Abstract
Progressive accumulation of DNA damage is causally involved in cellular senescence and organismal aging. The DNA damage kinase ATM plays a central role in maintaining genomic stability. ATM mutations cause the genetic disorder ataxia telangiectasia, which is primarily characterized by progressive neurodegeneration and cancer susceptibility. Although the importance of ATM function to protect against oxidative DNA damage and during aging is well described, the mechanism of ATM activation by these stimuli is not known. Here we identify ATM interactor (ATMIN) as an essential component of the ATM signaling pathway in response to oxidative stress and aging. Embryos lacking ATMIN (atminΔ/Δ) died in utero and showed increased numbers of cells positive for phosphorylated histone H2aX, indicative of increased DNA damage. atminΔ/Δ mouse embryonic fibroblasts accumulated DNA damage and prematurely entered senescence when cultured at atmospheric oxygen levels (20%), but this defect was rescued by addition of an antioxidant and also by culturing cells at physiological oxygen levels (3%). In response to acute oxidative stress, atminΔ/Δ mouse embryonic fibroblasts showed slightly lower levels of ATM phosphorylation and reduced ATM substrate phosphorylation. Conditional deletion of ATMIN in the murine nervous system (atminΔN) resulted in reduced numbers of dopaminergic neurons, as does ATM deficiency. ATM activity was observed in old, but not in young, control mice, but aging-induced ATM signaling was impaired by ATMIN deficiency. Consequently, old atminΔN mice showed accumulation of DNA damage in the cortex accompanied by gliosis, resulting in increased mortality of aging mutant mice. These results suggest that ATMIN mediates ATM activation by oxidative stress, and thereby ATMIN protects the aging brain by preventing accumulation of DNA damage.
Keywords: Brain, DNA Damage, DNA Repair, Neurodegeneration, Oxidative Stress
Introduction
Genomic instability plays a causative role in both cellular senescence and organismal aging (1, 2). Cytotoxic DNA lesions arise during normal metabolism due to the production of reactive oxygen species (ROS).2 At the cellular level, the persistence of foci of serine 139-phosphorylated H2aX (γH2aX) is often used to define the sites of DNA damage. γH2aX levels are increased in senescing cells due to the increased production of ROS combined with a reduced capacity to repair damaged DNA. Culturing of mouse embryonic fibroblasts (MEFs) in the presence of atmospheric oxygen levels of 20% leads to senescence, but the proliferative potential of MEFs can be extended by culturing them under physiological oxygen conditions (3%) (3). DNA damage has also been observed to increase severely during aging. Consequently, a high incidence of γH2aX foci is found in tissues from aging individuals and cells from patients with accelerated aging disorders such as Werner syndrome (4, 5). At the organismal level, certain tissues are particularly vulnerable to DNA damage such as the immune system where double-strand breaks are generated during somatic recombination, a process that involves ataxia telangiectasia mutated (ATM) function (6, 7). In the nervous system, which consumes ∼20% of the total inhaled oxygen, long-lived neurons have high rates of transcription and produce ROS that can also damage DNA. If unrepaired, such damage poses critical threats to genomic stability and cellular viability.
DNA damage leads to the activation of ATM, which is a member of the phosphoinositide 3-kinase (PI3K)-related protein kinase family that includes ATM and Rad3-related (ATR) and the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (8, 9). Once activated, ATM orchestrates the cellular response by phosphorylating numerous substrates in what is known as the DNA damage response (10). Activation of this signaling cascade is dependent on a heterotrimeric complex consisting of MRE11, Rad50, and NBS1 (MRN). MRN has been shown to sense the presence of chromatin changes and subsequently binds to and processes the damaged DNA ends facilitating recruitment of further substrates (11, 12). NBS1 serves as both a key substrate and an essential adapter required for ATM association at the break, facilitating the unwinding and nuclease activities of the MRN complex (13). The purpose of this intricate signaling network is to ensure cell survival by halting the cell cycle while the damage is repaired (14). Alternatively, in the presence of excessive damage, this pathway may culminate in the activation of programmed cell death to maintain genomic integrity.
ATM is inactivated in the genomic instability disorder ataxia telangiectasia (A-T), the classical hallmarks of which are neurodegeneration, cancer predisposition, premature aging, immunodeficiency, and genomic instability (15, 16). Cells from A-T patients exhibit chromosomal instability, premature senescence, and show severe sensitivity to agents that induce double-strand breaks (17, 18). Similarly to its role in proliferating cells, ATM is also required for the DNA damage response in postmitotic cells, with severe vulnerability to oxidative damage and a defective DNA damage response being responsible for the compromised organ homeostasis associated with A-T. The neurodegeneration in A-T patients manifests as progressive degeneration of Purkinje cells in the cerebellar cortex along with the gradual loss of dopaminergic neurons in the substantia nigra pars compacta (19). Although mouse mutants of ATM do not recapitulate the cerebellar degeneration, ATM deficiency has been shown to be associated with increased levels of reactive oxygen species in Purkinje cells, correlating with a progressive deterioration of the redox balance in mutant mice (20–23) and compromised antioxidant systems (24). Furthermore, an age-dependent reduction in the number of dopaminergic neurons present in the substantia nigra and striatum has been observed in ATM−/− mice, which was accompanied by severe gliosis (22) suggesting that ATM-deficient mice may model some of the neurological defects observed in A-T.
Studies using primary cells have reported that ATM deficiency is associated with premature aging attributed to the accumulation of oxidative damage in the brain (25). It is clear that diminished DNA repair combined with the chronic exposure to DNA damage is responsible for the neurodegeneration and premature aging associated with A-T. These observations are suggestive of a crucial role played by ATM in maintaining brain homeostasis.
