °C. Before the experiment, an aliquot of PIP2 solution was evaporated with a flow of argon gas and dissolved in the bath solution by sonication for 2–3 min. Solutions containing 2 mm K2ATP were prepared freshly each day. The pH of all solutions was adjusted to 7.35 with KOH prior to the experiments. Because the perfusion of NaCl did not shift the zero current level recorded in the presence of 10 μm CdCl2, NaCl was added to the bath solution without compensation for changes in the osmolarity and ionic strength. All experiments were performed at ambient temperature (23–25 °C). The data are expressed as means ± S.E. Statistics were performed using Student's t test.
RESULTS
Differences in Conformation between Kir3.2 Structures Obtained in Presence and Absence of Na+
The concatemer of polypeptide segments of cytoplasmic N and C termini of Kir3.2 (Fig. 1A) was expressed and crystallized in the absence of Na+ (Na+-free Kir3.2). The structure was solved by the molecular replacement method with the Kir3.2 structure obtained in the presence of Na+ (Na+-plus Kir3.2) as a search model (20). The electron density was continuous enough to trace CD and LM loops, which were partially disordered in the Na+-plus structure. The solution was brought to crystallographic refinement. Data collection and refinement statistics are presented in Table 1.
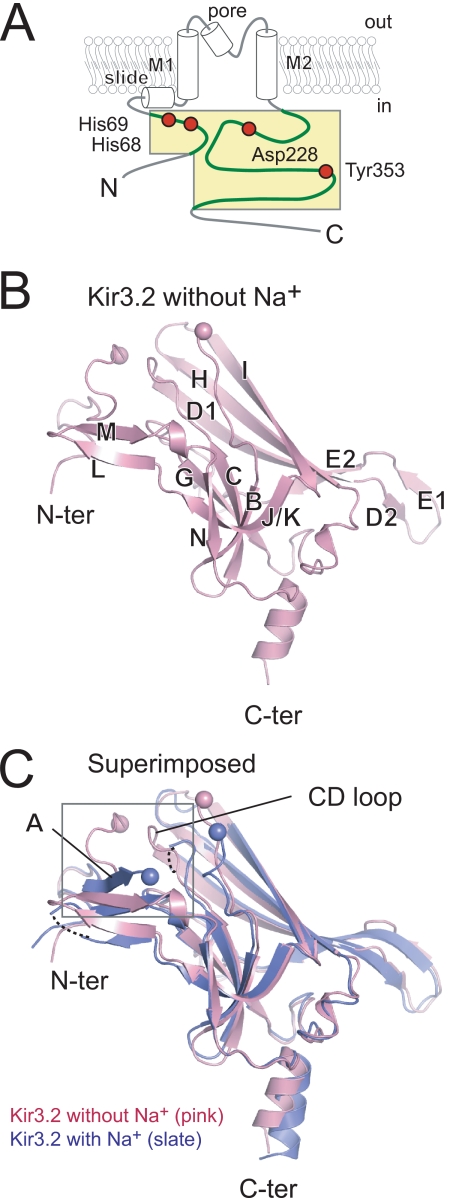
FIGURE 1.
Molecular architecture of Kir3.2 cytoplasmic domain in the absence of Na+. A, schematic presentation of Kir3.2. Both N and C termini of Kir3.2 are exposed to cytoplasm flanked by a transmembrane domain composed of slide, M1, pore, and M2 helices. The concatemer of the N and C termini of Kir3.2 (residues 53–74 and 200–381, respectively) comprises the soluble cytoplasmic core region. The region subjected for crystallization is colored yellow, and amino acids critically involved in the interaction between the N (N) and C (C) termini are indicated by red circles. B, monomer architecture of Kir3.2. A ribbon model of a monomer of the cytoplasmic domain of Kir3.2 in the absence of Na+ (pink) is presented with the position of the β strands. The Cα positions of the residues prior to or following the disordered region connecting the N (N-ter) and C (C-ter) termini are indicated by spheres. C, comparison of two Kir3.2 structures. Kir3.2 structure in the presence of Na+ (blue) is superimposed onto the Na+-free structure (pink). The βA strand is only observed in the Kir3.2 structure in the presence of Na+. The enlarged view shown in Fig. 2 is indicated by the square.
