Abstract
Crystal structures of the xenobiotic metabolizing cytochrome P450 2B4 have demonstrated markedly different conformations in the presence of imidazole inhibitors or in the absence of ligand. However, knowledge of the plasticity of the enzyme in solution has remained scant. Thus, hydrogen-deuterium exchange mass spectrometry (DXMS) was utilized to probe the conformations of ligand-free P450 2B4 and the complex with 4-(4-chlorophenyl)imidazole (4-CPI) or 1-biphenyl-4-methyl-1H-imidazole (1-PBI). The results of DXMS indicate that the binding of 4-CPI slowed the hydrogen-deuterium exchange rate over the B′- and C-helices and portions of the F-G-helix cassette compared with P450 2B4 in the absence of ligands. In contrast, there was little difference between the ligand-free and 1-PBI-bound exchange sets. In addition, DXMS suggests that the ligand-free P450 2B4 is predominantly open in solution. Interestingly, a new high resolution structure of ligand-free P450 2B4 was obtained in a closed conformation very similar to the 4-CPI complex. Molecular dynamics simulations performed with the closed ligand-free structure as the starting point were used to probe the energetically accessible conformations of P450 2B4. The simulations were found to equilibrate to a conformation resembling the 1-PBI-bound P450 2B4 crystal structure. The results indicate that conformational changes observed in available crystal structures of the promiscuous xenobiotic metabolizing cytochrome P450 2B4 are consistent with its solution structural behavior.
Keywords: Cytochrome P450, Mass Spectrometry (MS), Protein Folding, Protein Structure, X-ray Crystallography
Introduction
Cytochrome P450 (P450)4 dependent monooxygenases are involved in the biogenesis of sterols and hormones and oxidation of a broad range of xenobiotic compounds (1). Many individual mammalian cytochromes P450 can accept a wide variety of hydrophobic substrates of differing shapes and sizes and render them more hydrophilic for excretion or subsequent conjugation. In addition to their central role in drug clearance, the ability of mammalian cytochromes P450 to convert various inactive precursors to the respective bioactive compounds makes these enzymes of paramount importance for the healthcare and pharmaceutical industries (2–4).
Despite their broad range of substrates, the single domain fold of P450s is well conserved across families (5–9). Hence, the ability to adapt to molecules reflects the notable plasticity of many secondary structural elements (8, 9). P450s show the ability to form compact structures around small ligands or empty active sites, as evidenced by P450 2B4 complexed with 1-(4-chlorophenyl)imidazole (1-CPI) or 4-CPI (10, 11), P450 3A4 (12), and several proteins in the 2C subfamily (13, 14). Moreover, some P450s also appear able to alter their conformations to accommodate ligands of greater volume, as seen in P450 2B4 with bifonazole (15) or in P450 3A4 with erythromycin or with ketoconazole (16).
Our laboratory has utilized an engineered form of P450 2B4 termed P450 2B4dH5 (N-terminally modified and a C-terminal His tag) to gain insight into enzyme flexibility (10, 11, 15, 17–19). The interest in the P450 2B subfamily was prompted by some marked species differences in function that could be attributed to relatively few amino acid substitutions (20). Examination of crystal structures of P450 2B4dH reveals plastic regions able to accommodate binding of ligands of a wide range of sizes (15). Furthermore, these observations were supported by isothermal titration calorimetry (20), which provided early indications that P450 2B4dH can adopt different conformations in solution upon ligand binding, an ability also seen in P450 3A4 (16). However, direct observations of solution behavior have been lacking.
Although there is considerable information about static crystal structures of P450 2B4dH complexed with ligands and about thermodynamic parameters of ligand binding, much less is known concerning the plasticity of the dynamic regions of this enzyme in solution. Amide hydrogen-deuterium (H-D) exchange mass spectrometry (DXMS), which traditionally utilizes matrix-assisted laser desorption ionization (MALDI) or electrospray ionization (ESI), is an ideal method to determine such solution behavior (21–25). The ESI-based approach generally offers greater coverage of protein sequence and lower rate of back-exchange of deuterons than the MALDI-based approach. ESI DXMS has been utilized to investigate protein folding and unfolding (26–29), protein-ligand interactions (30–34), and dynamics of proteins under various solution conditions (34–38).