Hypomorphic mutations in MRN components NBS1 and MRE11 are associated with genetic disorders Nijmegan breakage syndrome (NBS) and the A-T-like disorder, respectively (26, 27). Although these disorders are distinct from A-T, they share many clinical phenotypes and cellular features reflecting the roles played by these proteins in regulating ATM function (28). Although neurogenesis is largely normal in the absence of ATM, mice with a neural-specific deletion of NBS1 develop microcephaly and apoptosis of postmitotic cerebellar neurons leading to ataxia (16). Thus the functions of ATM and NBS1 in the murine brain are not fully congruent.
Although NBS1/MRN is required for ATM activation by double-strand breaks, we have previously shown that an alternative cofactor, ATM interactor (ATMIN, also known as ASCIZ), is required for ATM signaling in response to agents that induce chromatin changes such as chloroquine or osmotic stress (29). ATM levels are reduced in the absence of ATMIN and downstream substrate phosphorylation is compromised in response to these stimuli. Thus NBS1 and ATMIN mediate ATM activation in response to distinct stimuli (30). Although ATM activation in cultured cells by chloroquine or osmotic stress is rather artificial, we now identify oxidative stress as a stimulus that requires ATMIN for full ATM activation. ATMIN-deficient cells proliferate normally under physiological conditions but enter senescence prematurely in response to oxidative damage, similar to what was previously reported for ATM-deficient cells. Similarly, mice with CNS-specific deletion of ATMIN are growth retarded, and exhibit cortical neurodegeneration as well as a significant loss of dopaminergic neurons in the substantia nigra. Taken together, these results suggest that ATMIN contributes to some of the neuroprotective functions of ATM.
EXPERIMENTAL PROCEDURES
Mice
Mice with a targeted mutation of the atmin gene were generated by insertion of lox P sequences upstream and downstream of exon 4 of atmin (ATMINF) in ES cells, injection of ATMINWT/F ES cells into blastocysts, and mating of the resulting chimera. After germline transmission of the targeted locus, the ATMINF/+ was removed using germline deleting 3-phosphoglycerate kinase-cre transgenic mice. Heterozygous atminΔ/+ mice are viable and fertile. For cre-mediated deletion of atmin in the CNS, ATMINF/F mice were crossed with mice expressing cre under the control of the rat nestin promoter, designated ATMINΔN. Animals were maintained and bred in specific pathogen-free facilities. Nestin-cre efficiency and genotyping of mice was determined using a PCR based assay using primers specific for the floxed exon 4, deleted exon 4, and WT atmin alleles using the primers: Lox6133F, 5′-TCAGCATCTTCTCCAGAGAGACAG-3′, Lox6617R, 5′CACATGTGTACAGCACATTCATTG-3′, Lox10252R, 5′-CTCAGGGTACACATACTATGCTTGC-3′.
Cell Culture and Treatment
MEFs were derived from E12.5 embryos resulting from heterozygous atminΔ/+ intercrosses as described previously (50). ATMIN+/+ and ATMINΔ/Δ fibroblasts were cultured in DMEM supplemented with 10% FCS. Cells were cultured at 37 °C with 5% CO2 and either 3 or 20% oxygen. In some cases cells were treated with 100 μm NAC during the course of the experiment or with 10 μm paraquat for 10 h. IR experiments were performed using a Cs137 Gamma Irradiator at 2.1 gray/min. For cell cycle profiling, cells were stained with Hoechst and analyzed using a BD Biosciences FACScan. Cell viability was determined by trypan blue exclusion.
Western Blotting
Cells and tissues were extracted in RIPA lysis buffer (New England Biolabs) supplemented with protease inhibitors (Sigma). Western blots were performed using standard procedures. Protein samples were separated by SDS-PAGE, and subsequently transferred onto PVDF membranes. The following antibodies were used: pS1981-ATM (10H11.E12 Cell signaling), ATM (2C1, Santa Cruz), p53 (DO-1, Santa Cruz), pS15-p53 (Cell signaling), P21 (C19, Santa Cruz), pS824-Kap1 (Bethyl Laboratories), Kap1 (Bethyl Laboratories), β-actin (A5060, Sigma), pS957-SMC1 (5D11G5, Millipore), SMC1 (AB3908, Millipore), pT68-Chk2 (Cell Signaling), Chk2 (clone 7, Millipore), and HRP-conjugated goat anti-mouse/rabbit IgG (Sigma). All primary antibodies were used at 1:1000 dilution and secondary antibodies at 1:2000.
Immunohistochemistry and in Situ Hybridization
Tissue was fixed overnight in 10% neutral buffered formalin, briefly washed with PBS, and transferred into 70% ethanol, processed, and embedded into paraffin. Sections were cut at 4 μm for hematoxylin and eosin staining and immunohistochemistry. Antibodies used were tyrosine hydroxylase (ab152, Millipore, 1:300), glial fibrillary acidic protein (Z0334, Dako, 1:750), ionized calcium-binding adaptor molecule (IBA1) (019-19741, WAKO, Germany, 1:25), NeuN (MAB377, Millipore, 1:400), αM (MCA74, Serotec, 1:5000), αX (clone N418, Pierce, 1:200), P53 (NCL-p53-CM5p, Novacastra, 1:300), pS957-pSMC1 (5D11G5, Millipore, 1:100), pS139-γH2aX (ab2893, Abcam, 1:100), pS1981-ATM (ab2888, Abcam, 1:250), and ATMIN (JCRO1, made in house at Cancer Research UK, 1:300). Samples analyzed for ATMIN RNA by in situ hybridization were processed as described above by the Experimental Pathology Laboratory service lab at the London Research Institute.