The cytoplasmic domain of the Na+-free Kir3.2 is a β strand-rich immunoglobulin-like domain (Fig. 1B, left panel). The positions of backbone Cα atoms in the Na+-free Kir3.2 structure are similar to those in the Na+-plus Kir3.2 (root mean square deviation, 2.0 Å) (Fig. 1B, right panel). A large difference in the region connecting N and C termini and small differences in CD, DE, HI, and LM loops contribute to an increase (271 Å2) in the buried surface area between subunits in the tetrameric assembly of the Na+-free Kir3.2. However, there was no apparent difference in the subunit interaction between the two Kir3.2 structures. The top of the cytoplasmic pore consists of HI loops from four subunits. This loop is positioned differently in different Kir channels, and it is thought to function as a part of the gate (5, 19). Both of the Kir3.2 crystal structures possess a wide opening at the entrance of cytoplasmic pore, enough to allow partially hydrated cations to pass through the pore.
A prominent difference between the two Kir3.2 structures is observed at the interface between the N and C termini (Fig. 1B, square). The side chain position of Asp-228 in the Na+-plus Kir3.2 is equivalent to that in other Kir channel structures (Fig. 2A) (4–6, 17–19, 34). However, the Cγ atom of the carboxyl group of Asp-228 in Na+-free Kir3.2 was rotated counterclockwise by ∼70 ° compared with that in the Na+-plus Kir3.2. The positions of a carboxyl group of Asp-228 and a water molecule (wat2 in A) in the Na+-plus Kir3.2 were substituted by a water molecule (wat1 in B) and a carboxyl group of Asp-228, respectively, in the Na+-free Kir3.2. This unique side chain position of Asp-228 in the Na+-free Kir3.2 suggests that the pivoting movement of Asp-228 affects the conformation of the N terminus and the CD loop.
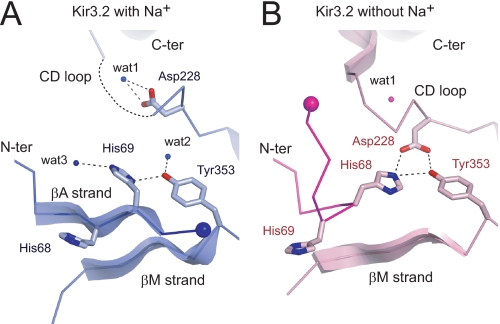
FIGURE 2.
Enlarged view of the interface between N terminus and CD loop of Kir3.2 cytoplasmic domain. Ribbon models of the structural elements at the interface between the N terminus (N-ter) and the CD loop of the C terminus (C-ter) of Kir3.2 in the presence (A) or the absence (B) of Na+ are shown in blue and pink, respectively. The residues important in interactions are shown as stick models. Water (wat) molecules are indicated by small spheres. Large spheres indicate a Cα atom at the terminal residue of the N terminus in each structure. The N termini in the two structures are shown in dense colors.
In the Na+-plus Kir3.2, an imidazole ring of His-69 contacts with the hydroxyl group of Tyr-353 with a distance of 3.0 Å (Fig. 2A). This interaction is involved in the hydrogen bond network with water molecules (wat2 and wat3 in A), partially contributing to the stabilization of the conformation of a β (βA) strand (amino acids 66–70), which is conserved among all Kir channel structures (4–6, 18, 19, 34). On the other hand, in the Na+-free Kir3.2, a βA strand is missing, and an imidazole ring of His-68 approaches to within 2.9 Å of a carboxyl group of Asp-228 on the CD loop, possibly forming an ionic bond (Fig. 2B). In addition to this electrostatic interaction, a hydroxyl group of Tyr-353 interacts with His-68 and Asp-228 with distances of 3.4 and 2.7 Å, respectively. Therefore, the direct binding of the N terminus and the CD loop appears to influence the conformation at the interface between them.