In this study, ESI-based DXMS was used for the detection of ligand-induced conformational changes of P450 2B4dH and identification of specific regions of the enzyme affected by ligand binding. Evidence from the previously determined structures of P450 2B4dH and a novel ligand-free crystal structure solved at 1.8 Å resolution allows interpretation of conformational dynamics associated with ligand binding.
EXPERIMENTAL PROCEDURES
Materials
Imidazole compounds and porcine pepsin were from Sigma. CYMAL-5 was from Anatrace (Maumee, OH). Molecular weight cutoff (50 kDa) Amicon ultrafiltration devices were from Millipore (Billerica, MA). The Wizard III crystal screen was obtained from Emerald Biosystems (Bainbridge Island, WA). Nickel-nitrilotriacetic acid affinity resin was from Qiagen (Valencia, CA). Macroprep CM-Sepharose cation-exchange resin was from Bio-Rad. TOPP3 cells were from Stratagene (La Jolla, CA).
Protein Expression and Purification
Cytochrome P450 2B4dH was expressed in TOPP3 cells as described previously (39) and purified by the same protocol. The pellet was resuspended in 10% of the original culture volume in buffer containing 20 mm potassium phosphate (pH 7.4 at 4 °C), 20% (v/v) glycerol, 10 mm 2-mercaptoethanol (BME), and 0.5 mm phenylmethanesulfonyl fluoride (PMSF). The resuspended cells were further treated with lysozyme (0.3 mg/ml) and stirred for 30 min, followed by a brief centrifugation for 30 min at 7000 rpm in a JA-14 rotor in a Beckman Coulter Avanti J-26 XPI centrifuge. After decanting the supernatant, spheroplasts were resuspended in 5% of the original culture volume in buffer containing 500 mm potassium phosphate (pH 7.4 at 4 °C), 20% (v/v) glycerol, 10 mm BME, and 0.5 mm PMSF and were sonicated for three times for 45 s on ice. The membrane pellet was separated by centrifugation for 10 min at 7000 rpm, and CYMAL-5 was added to the supernatant at a final concentration of 4.8 mm. This was allowed to stir for 30 min at 4 °C prior to ultracentrifugation for 45 min at 41,000 rpm using a fixed-angle Ti 50.2 rotor in a Beckman Coulter Optima l-80 XP ultracentrifuge. The P450 concentration was measured using the reduced CO difference spectra from the resulting supernatant (40, 41).
The supernatant was applied to a nickel-nitrilotriacetic acid column. The column was washed with lysis buffer containing 100 mm potassium phosphate (pH 7.4 at 4 °C), 100 mm NaCl, 20% (v/v) glycerol, 10 mm BME, 0.5 mm PMSF, 4.8 mm CYMAL-5, and 1 mm histidine, and the protein was eluted using buffer containing 10 mm potassium phosphate (pH 7.4 at 4 °C), 100 mm NaCl, 20% (v/v) glycerol, 10 mm BME, 0.5 mm PMSF, 4.8 mm CYMAL-5, and 60 mm histidine. The P450-containing fractions were pooled and were loaded onto a Macroprep CM-Sepharose column. The cation-exchange column was washed using low salt buffer, and the protein was eluted with high salt buffer containing 50 mm potassium phosphate (pH 7.4 at 4 °C), 500 mm NaCl, 20% (v/v) glycerol, 1 mm EDTA, and 0.2 mm DTT. The P450 fractions were pooled, and the concentration was measured using the reduced CO-difference spectra.