Immunofluorescence
Primary MEFs were adhered onto slides and treated as indicated and fixed with 4% PFA. Antibodies used were pS139-γH2aX (JBW301, Millipore, 1:400), pS1981-ATM (10H11.E12, Cell signaling, 1:200), pS957-pSMC1 (5D11G5, Millipore, 1:100), and FITC/Cy3-conjugated goat anti-mouse/rabbit IgG (H&L, Jackson, 1:400). For detection of senescence-associated β-galactosidase, cells were fixed and stained according to the manufacturer's instructions using the histochemical staining kit (CS0030, Sigma).
Open Field
Open field activity was assessed at 6–8 weeks of age using the HVS Image tracking software (HVS Image 2100, Hampton, UK). Mice were individually placed in a square wooden arena (45 × 45 × 30 cm) that had a base covered in sawdust. Each mouse was released into a corner of the box and was allowed to explore for 5 min. The tracking system recorded path length, % time moving, and % time spent in the center.
Rotarod
The rotarod apparatus (Accelerating Model, Ugo Basile, Italy) was used to measure fore- and hindlimb motor coordination and balance. Mice were trained at 6–8 weeks of age and received three trials per day with an inter-trial interval of 1 h for 3 consecutive days. The rod accelerated from 5 to 60 rpm over a period of 600 s and the latency to fall was recorded.
RESULTS
ATMIN-deficient Cells Exhibit Increased Susceptibility to Oxidative DNA Damage
To understand the physiological function of ATMIN, we generated an ATMIN conditional allele that allows for tissue-specific ATMIN inactivation. Exon 4 of ATMIN, which encodes a large proportion of the ATMIN protein (amino acids 217–818), was flanked with loxP sites (supplemental Fig. S1A). Floxed ATMIN mice (ATMINF) were crossed with mice expressing cre recombinase under the control of the 3-phosphoglycerate kinase promoter (31), which resulted in germline deletion of ATMINF, yielding the ATMINΔ allele. Complete absence of ATMIN in ATMINΔ/Δ homozygous mice resulted in embryonic lethality at around E16.5, at which stage ATMIN-mutant embryos exhibited drastic craniofacial defects and midbrain exencephaly (Fig. 1A and supplemental Fig. S1, B–D). As ATMIN is a regulator of ATM function, we investigated the extent of DNA damage in ATMIN-mutant embryos. ATMINΔ/Δ mice exhibited an increased accumulation of Ser139-phosphorylated H2aX (pS139-γH2aX) in the midbrain, suggesting the presence of persistent DNA damage during development in the absence of ATMIN.
FIGURE 1.
Accumulation of DNA damage in ATMIN-deficient embryos and cells. A, H&E staining of sagittal sections through E12.5 ATMIN+/+ and ATMINΔ/Δ embryonic brains and high magnification of the indicated region. Lower panel shows pS139-γH2aX staining of a representative region and the histogram shows quantification of cells with pS139-γH2aX foci staining, n = 3. B, ATMIN+/+ and ATMINΔ/Δ fibroblasts were cultured at 3 or 20% oxygen for either 2 (P2) or 4 (P4) passages and a growth curve was performed to monitor cell proliferation. C, cells were cultured as above for 4 passages followed by fixation with 4% PFA and staining for pS139-γH2aX. The percentage of positive cells was quantitated.
To study the role of ATMIN in physiological DNA damage more closely, we isolated primary MEFs from ATMINΔ/Δ and control E12.5 embryos, and cultured them for up to 4 passages at both physiological (3%) and atmospheric (20%) oxygen levels. ATMIN-deficient fibroblasts showed normal cell cycle profiles (supplemental Fig. S2) and proliferated normally at early passage (P2) (Fig. 1B). However, following continued culture to P4, mutant MEFs displayed reduced proliferation both at 3 and 20% oxygen (Fig. 1B). Thus the absence of ATMIN appears to reduce the proliferation of MEFs after prolonged culture. Quantification of the extent of DNA damage by immunofluorescence using pS139-γH2aX and another marker of double breaks, pS957-SMC1, revealed minimal DNA damage at early passage in ATMINΔ/Δ and control MEFs, but passage 4 (P4) ATMIN-deficient MEFs showed higher levels of DNA damage when cultured in a high oxygen environment (Fig. 1C and supplemental Fig. S3).
Oxidative stress is believed to contribute to cellular aging (32) and the accumulation of DNA damage often precedes the entry of cells into a state of senescence. Consequently, we observed that ATMIN deficiency resulted in increased levels of senescent cells as marked by the higher levels of senescence-associated β-galactosidase staining in mutant MEFs (Fig. 2A). p21 levels are known to be up-regulated during senescence (33), and we observed elevated p21 levels when ATMIN-deficient MEFs were cultured at high oxygen concentrations (Fig. 2B). Because the premature entry into senescence of ATMIN-deficient MEFs was triggered by increased oxygen concentrations, we assessed whether the premature senescence was due to oxidative DNA damage. The thiol-containing antioxidant N-acetyl-l-cysteine (NAC) is readily taken up by cells and has been shown to detoxify reactive oxygen species (34). Dietary supplementation with NAC has also been shown to suppress development of lymphomas in ATM-deficient mice (35). Culturing atminΔ/Δ MEFs in the presence of 100 μm NAC rescued the increased senescence of mutant MEFs indicating a function for ATMIN in the prevention of senescence by inhibiting the accumulation of oxidative damage (Fig. 2C).
FIGURE 2.