Functional Implications of Role of Ionic Bond between N Terminus and CD Loop in Kir3.2 Channel Gating
Because neither Gβγ nor Na+ was present in the crystallization conditions of the Na+-free Kir3.2 cytoplasmic domain, the ionic bond between His-68 and Asp-228 could be involved in the control of the sensitivity of the channel to PIP2. To test this possibility, we measured channel activity in inside-out membrane patches from HEK293T cells expressing either Kir3.2 WT or mutant channels (Fig. 3A). As noted elsewhere (27), the increase in current amplitude induced by PIP2 in Kir3.2 WT was small, 3.3 ± 1.0% of that evoked by 10 μm GTPγS (n = 5) (Fig. 3, A and C). The substitution of an asparagine for Asp-228 (D228N) resulted in a significant increase in the PIP2-induced current (39.8 ± 7.7%, n = 5), indicating that the D228N mutant is more sensitive to PIP2.
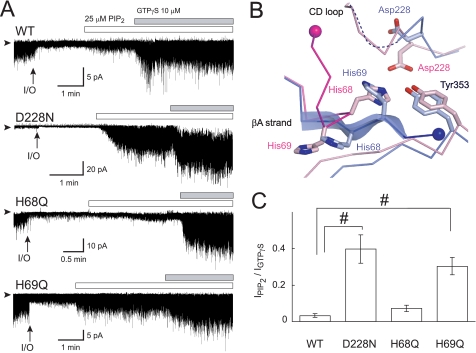
FIGURE 3.
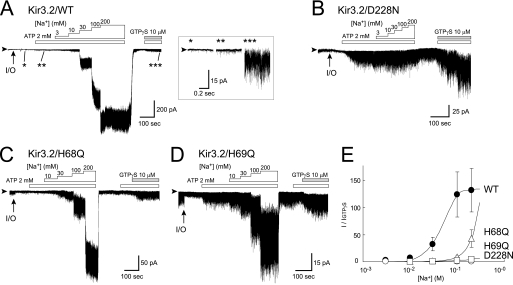
Effects of amino acid substitution on PIP2-induced channel activation. A, effects of PIP2 and GTPγS on currents of Kir3.2 WT, D228N, H68Q, and H69Q mutant channels. The channels were expressed in HEK cells and examined in inside-out membrane patches. Experiments were conducted in symmetric 135 mm K+ solutions with a holding potential of −100 mV. The protocol of perfusion of substances to the intracellular side of the patch membrane is indicated above the current traces. Arrowheads indicate the zero current levels, and arrows (I/O) indicate the time point when the patch was excised. B, comparison of residues involved in the interaction between the N terminus and the CD loop. The Cα traces involved in this interaction in the Na+-plus and Na+-free Kir3.2 structures are colored blue and pink, respectively. The residues critical for the interaction are depicted as stick models. The ends of the N terminus connecting to the slide helix are shown as spheres. C, the summary of PIP2-induced current amplitudes of WT (n = 5), D228N (n = 5), H68Q (n = 7), and H69Q mutants (n = 9) channels (means ± S.E.), which were normalized against that activated by 10 μm GTPγS (#, p < 0.01).