DXMS Analysis, Optimization of the Fragmentation Conditions
Prior to deuteration studies, test digests prepared with undeuterated buffer in varying concentrations of the denaturant guanidine hydrochloride (GdnHCl) were made to optimize proteolysis conditions for maximal peptide coverage (42). In short, 5 μl of stock solution of P450 2B4dH at 226 μm was diluted with 15 μl of the high salt CM elution buffer described above and quenched with 30 μl of 0.8% (v/v) formic acid containing various concentrations of GdnHCl (0.8, 1.6, 3.2, and 6.4 m) at 0 °C. This quenching step reduces the amount of H-D exchange with a decrease in pH to 2.2–2.5 in addition to denaturing the protein prior to pepsin proteolysis with GdnHCl and acidic conditions. The quenching denaturation process was allowed to proceed on ice for 30 s, after which the sample was frozen on dry ice. The frozen sample was stored at −80 °C until it was transferred to the dry ice-containing sample basin of the cryogenic autosampler module of the DXMS apparatus. Samples were thawed on ice and immediately passed over a protease column (66-μl bed volume) filled with porcine pepsin at a flow rate of 100 μl/min with 0.05% trifluoroacetic acid. The duration of digestion was 40 s. The proteolytic products were directly collected by a C18 column (Vydac catalog no. 218MS5105) and then eluted with a linear gradient of 0.046% (v/v) trifluoroacetic acid, 6.4% (v/v) acetonitrile to 0.03% (v/v) trifluoroacetic acid, 38.4% (v/v) acetonitrile for 30 min. The column effluent was analyzed on an LCQ Classic (Thermo Finnigan, Inc.) electrospray ion trap-type mass spectrometer and an electrospray Q-TOF mass spectrometer (Micromass) and data acquisition in either data-dependent tandem mass spectrometric mode or MS1 profile mode. Determination of pepsin-generated peptides from the resulting MS/MS data sets was facilitated through the use of SEQUEST (Thermo Finnigan, Inc.). This set of peptides was then further examined by specialized software, DXMS Explorer (Sierra Analytics Inc., Modesto, CA), and all data processing was the same as described previously (42). The peptide coverage maps for the different concentrations of GdnHCl were compared, and the condition with the best coverage map was used for deuterium exchange experiments.
Deuterium Exchange Experiments
Ligand-bound P450 2B4dH samples were prepared by mixing 2.7 μl of 15 mm 4-CPI (final concentration of 270 μm) or 2.7 μl of 15 mm 1-PBI (final concentration of 270 μm) with 150 μl of 226 μm P450 2B4dH and incubated at 0 °C for 30 min. Ligand-free samples were prepared by mixing 150 μl of P450 2B4dH with 0.27 μl of methanol, making the final volume equivalent to the ligand-bound samples. All protein samples, with the exception of the equilibrium-deuterated control, and buffers were pre-chilled on ice and prepared in the cold room. Three types of hydrogen-deuterium exchange samples were prepared as follows: nondeuterated, deuterated, and fully deuterated. The nondeuterated sample was processed exactly as described under “DXMS Analysis, Optimization of the Fragmentation Conditions.” The fully deuterated sample represents the “maximum” H-D exchange for a certain time period, which in these experiments was a period of 12 h where the samples were allowed to exchange at room temperature in D2O buffer (1% (v/v) formic acid). The deuterated samples represent different incubation times prior to the quenching of the exchange process. All samples were initiated by mixing 5 μl of ligand-free sample or ligand-bound sample with 15 μl of D2O buffer. D2O buffer contained 50 mm potassium phosphate, pH 7.4, 500 mm NaCl, 20% glycerol (v/v), 1 mm EDTA, 0.2 mm DTT, pH 7.4. The samples were incubated at 0 °C for 10, 100, 1000, 10,000, and 100,000 s and 39 °C for 10,000 and 100,000 s. The deuterium exchange was quenched by adding 30 μl of ice-cold quench solution (1.6 m GdnHCl in 0.8% formic acid) that acidified the sample from pH 2.2 to 2.5 as described previously (43). The deuterated samples were then transferred to ice-cooled autosampler vials, frozen on dry ice, and stored at −80 °C. The pepsin digestion, chromatography, and the mass spectral acquisition proceeded as described under “DXMS Analysis, Optimization of the Fragmentation Conditions.” Data processing and reduction of H-D exchange experiments utilized DXMS data reduction software (Sierra Analytics, Modesto, CA). Back-exchange occurs right after the addition of quench, and corrections for it were determined via the methods of Zhang and Smith (44)
where m(P), m(N), and m(F) are the centroid value of the partially deuterated, nondeuterated, and fully deuterated peptide, respectively.