ATMINΔ/Δ fibroblasts are highly susceptible to oxidative damage and exhibit aberrant DNA damage signaling. A, ATMIN+/+ and ATMINΔ/Δ fibroblasts were cultured at 3 or 20% oxygen for 1 passage (P1) or up to 4 passages (P4). Cells were fixed and stained for senescence-associated β-galactosidase. The histogram shows quantification of staining. B, ATMIN+/+ and ATMINΔ/Δ fibroblasts were treated as in A for 48 h following which cells were harvested and proteins were blotted for p21. C, ATMIN+/+ and ATMINΔΔ fibroblasts were treated as in A for 4 passages either in the absence or presence of 100 μm NAC. Cells were fixed and stained with senescence-associated β-galactosidase and the percentage of positive cells was counted. The histogram shows quantification from two experiments. D, ATMIN+/+ and ATMINΔΔ fibroblasts were cultured at 20% oxygen for 4 passages followed by fixation with 4% PFA and staining with pS139-γH2aX and pS1987-ATM. Colocalizing and noncolocalizing foci are indicated by yellow and white arrowheads, respectively. The histogram shows quantification of the percentage of pS139-γH2aX-positive cells that do not have colocalizing pS1987-ATM foci. E, ATMIN-deficient fibroblasts display reduced DNA damage signaling in response to treatment with paraquat. Cells were treated with 10 μm paraquat for 10 h and cell extracts were blotted for pS1987-ATM, ATM, pS824-Kap1, Kap1, pS15-P53, P53, pT68-Chk2, Chk2, and β-actin.
Next, we investigated DNA damage signaling in ATMIN-deficient MEFs. In control MEFs cultured at high oxygen concentrations up to passage 4, active ATM phosphorylated at serine 1987 (pS1987-ATM) was efficiently recruited to sites of DNA damage marked by pS139-γH2aX. In contrast, in ATMIN-deficient cells, pS1987-ATM failed to be recruited to 35% of pS139-γH2aX foci (Fig. 2D). It is noteworthy that whereas the numbers of pS139-γH2aX foci was greatly increased, the total number of P-ATM foci was not altered in atminΔ/Δ MEFs (Fig. 2D), suggesting that ATM recruitment to some sites of DNA damage was compromised.
As ATMIN appeared to play a role in ATM function in response to chronic oxidative damage, we next looked at a possible role for ATMIN in signaling in response to acute oxidative damage. In cells, the herbicide paraquat gets converted into free radicals, which react with molecular oxygen producing superoxide anions and hydrogen peroxide (36). Paraquat treatment induced ATM activation as judged by phosphorylation at serine 1987, but several other aspects of ATM signaling induced by paraquat were unexpected. Strikingly total ATM protein levels were reduced by paraquat treatment in both control and ATMIN-mutant cells. Quantification revealed a slight decrease in the extent of phosphorylation of some ATM substrates such as pS824-Kap1 and pS18-p53 (Fig. 2E and Table 1). These results suggest that ATMIN appears to be largely dispensable for ATM signaling during acute oxidative stress induced by paraquat, but conversely ATMIN is required for ATM signaling triggered by chronic oxidative stress.
TABLE 1.
Quantification of extent of ATM signaling in response to paraquat treatment
| Phosphorylated/ unphosphorylated ratio | ATMINΔ/Δ |
ATMIN+/+ |
||
|---|---|---|---|---|
| Untreated | Paraquat | Untreated | Paraquat | |
| ATM | 0.49 | 1.27 | 0.04 | 3.95 |
| Kap1 | 0.09 | 1.16 | 0.04 | 1.77 |
| p53 | 0.02 | 0.68 | 0.04 | 1.77 |
| Chk2 | 0.00 | 0.26 | 0.00 | 0.35 |
Morbidity of Aging ATMINΔN Mice
The nervous system is highly susceptible to oxidative damage as the brain consumes high amounts of oxygen (37). The accumulation of free radicals as a by-product of oxidative stress has been implicated in the neurodegenerative changes associated with A-T as well as during aging.
Hence to further investigate ATM regulation by ATMIN, floxed atmin (atminF) mice were crossed with Nestin-cre mice, which have previously been shown to result in gene inactivation in the central nervous system (38). These mice were designated ATMINΔN mice. ATMIN was highly expressed throughout the mouse brain, but atminF deletion was efficient as judged by in situ hybridization and immunohistochemistry (Fig. 3A and supplemental Fig. 4). ATMINΔN mice were viable and were born with Mendelian frequency but showed a reduction in weight that was apparent from the first weeks after birth onwards (Fig. 3, B and C). Both male and female adult ATMINΔN mice were fertile and appeared healthy.
FIGURE 3.
Characterization of ATMINΔN mice. A, in situ hybridization for ATMIN demonstrates its normal localization throughout the cortex, hippocampus, and dentate gyrus. The signal is significantly diminished in the brains of ATMINΔN mice. B, representative pictures of ATMINF/F and ATMINΔN mice at 3 months of age demonstrating severe growth retardation. C, ATMINΔN mice exhibit reduced body weight from birth. A cohort of ATMINF/F and ATMINΔN mice were weighed weekly over a period of 7 weeks. n = at least 10 mice per genotype. D, Kaplan-Meier curve for survival of ATMINΔN mice over a period of 18 months. E, sections through the substantia nigra pars compacta of ATMINF/F and ATMINΔN mice were stained with anti-tyrosine hydroxylase antibody to measure the number of dopaminergic neurons in this region. The density of tyrosine hydroxylase-positive cells was counted and corrected using Abercrombie correction.