The amino acids corresponding to His-68 of Kir3.2 are not conserved among Kir channels (see Fig. 5B). However, although the substitution of a glutamine for His-68 (H68Q) produced a functional channel, mutants where His-68 were replaced by either glycine, cysteine, glutamate, tyrosine, or tryptophan dramatically reduced functional channel expression. The H68Q mutant showed a low basal activity associated with PIP2 (7.2 ± 1.6%, n = 7), which was as low as that observed in the WT (Fig. 3, A and C). This low sensitivity of the H68Q mutant to PIP2 in the absence of channel activators suggests that the contribution of His-68 to the control of the association with PIP2 is insignificant in the functional channel. When N termini of the two Kir3.2 structures were compared an imidazole ring of His-69 in the Na+-plus Kir3.2 was found at the position equivalent to His-68 in the Na+-free structure (Fig. 3B). In this case, it could be His-69 that geometrically made an ionic bond with Asp-228. The basal current amplitude associated with PIP2 of the mutant Kir3.2 which substituted a glutamine for His-69 (H69Q) was high (30.4 ± 4.8%, n = 9). The introduction of a cysteine for His-69 also resulted in PIP2 increasing the basal current (30.6 ± 8.4%, n = 3) (supplemental Fig. S1). Therefore, whereas His-68 binds to Asp-228 in the Na+-free Kir3.2 crystal structure, in the functional channel it is His-69 that is more likely to be involved in the control of the interaction with PIP2, presumably by forming the ionic bond with Asp-228.
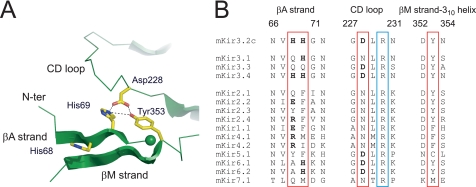
FIGURE 5.
Model of cytoplasmic domain of Kir3. 2 insensitive to PIP2 and comparison of amino acid sequences of the interface between the N and C terminus. A, structural model of cytoplasmic domain of Kir3.2 insensitive to PIP2. Based on the functional analyses, we propose a model where His-69 on the N terminus directly interacts with Asp-228 on the CD loop. In addition to the electrostatic interaction, a hydroxyl group of Tyr-353 may contact with the side chains of these two residues. B, alignments of the amino acid sequences at the interface between the N and the C termini in mouse Kir channels. The residues involved in the direct association and a conserved arginine in the CD loop, which is thought to be crucial for the interaction with PIP2, are enclosed by red and blue squares, respectively. Charged residues in the squares are indicated in boldface. Numbers above the alignments correspond to amino acid numbers in the longest isoform of mouse Kir3.2.
Functional Implications of Residues Forming Ionic Bond in Na+-dependent Activation
Next, we examined the effect of the mutations on Na+-dependent activation. In these experiments, we used ATP instead of PIP2 to produce PIP2 at the inner leaflet of patch membranes (14, 15, 27). Intracellular Na+ activated the Kir3.2 WT channel more effectively than GTPγS, with a 130 (± 40)-fold increase with 200 mm Na+ (n = 5) (Fig. 4A) (27). The activation by Na+ was concentration-dependent with a Hill coefficient of 3.0 ± 0.6 and a half-effective concentration (EC50) value of 56.8 ± 7.6 mm (Fig. 4E). The Kir3.2 D228N mutant was defective for Na+-dependent activation (Fig. 4B). On the other hand, both H68Q and H69Q mutant channels were activated by Na+ in a concentration-dependent manner (Fig. 4, C and D), but the EC50 values were increased at least 3-fold compared with the WT (Fig. 4E). Furthermore, the amplitude of channel currents supported by H68Q and H69Q mutants elicited by 200 mm Na+ were only 30-fold and 3-fold, respectively, greater than the current induced by 10 μm GTPγS. In both cases, the currents elicited by GTPγS were not significantly different from that of the WT. Thus, both mutations appear to reduce the sensitivity to Na+. Because H69Q and H69C mutants showed similar susceptibilities to PIP2 and Na+ (supplemental Fig. S1), the effect of these point mutations at position 69 might be due to the elimination of the side chain of histidine, rather than to the particular characteristics of the introduced residues.
FIGURE 4.
Effects of amino acid substitution on Na+-dependent channel activation. The effect of Na+ on currents of Kir3.2 WT (A), D228N (B), H68Q (C), and H69Q (D) mutant channels were examined. Experimental conditions were as presented in Fig. 3. ATP was applied to induce PIP2 at the inner leaflet of patch membrane. The Kir3.2 WT currents in the absence of channel activators (*) and in the presence of ATP alone (**) or ATP and GTPγS (***) are enlarged in the inset of A. The currents activated by various concentrations of Na+ were normalized against that activated by 10 μm GTPγS (E). N-ter, N terminus.