Crystallization and Data Collection
The pooled protein fractions from the ion-exchange column were concentrated to 550–600 μm and supplemented with 4.8 mm CYMAL-5 and 0.028% (w/v) 3α,7α,12α-tris[(β-d-maltopyranosyl)ethyloxy]cholane (45) before crystallization. Crystal screening was performed by sitting drop vapor diffusion method using the Emerald Biosystems Wizard III screen. Crystals of P450 2B4dH were obtained at 18 °C after incubating the protein in a 1:1 ratio with the precipitant containing 14.4% (w/v) PEG 8000, 0.08 m cacodylate, pH 6.5, 0.6 m calcium acetate, and 20% (v/v) glycerol. Crystals were transferred to mother liquor containing 20% (v/v) glycerol before being flash-frozen in liquid nitrogen. Crystallographic data were collected remotely at Stanford Synchrotron Radiation Lightsource beam line 7-1 (46) using 1° oscillations over 180 frames and 20-s exposures. Crystals diffracted to 1.8 Å, and the protein crystallized in the P31 space group. Data were integrated using iMOSFLM (47) and scaled via SCALA (48) in CCP4i.
Structure Determination and Refinement
The structure of P450 2B4dH was determined by molecular replacement using the previously determined P450 2B4dH-1-CPI structure (PDB entry 2Q6N) as a search model in Phaser (49). Matthews coefficient determination suggested the presence of two molecules in the asymmetric unit. The output model from Phaser was submitted to rigid body and restrained refinement in REFMAC (50), and the molecular model was built with COOT (51) using electron density maps Fo − Fc and 2Fo − Fc contoured at 3σ and 1σ, respectively. Water molecules were placed manually, and iterative model building and refinement was continued until the completion of the model.
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations were performed with starting coordinates of the closed ligand-free P450 2B4dH structure (PDB entry 3MVR) using the molecular dynamics software package GROningen MAchine for Chemical Simulation (GROMACS) version 4.07 (52). Residues 20–27, 473, 474, and 492–495, not found in the ligand-free crystal structure of P450 2B4dH, were added using the homology modeling program MODELLER and the complete amino acid sequence of the protein (53). The topology files used in the energy minimization and MD simulation were modified to reflect the cysteinyl ligation to heme (54, 55). The P450 2B4dH structure was immersed in a simulated water box with 120 Å sides and containing ∼80,000 waters, corresponding to twice the length of the longest diagonal of the protein (∼65 Å). The structure was energy-minimized by the method of steepest descent to remove van der Waals contact between overlapping waters and the amino acids of the protein. Simulations were run with Berendsen temperature and pressure coupling (also known as “bath”) (56) at a simulated temperature of 300 K using the GROMOS 53a6 force field (57) and periodic boundary conditions in all directions. Electrostatics of the system were measured using the particle-mesh Ewald method (58). The simulations were then conducted using a Linux cluster at the University of Washington (Seattle). During the first 250 ps of the MD simulation, the protein was position-restrained to allow the waters to fill in the cavities. After that, the MD simulation was continued without restraints for another 15 ns.
Figures
All protein model figures were generated using PyMOL (59). All chemical structures were created using MarvinSketch version 5.2.6, 2009, ChemAxon.