To investigate more closely a potential function of ATMIN during neurogenesis, motor activity and anxiety levels of the ATMIN mice were assessed using the open field task. ATMIN-deficient mice were hyperactive as they moved a significantly greater distance than their littermate controls (t13 = 2.2, p < 0.05) but spent only slightly more time moving around the arena (t13 = 1.5, not significant; Table 2). Another aspect of this task is to assess anxiety levels. Mice that are more anxious will spend significantly less time in the bright center of the box compared with the darker, safer periphery of the box. We found that ATMINΔN mice had normal anxiety levels as there was no significant difference between genotypes in the time they spent in the center of the arena (t13 = 1.6, not significant; Table 2).
TABLE 2.
Movement and anxiety behavior of ATMIN mice
| ATMINF/F (n = 8) | ATMINΔ/N (n = 7) | |
|---|---|---|
| Open field behavior | ||
| Path length (m) | 11.6 ± 2.1 | 16.7 ± 0.7 |
| % Time moving | 70.7 ± 5.8 | 79.9 ± 0.7 |
| % Time in center | 24.2 ± 5.5 | 13.5 ± 3.6 |
| Rotating rod performance | ||
| Day 1 | 120 ± 13 | 163 ± 36 |
| Day 2 | 201 ± 23 | 225 ± 56 |
| Day 3 | 230 ± 27 | 285 ± 57 |
We next tested motor coordination using the accelerating rotarod. Mice were trained with 3 trials/day for 3 days and the latency to fall from the rod was recorded. Both groups of mice improved over the course of the training period and although there was a trend for the ATMIN mice to have enhanced performance in this task it did not reach significance (effect of day, F2,26 = 29.0, p < 0.001; effect of genotype, F1,13 = 3.3, p = 0.09; genotype × day interaction, F2,26 = 0.8, not significant; Table 2). These results show that the ATMIN-deficient mice are hyperactive but have normal motor coordination and anxiety levels, arguing against a profound function of ATMIN during brain development.
However after 1 year ATMINΔN mice showed increased morbidity and died significantly earlier than their littermates (Fig. 3D). Thus absence of ATMIN in the brain reduces the lifespan of aging mice.
ATMIN Deficiency in the CNS Causes Neuronal Loss
To determine the cause of premature death of ATMINΔN mice, we examined the brains of ATMINΔN mice. The overall morphology and gross histology of ATMINΔN brains was normal, arguing against an important role for ATMIN in neurogenesis (supplemental Fig. S5). Dopaminergic neurons in the substantia nigra have a high metabolic rate and are hence particularly vulnerable to the accumulation of oxidative DNA damage. ATM deficiency in mice leads to a degeneration of this population of cells. We observed a partial, but significant reduction in the number of tyrosine-hydroxylase positive dopaminergic neurons present in the substantia nigra pars compacta in ATMINΔN mice (Fig. 3E). This indicates that the integrity of these neurons is partially dependent on the presence of ATMIN.
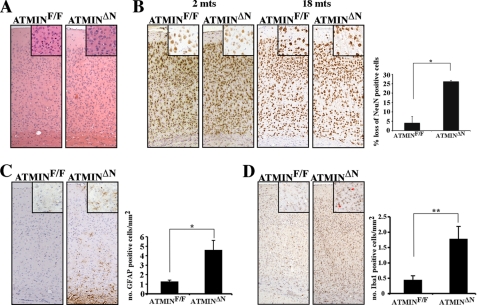
Whereas young ATMINΔN mice appeared healthy, aging mutant mice died earlier, indicating that ATMIN may counteract the aging process. To determine the cause of premature death of ATMINΔN mice, we examined the brains of ATMINΔN mice at young (2 months) and old (18 months) age. Because ATMIN was highly expressed in the cortex, we decided to look specifically at the consequence of ATMIN deletion in this region. The overall cellularity throughout the cortex of young ATMIN-deficient brains was normal (Fig. 4A). However, the number of NeuN-positive neurons was significantly decreased in the cortex of aged surviving ATMINΔN mice (Fig. 4B). Around 26% of the neurons were lost in the aged mutant cortex, compared with only 4% loss in control mice (Fig. 4B). In the ATMIN-deficient cortex considerable glial fibrillary acidic protein staining was detectable indicative of reactive gliosis in this region (Fig. 4C). Microglial activation often accompanies the induction of oxidative stress in mouse models of neurodegenerative disease (39, 40). Consequently, increased Iba1 staining, which marks activated microglial cells, was present throughout the cortex of old ATMINΔN mice (Fig. 4D). The hallmarks of cellular stress were not confined to the cortex. Cells expressing αM and αX integrin subunits, which mark activated and phagocytic microglia, respectively, were both significantly up-regulated throughout the midbrain of ATMINΔN mice (supplemental Fig. S6, A–C). Additionally, reactive astrocytic gliosis was also identified throughout the midbrain of ATMIN-deficient mice (supplemental Fig. S6D). These results suggest that ATMIN deficiency causes neuronal demise and subsequent premature death.
FIGURE 4.
Increased neurodegeneration in ATMINΔN brains. A, H&E stain of coronal sections through the midbrain of 18-month-old ATMINF/F and ATMINΔN mice showing similar morphology. B, sections from 2- and 18-month-old ATMINF/F and ATMINΔN mice were stained with NeuN to quantify neuronal density. The percentage loss in neuronal density through the cortex of ATMINF/F and ATMINΔN mice was calculated. C, sections from 18-month-old mice were stained with anti-glial fibrillary acidic protein antibody to measure astrocytic gliosis and with anti-Iba1 antibody for activated microglia (D). All numbers were corrected using the Abercrombie method. The histograms represent the quantification of staining from three mice.