DISCUSSION
Role of Ionic Bond between N Terminus and CD Loop in Conformational Changes in Cytoplasmic Domain of Kir3.2
In crystal structures of full-length Kir channels (4–6, 17), the channels are presumed to be in the closed state, with the physical constraints for ion permeation in the transmembrane domain. The conformation of the cytoplasmic domain in these complete structures is comparable with those of isolated cytoplasmic domains lacking the transmembrane domain (18–20). Thus, it could be speculated that, in the closed state, the attachment of the transmembrane domain had little effect upon the structure of the cytoplasmic domain. The association of Gβγ and Na+ with the cytoplasmic domain activates KG channels by increasing their PIP2 sensitivity and do not activate the channels by themselves (7–9). This mode of regulation implies that their conformational changes may be restricted to the cytoplasmic domain. The structure of the Na+-free Kir3.2 would correspond to the closed state, and local structural differences between Na+-free and Na+-plus Kir3.2 structures would be consistent with this mode of regulation of the channel. Potentially, the structural difference results from the rotational movement of Asp-228, which couples to the movement of the CD loop (20). Gly-227 adjacent to Asp-228 tends to be conserved within the Kir channel family (Fig. 5B), thereby suggesting that Gly-227 underlies the conformational flexibility of the CD loop. Furthermore, the position of Asp-228 in the Na+-free Kir3.2 would be stabilized by the positively charged histidine on the N terminus. Although His-68 is involved in the ionic bond with Asp-228 in the crystal structure (Fig. 2), His-69 appears to contact with Asp-228 in the full channel (Fig. 3). This discrepancy between which histidine acts as a partner for the ionic bond with Asp-228 can be attributed to the effect of the region linking cytoplasmic N and C termini. It is also possible that the precise position of Asp-228 in the Na+-free Kir3.2 is influenced by conformational changes in the N terminus and/or the linker induced by crystallization. In this case, the CD loop including Asp-228 and the βA strand on the N terminus are structural elements that can change their conformation. Although the absence of a structural model for the open state at the atomic level hampers any rigorous assessment of how the cytoplasmic domain changes its conformation during gating, it might link to the sequential conformational changes in the cytoplasmic domain to control the gate. This is consistent with the results from crystallographic and biochemical analyses of the bacterial Kir channel KirBac3.1 of which the N terminus and the CD loop alter their conformation during gating (17, 35). Thus, we propose a structural model of Kir3.2 in the closed state with an ionic bond between His-69 on the N terminus and Asp-228 on the CD loop (Fig. 5A). This ionic bond would be a crucial structural element for Kir3.2 channel activity as a constraint on the conformational changes in the cytoplasmic domain leading to channel opening.
In Na+-sensitive Kir4.1/Kir5.1 channels, the aspartate that corresponds to Asp-228 in Kir3.2 was proposed to reduce PIP2 sensitivity by forming a bond with the conserved arginine in the same CD loop (Arg-230 in Kir3.2, Fig. 5B) rather than His-69 on the N terminus (30). This proposal could explain how Na+ increases PIP2 sensitivity in both the KG channel and the Kir4.1-Kir5.1 complex. However, these two Kir channels are markedly different in their basal activity; the KG channel is constitutively inactive and the Kir4.1/Kir5.1 channel is constitutively active (36). Thus, the mechanism to maintain the channel in the closed state cannot be accounted for by a bond between the aspartate and the arginine on the CD loop.