Increased DNA Damage Signaling throughout the Cortex of ATMINΔN Mice
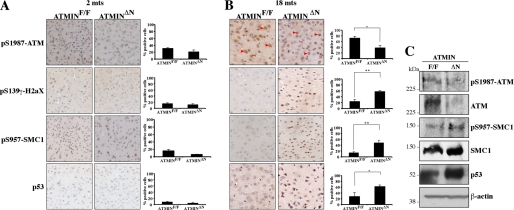
DNA damage accumulates in the aging brain and has also been suggested to contribute to several neurodegenerative disorders (41). Furthermore, ATM activation has been shown to increase with age (42). Having observed that ATMIN deficiency was associated with defects in the response to oxidative DNA damage in fibroblasts, we investigated ATM signaling in young and aged mice. In the cortices of young adult mice, there were only few cells exhibiting signs of DNA damage as judged by H2aX phosphorylation (pS139-γH2aX). Accordingly, there were moderate levels of active phosphorylated ATM (pS1987-ATM), and the numbers of cells positive for the ATM substrate phosphorylated SMC1 (pS957-SMC1) and expressing p53 protein were low. Importantly, there were no significant differences between control and ATMIN-mutant young mice (Fig. 5A). As previously described, ATM activation increased with age in control animals, however, this occurred to a lesser extent in ATMINΔN mice, indicating that ATMIN contributes to ATM activation during organismal aging. This deficit in ATM activation was accompanied by substantial increases in neurons positive for pS139-γH2aX (Fig. 5B and supplemental Fig. S7). Probably as a result of persistent DNA damage, pS957-SMC1 levels were increased and p53 protein was stabilized (Fig. 5B). The decrease in the levels of pS1987-ATM and the increase in SMC1 phosphorylation and p53 protein were confirmed by Western blotting using extracts isolated from control and mutant cortices (Fig. 5C). Because ATM deficiency in mice results in accumulation of oxidative DNA damage (43), these findings are consistent with the notion that ATMIN contributes to ATM activation and function in the aging brain.
FIGURE 5.
Aging related DNA damage signaling in ATMIN-deficient mice. Mice were sacrificed at 2 (A) or 18 months (B) of age and coronal sections through the cortex were cut. Representative sections were stained with pS1987-ATM, pS957-SMC1, pS139-γH2aX, and p53 and the percentage of positive cells in an equivalent area was counted. Cells positive for pS1987-ATM are highlighted with red arrowheads. The histograms represents the mean percentage of positive cells from three to five mice. C, the brains of the aged mice in B were lysed and proteins were blotted for pS1987-ATM, ATM, pS957-SMC1, SMC1, pS15-p53, p53 and β-actin.
DISCUSSION
This study demonstrates a role for ATMIN in regulating ATM function in counteracting oxidative DNA damage. ATMIN is required for ATM signaling in response to oxidative damage in both cultured cells and in the nervous system. ATMINΔN mice partially phenocopy many of the defects associated with ATM deletion. Similarly to ATM mutant mice, mice lacking the cofactor ATMIN in neurons develop a significant reduction in the number of dopaminergic neurons in the substantia nigra. Furthermore, ATMINΔN mice exhibit progressive cortical degeneration. Taken together, these results suggest that in the nervous system, ATMIN contributes to DNA damage signaling through regulation of ATM.
ATMIN Is Required for Signaling the Presence of Oxidative Damage
Mice deficient in components of the DNA damage response are particularly vulnerable to genomic instability. Seckel mice, which carry a mutation in the ATR gene, exhibit activation of the DNA damage response in the developing embryo (44). This has been attributed to the accumulation of extensive replicative stress during embryogenesis when proliferation is widespread. We have observed that germline deletion of ATMIN is lethal at around E16.5 with mice exhibiting a number of craniofacial defects and severe midbrain exencephaly (Fig. 1A and supplemental Fig. S1). It is noteworthy that deletion of the other ATM cofactor NBS1 also results in early embryonic lethality (45). Thus both ATMIN and NBS1 must have ATM-independent functions.
We decided to look at the cause of death by examining ATMINΔ/Δ embryos earlier on during gestation. At E12.5 ATMINΔ/Δ mice showed high levels of pS139-γH2aX staining in the midbrain indicating the presence of persistent DNA damage (Fig. 1A). This suggested that ATMIN is required to maintain genomic stability during embryonic development. To study the precise requirement for ATMIN more closely, we isolated MEFs from E12.5 embryos and cultured them at both physiological and high oxygen conditions (Figs. 1C and 2A). ATMIN-deficient fibroblasts exhibited severe vulnerability to oxidative stress, accumulating higher levels of DNA damage before entering into a state of premature growth arrest. ATM-deficient fibroblasts also possess a limited proliferation potential in culture, entering senescence at an earlier passage than normal cells (18, 25). The protein levels of the CDK inhibitor p21 are elevated in ATM-deficient cells (46), and we found the same to be true for ATMIN deficiency (Fig. 2B). The rescue of premature senescence by culturing ATMIN-deficient fibroblasts in the presence of 100 μm NAC (Fig. 2C) suggests a protective role for the cofactor in regulating ATM activation in response to oxidative damage.
Regulation of ATM Signaling following Chronic Oxidative Damage
We have previously demonstrated that ATMIN is required for ATM substrate phosphorylation in response to signals that induce changes in chromatin structure such as chloroquine (29). We now demonstrate that ATMIN contributes to ATM signaling in response to chronic oxidative damage (Fig. 2D). Having observed that the extent of pathway activation is compromised in the absence of ATMIN, we looked at the long-term consequence of this reduced response in the brain. Postmitotic neurons in the CNS accumulate excessive damage throughout their extended life. ATM levels were reduced in the cortex of 18-month-old ATMIN-deficient mice, however, substrate phosphorylation was increased (Fig. 5, A-C). This most likely reflects the activation of parallel pathways such as ATR signaling by the accumulated DNA damage. It is likely that the increased DNA damage throughout the cortex of atminΔN animals is causally involved in neurodegeneration and subsequent death.
ATMIN Deletion Phenocopies Other Genomic Instability Syndromes
A functional MRN complex is known to be required for coordinating efficient detection, signaling, and repair of double-strand breaks by mediating ATM recruitment to the site of damage via association with the C terminus of NBS1 (47). The primary neurological phenotype associated with A-T is the progressive degeneration of Purkinje neurons in the cerebellum leading to early-onset ataxia. This is thought to be a direct consequence of a diminished response to DNA damage. Mutations in components of the MRN complex are also often associated with genomic instability disorders. MRE11 deletion mimics several indices that are associated with ATM deficiency including progressive cerebellar degeneration (27). Moreover, defective DNA damage signaling has been shown to be responsible for damage to both the cerebellum and cerebral cortex in several other human neurodegenerative disorders (48). ATMIN deficiency also recapitulates some of the features that are associated with such syndromes (Fig. 3). ATMINΔN mice are severely growth retarded, as has been observed for mice carrying germline deletions in ATM and ATR as well as a CNS specific deletion of NBS1. This weight loss has been attributed to the accumulation of DNA damage and strengthens the suggestion that all these disorders are caused by compromised DNA damage signaling.
Mouse models of A-T manifest a predisposition to cancer causing subsequent early death, which prevents the analysis of ATM function in the aging brain. Notwithstanding, mouse models of ATM deficiency do possess mild degenerative changes in the cerebral cortex and other brain regions (22, 49). In line with this, upon closer examination we observed that ATMINΔN mice also exhibit progressive neurodegeneration (Fig. 4). These results further strengthen the suggestion that ATMIN plays a preventative role against the accumulation of DNA damage and thus neurodegeneration in the mouse brain.
An additional feature of human A-T that is recapitulated in both ATM- and ATMIN-deficient mice is the reduction in the number of tyrosine hydroxylase positive, dopaminergic nigro-striatal neurons (Fig. 3E) (21, 22). As with many other neurodegenerative disorders including Parkinson disease, oxidative stress has been implicated as the primary cause of cell loss in this metabolically active compartment. This is evidenced by both the reduced antioxidant capacity, as well as the increased levels of reactive oxygen species in the brains of affected individuals.
The pathological changes observed in the brains of ATMINΔN mice are most likely attributable to diminished ATM levels, causing both the neurodegenerative changes observed throughout the cortex and degeneration of dopaminergic neurons in the substantia nigra. Taken together, our results are in agreement with the notion that ATMIN contributes to physiological ATM function in the CNS.
Supplementary Material
Acknowledgments
We are grateful to the Animal Unit, Equipment Park, FACS Lab, and the Experimental Pathology Lab at the London Research Institute for technical help and advice on histology. We thank S. Marino and G. P. Schiavo for critical reading of the manuscript.
This work was supported by a grant from The London Research Institute from Cancer Research UK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- ROS
- reactive oxygen species
- NBS
- Nijmegan breakage syndrome
- ATM
- ataxia telangiectasia mutated
- ATMIN
- ATM interactor
- NAC
- N-acetyl-l-cysteine
- MEF
- mouse embryonic fibroblast
- ATR
- ATM and Rad3-related
- MRN
- MRE11, Rad50, and NBS1
- pS
- phosphorylated serine.
REFERENCES
- 1.Chen J. H., Hales C. N., Ozanne S. E. (2007) Nucleic Acids Res. 35, 7417–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimes A., Chandra S. B. (2009) Cancer Res. Treat. 41, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J. (2003) Nat. Cell Biol. 5, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faragher R. G., Kill I. R., Hunter J. A., Pope F. M., Tannock C., Shall S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 12030–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sedelnikova O. A., Horikawa I., Zimonjic D. B., Popescu N. C., Bonner W. M., Barrett J. C. (2004) Nat. Cell Biol. 6, 168–170 [DOI] [PubMed] [Google Scholar]
- 6.Lumsden J. M., McCarty T., Petiniot L. K., Shen R., Barlow C., Wynn T. A., Morse H. C., 3rd, Gearhart P. J., Wynshaw-Boris A., Max E. E., Hodes R. J. (2004) J. Exp. Med. 200, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reina-San-Martin B., Chen H. T., Nussenzweig A., Nussenzweig M. C. (2004) J. Exp. Med. 200, 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakkenist C. J., Kastan M. B. (2004) Cell 118, 9–17 [DOI] [PubMed] [Google Scholar]
- 9.Durocher D., Jackson S. P. (2001) Curr. Opin. Cell Biol. 13, 225–231 [DOI] [PubMed] [Google Scholar]
- 10.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 11.Reina-San-Martin B., Nussenzweig M. C., Nussenzweig A., Difilippantonio S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. (2003) EMBO J. 22, 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J. H., Ghirlando R., Bhaskara V., Hoffmeyer M. R., Gu J., Paull T. T. (2003) J. Biol. Chem. 278, 45171–45181 [DOI] [PubMed] [Google Scholar]