The structures of whole Kir channels (4–6, 17) indicate that the N terminus in one subunit associates with the C terminus of the adjacent subunit in functional channel assembly. This implies that the ionic bond between the N terminus and the CD loop in Kir3.2 is an intersubunit interaction. The pair of amino acids forming the bond in Kir3.2 is conserved in Kir3.4, Kir6.1, and Kir6.2, and His-69 in Kir3.2 is also observed in Kir3.1 (Fig. 5B). This suggests that the electrostatic interaction between the N terminus and the CD loop may control the conformational changes in the cytoplasmic domain not only in the homomeric KG channels and KATP channels, but also in the heteromeric KG channels assembled with Kir3.1 and other KG channel subunits.
Functional Implications of Residues Involved in Ionic Bond
The Asp-228 in the CD loop of Kir3.2 (27, 28) and the corresponding aspartates in Kir3.4 (16) and Kir5.1 (30) are known to be involved in Na+-dependent activation of these Kir channels. Furthermore, the histidines equivalent to His-233 in Kir3.2 in the same CD loops in Kir3.4 and Kir5.1 appear to participate in the coordination of Na+ (30). The order of ion selectivity for Kir3.2 activation by monovalent cations is Na+ ≫ Li+ ≫ K+ = Cs+ (data not shown) (13), which corresponds to Eisenman's selectivity sequence X (37). This suggests that the Na+-binding site of Kir3.2 consists of an anionic and relatively compact pocket. However, the point mutations at two histidine residues on the N terminus reduced the Na+-sensitivity of Kir3.2 (Fig. 4 and supplemental Fig. S1). This may be either an indirect effect upon Na+-binding to the site largely contributed to by the CD loop or the conformational changes following the association with Na+ (38). Alternatively, the N terminus might provide additional Na+-binding sites associated with channel activation. Because Na+ and water molecules have an equal number of electrons, it is difficult to distinguish between them on the electron density map. A high resolution structure of Kir3.2 in the presence of a maximal or supramaximal concentration of Na+ would help to address the question of how Na+ interacts with Kir3.2.
On the other hand, the histidine residues in Kir3.1 and Kir3.4 (His-57 and His-64, respectively), which correspond to His-69 in the N terminus of Kir3.2, are involved in channel activation by Gβγ (25). Furthermore, the equivalent histidine in Kir3.4 participates in the inhibition of channel activity by intracellular acidification (39). Based on our observations, the effect of lowering pH on the reduction of the macroscopic current of Kir3.4 could be attributed to facilitation of the formation of the ionic bond between His-64 and Asp-223 on its CD loop. Thus, these reports strongly support our idea that electrostatic interaction between N and C termini is a mechanism crucial for the control of Kir3.2 channel activity.
Activator-induced Conformational Changes in Cytoplasmic Domain
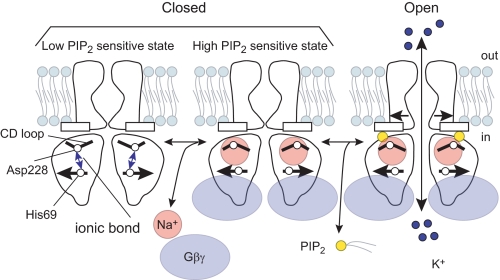
The CD loop possesses an arginine that is conserved among the Kir channel family, and it is thought to be essential for PIP2-dependent channel activation (Fig. 5B) (16, 22). In addition, some of the Andersen-Tawil syndrome mutations in the CD loop of Kir2.1 cause a reduction in the sensitivity to PIP2 (22, 23). The direct interaction between the N terminus and the CD loop, therefore, seems crucial for lowering the sensitivity to PIP2 by constraining the conformation of the CD loop via the control of the pivoting movement of Asp-228 (Fig. 6). However, whereas Kir3.2 mutants that lacked the ionic bond showed increased sensitivity to PIP2, they were also further activated by Na+ and/or Gβγ (Figs. 3 and 4 and supplemental Fig. S1). Thus, the disruption of this ionic bond is not the only effect of the activators. The coordination of Na+ (30) and multiple contacts with Gβγ (24, 26, 40) would also be required for channel activation. Therefore, in addition to the disruption of the ionic bond, the activators appear to induce other conformational changes to further facilitate the sensitivity to PIP2 or stabilize the open channel conformation. Taken together, the intersubunit ionic bond between the N terminus and the CD loop seems to be characteristic of KG channel subunits and acts as an inhibitory mechanism for channel activation (Fig. 6). The two different KG channel activators, Gβγ and Na+, would disrupt the bond, increase the sensitivity to PIP2, and open the gate.
FIGURE 6.
Model of conformational changes in the cytoplasmic domain associated with Kir3.2 channel gating. In the resting state, the N terminus and the CD loop of the cytoplasmic domain interact via an ionic bond. The bond constrains the conformation of the CD loop to maintain the channel in a state less sensitive to PIP2. The channel activators, such as Na+ and Gβγ, disrupt the ionic bond and allow Kir3.2 to interact with PIP2, leading to the opening of Kir3.2.
Supplementary Material
Acknowledgments
We thank Tomohito Okada, Yukiko Nishida, and Chizuru Tsuzuki for technical assistance. We are grateful to Dr. Ian Findlay (EA 4433, Faculté des Sciences, Université de Tours) for critical reading of this manuscript.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 3AGW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PIP2
- phosphatidylinositol-(4,5)-bisphosphate.
REFERENCES
- 1.Hibino H., Inanobe A., Furutani K., Murakami S., Findlay I., Kurachi Y. (2010) Physiol. Rev. 90, 291–366 [DOI] [PubMed] [Google Scholar]
- 2.Yamada M., Inanobe A., Kurachi Y. (1998) Pharmacol. Rev. 50, 723–760 [PubMed] [Google Scholar]
- 3.Lüscher C., Slesinger P. A. (2010) Nat. Rev. Neurosci. 11, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. (2003) Science 300, 1922–1926 [DOI] [PubMed] [Google Scholar]
- 5.Nishida M., Cadene M., Chait B. T., MacKinnon R. (2007) EMBO J. 26, 4005–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilgemann D. W., Ball R. (1996) Science 273, 956–959 [DOI] [PubMed] [Google Scholar]
- 8.Bichet D., Haass F. A., Jan L. Y. (2003) Nat. Rev. Neurosci. 4, 957–967 [DOI] [PubMed] [Google Scholar]
- 9.Logothetis D. E., Jin T., Lupyan D., Rosenhouse-Dantsker A. (2007) Pflugers Arch. 455, 83–95 [DOI] [PubMed] [Google Scholar]
- 10.Logothetis D. E., Kurachi Y., Galper J., Neer E. J., Clapham D. E. (1987) Nature 325, 321–326 [DOI] [PubMed] [Google Scholar]
- 11.Ito H., Tung R. T., Sugimoto T., Kobayashi I., Takahashi K., Katada T., Ui M., Kurachi Y. (1992) J. Gen. Physiol. 99, 961–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuveny E., Slesinger P. A., Inglese J., Morales J. M., Iñiguez-Lluhi J. A., Lefkowitz R. J., Bourne H. R., Jan Y. N., Jan L. Y. (1994) Nature 370, 143–146 [DOI] [PubMed] [Google Scholar]
- 13.Lesage F., Guillemare E., Fink M., Duprat F., Heurteaux C., Fosset M., Romey G., Barhanin J., Lazdunski M. (1995) J. Biol. Chem. 270, 28660–28667 [DOI] [PubMed] [Google Scholar]
- 14.Sui J. L., Chan K. W., Logothetis D. E. (1996) J. Gen. Physiol. 108, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C. L., Feng S., Hilgemann D. W. (1998) Nature 391, 803–806 [DOI] [PubMed] [Google Scholar]
- 16.Zhang H., He C., Yan X., Mirshahi T., Logothetis D. E. (1999) Nat. Cell Biol. 1, 183–188 [DOI] [PubMed] [Google Scholar]
- 17.Clarke O. B., Caputo A. T., Hill A. P., Vandenberg J. I., Smith B. J., Gulbis J. M. (2010) Cell 141, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 18.Nishida M., MacKinnon R. (2002) Cell 111, 957–965 [DOI] [PubMed] [Google Scholar]