- 14.Lukas J., Lukas C., Bartek J. (2004) DNA Repair 3, 997–1007 [DOI] [PubMed] [Google Scholar]
- 15.Chun H. H., Gatti R. A. (2004) DNA Repair 3, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 16.Frappart P. O., McKinnon P. J. (2006) Neuromol. Med. 8, 495–511 [DOI] [PubMed] [Google Scholar]
- 17.Kühne M., Riballo E., Rief N., Rothkamm K., Jeggo P. A., Löbrich M. (2004) Cancer Res. 64, 500–508 [DOI] [PubMed] [Google Scholar]
- 18.Shiloh Y., Tabor E., Becker Y. (1982) Exp. Cell Res. 140, 191–199 [DOI] [PubMed] [Google Scholar]
- 19.Boder E. (1985) Kroc Found. Ser. 19, 1–63 [PubMed] [Google Scholar]
- 20.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J. N., Ried T., Tagle D., Wynshaw-Boris A. (1996) Cell 86, 159–171 [DOI] [PubMed] [Google Scholar]
- 21.Eilam R., Peter Y., Elson A., Rotman G., Shiloh Y., Groner Y., Segal M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12653–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilam R., Peter Y., Groner Y., Segal M. (2003) Neuroscience 121, 83–98 [DOI] [PubMed] [Google Scholar]
- 23.Elson A., Wang Y., Daugherty C. J., Morton C. C., Zhou F., Campos-Torres J., Leder P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13084–13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamsler A., Daily D., Hochman A., Stern N., Shiloh Y., Rotman G., Barzilai A. (2001) Cancer Res. 61, 1849–1854 [PubMed] [Google Scholar]
- 25.Metcalfe J. A., Parkhill J., Campbell L., Stacey M., Biggs P., Byrd P. J., Taylor A. M. (1996) Nat. Genet. 13, 350–353 [DOI] [PubMed] [Google Scholar]
- 26.Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 27.Stewart G. S., Maser R. S., Stankovic T., Bressan D. A., Kaplan M. I., Jaspers N. G., Raams A., Byrd P. J., Petrini J. H., Taylor A. M. (1999) Cell 99, 577–587 [DOI] [PubMed] [Google Scholar]
- 28.Shull E. R., Lee Y., Nakane H., Stracker T. H., Zhao J., Russell H. R., Petrini J. H., McKinnon P. J. (2009) Genes Dev. 23, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanu N., Behrens A. (2007) EMBO J. 26, 2933–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanu N., Behrens A. (2008) Cell Cycle 7, 3483–3486 [DOI] [PubMed] [Google Scholar]
- 31.Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. (1998) Transgenic Res. 7, 105–112 [DOI] [PubMed] [Google Scholar]
- 32.Muller F. L., Lustgarten M. S., Jang Y., Richardson A., Van Remmen H. (2007) Free Radic. Biol. Med. 43, 477–503 [DOI] [PubMed] [Google Scholar]
- 33.Dulić V., Beney G. E., Frebourg G., Drullinger L. F., Stein G. H. (2000) Mol. Cell. Biol. 20, 6741–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Flora S., Izzotti A., D'Agostini F., Balansky R. M. (2001) Carcinogenesis 22, 999–1013 [DOI] [PubMed] [Google Scholar]
- 35.Reliene R., Schiestl R. H. (2007) J. Nutr. 137, 229S–232S [DOI] [PubMed] [Google Scholar]
- 36.Farrington J. A., Ebert M., Land E. J., Fletcher K. (1973) Biochim. Biophys. Acta 314, 372–381 [DOI] [PubMed] [Google Scholar]
- 37.Dringen R., Gutterer J. M., Hirrlinger J. (2000) Eur. J. Biochem. 267, 4912–4916 [DOI] [PubMed] [Google Scholar]
- 38.Kramer E. R., Knott L., Su F., Dessaud E., Krull C. E., Helmbacher F., Klein R. (2006) Neuron 50, 35–47 [DOI] [PubMed] [Google Scholar]
- 39.Mattson M. P. (2000) Nat. Rev. Mol. Cell Biol. 1, 120–129 [DOI] [PubMed] [Google Scholar]
- 40.Peng J., Stevenson F. F., Oo M. L., Andersen J. K. (2009) Free Radic. Biol. Med. 46, 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovell M. A., Markesbery W. R. (2007) Nucleic Acids Res. 35, 7497–7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panda S., Isbatan A., Adami G. R. (2008) Mech. Ageing Dev. 129, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barlow C., Dennery P. A., Shigenaga M. K., Smith M. A., Morrow J. D., Roberts L. J., 2nd, Wynshaw-Boris A., Levine R. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9915–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murga M., Bunting S., Montaña M. F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P. J., Nussenzweig A., Fernandez-Capetillo O. (2009) Nat. Genet. 41, 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumon-Jones V., Frappart P. O., Tong W. M., Sajithlal G., Hulla W., Schmid G., Herceg Z., Digweed M., Wang Z. Q. (2003) Cancer Res. 63, 7263–7269 [PubMed] [Google Scholar]
- 46.Xu Y., Ashley T., Brainerd E. E., Bronson R. T., Meyn M. S., Baltimore D. (1996) Genes Dev. 10, 2411–2422 [DOI] [PubMed] [Google Scholar]
- 47.Falck J., Coates J., Jackson S. P. (2005) Nature 434, 605–611 [DOI] [PubMed] [Google Scholar]
- 48.Barzilai A., Biton S., Shiloh Y. (2008) DNA Repair 7, 1010–1027 [DOI] [PubMed] [Google Scholar]
- 49.Liu N., Stoica G., Yan M., Scofield V. L., Qiang W., Lynn W. S., Wong P. K. (2005) Lab. Invest. 85, 1471–1480 [DOI] [PubMed] [Google Scholar]
- 50.Behrens A., Sibilia M., Wagner E. F. (1999) Nat. Genet. 21, 326–329 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.