- 19.Pegan S., Arrabit C., Zhou W., Kwiatkowski W., Collins A., Slesinger P. A., Choe S. (2005) Nat. Neurosci. 8, 279–287 [DOI] [PubMed] [Google Scholar]
- 20.Inanobe A., Matsuura T., Nakagawa A., Kurachi Y. (2007) Channels 1, 39–45 [PubMed] [Google Scholar]
- 21.Plaster N. M., Tawil R., Tristani-Firouzi M., Canún S., Bendahhou S., Tsunoda A., Donaldson M. R., Iannaccone S. T., Brunt E., Barohn R., Clark J., Deymeer F., George A. L., Jr., Fish F. A., Hahn A., Nitu A., Ozdemir C., Serdaroglu P., Subramony S. H., Wolfe G., Fu Y. H., Ptácek L. J. (2001) Cell 105, 511–519 [DOI] [PubMed] [Google Scholar]
- 22.Lopes C. M., Zhang H., Rohacs T., Jin T., Yang J., Logothetis D. E. (2002) Neuron 34, 933–944 [DOI] [PubMed] [Google Scholar]
- 23.Donaldson M. R., Jensen J. L., Tristani-Firouzi M., Tawil R., Bendahhou S., Suarez W. A., Cobo A. M., Poza J. J., Behr E., Wagstaff J., Szepetowski P., Pereira S., Mozaffar T., Escolar D. M., Fu Y. H., Ptácek L. J. (2003) Neurology 60, 1811–1816 [DOI] [PubMed] [Google Scholar]
- 24.Finley M., Arrabit C., Fowler C., Suen K. F., Slesinger P. A. (2004) J Physiol 555, 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He C., Yan X., Zhang H., Mirshahi T., Jin T., Huang A., Logothetis D. E. (2002) J. Biol. Chem. 277, 6088–6096 [DOI] [PubMed] [Google Scholar]
- 26.He C., Zhang H., Mirshahi T., Logothetis D. E. (1999) J. Biol. Chem. 274, 12517–12524 [DOI] [PubMed] [Google Scholar]
- 27.Ho I. H., Murrell-Lagnado R. D. (1999) J. Biol. Chem. 274, 8639–8648 [DOI] [PubMed] [Google Scholar]
- 28.Ho I. H., Murrell-Lagnado R. D. (1999) J. Physiol. 520, 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui J. L., Petit-Jacques J., Logothetis D. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenhouse-Dantsker A., Sui J. L., Zhao Q., Rusinova R., Rodríguez-Menchaca A. A., Zhang Z., Logothetis D. E. (2008) Nat. Chem. Biol. 4, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project N (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 33.Inanobe A., Horio Y., Fujita A., Tanemoto M., Hibino H., Inageda K., Kurachi Y. (1999) J. Physiol. 521, 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegan S., Arrabit C., Slesinger P. A., Choe S. (2006) Biochemistry 45, 8599–8606 [DOI] [PubMed] [Google Scholar]
- 35.Gupta S., Bavro V. N., D'Mello R., Tucker S. J., Vénien-Bryan C., Chance M. R. (2010) Structure 18, 839–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pessia M., Tucker S. J., Lee K., Bond C. T., Adelman J. P. (1996) EMBO J. 15, 2980–2987 [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenman G. (1962) Biophys. J. 2, 259–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colquhoun D. (1998) Br. J. Pharmacol. 125, 924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao J., Wu J., Chen F., Wang X., Jiang C. (2003) J. Biol. Chem. 278, 7091–7098 [DOI] [PubMed] [Google Scholar]
- 40.Ivanina T., Rishal I., Varon D., Mullner C., Frohnwieser-Steinecke B., Schreibmayer W., Dessauer C. W., Dascal N. (2003) J. Biol. Chem. 278, 29174–29183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.