RESULTS
Deuterium Incorporation into Ligand-free P450 2B4dH
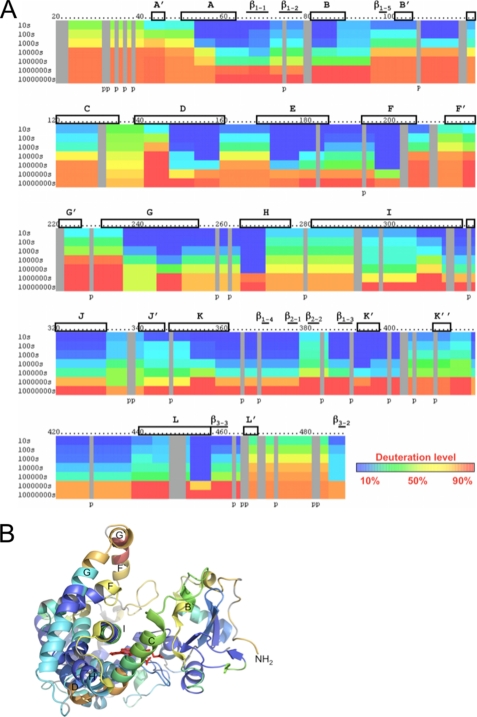
The DXMS profile of P450 2B4dH is shown in Fig. 1A and mapped onto the existing ligand-free open structure in Fig. 1B. The greatest solvent accessibility over time occurs in the previously defined plastic regions of the protein consisting of the B′-, C-, F-, F′-, G′-, and G-helices and their associated loops (15). The C-D loop shows fairly fast exchange rates as well. The remainder of the protein shows slower deuteration rates indicating less movement in solution.
FIGURE 1.
Hydrogen-deuterium exchange of ligand-free P450 2B4dH. A, deuteration level of ligand-free P450 2B4dH is shown with the lowest levels in blue to highest levels in red, as indicated by the color bar. Each bar under the primary sequence is divided into rows corresponding to each time point from 10 to 10,000,000 s (top to bottom). The deuterated samples of the last two time points, 1,000,000 and 10,000,000 s, were incubated at 39 °C for 10,000 and 100,000 s, respectively. As shown in previous studies (79–81), incubation at 39 °C for 10,000 or 100,000 s is equivalent to incubation at 0 °C for 1,000,000 or 10,000,000 s, respectively. Boxes and bars represent helices and β-sheets, respectively, as seen in the open ligand-free structure (PDB code 1PO5) (19). B, deuteration levels after 1000 s were mapped onto the P450 2B4dH open structure, and helices are labeled as reported previously (19).
Deuterium Exchange Shows Differential Solvent Exposure upon Binding of Imidazole Inhibitors of Different Size
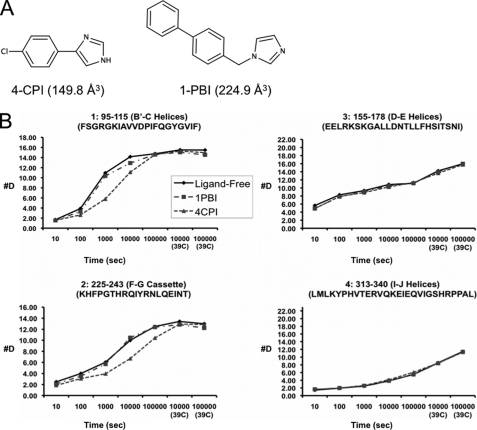
Crystal structures of P450 2B4dH complexed with 4-CPI (PDB code 1SUO) and with 1-PBI (PDB codes 3G5N and 3G93) show significant shifts in the plastic regions of the protein compared with the open structure of the enzyme (PDB code 1PO5). The structures are closed (4-CPI) or intermediately open (1-PBI) relative to the range of conformations adopted in crystal structures (11, 15, 18, 19). These compounds bind tightly to the protein with KS values for 4-CPI and 1-PBI of 0.04 and 0.23 μm, respectively (18, 60). Using this information and the equation for “tight binding” solved for [ES], P450 2B4dH showed saturation of greater than 99% for 4-CPI and greater than 95% for 1-PBI at experimental concentrations of protein and ligand (18 and 22 μm, respectively, after quenching) (61). In the presence of 4-CPI (Fig. 2A), deuteration of some of the plastic regions of P450 2B4dH showed significant slowing over the course of the experiment compared with ligand-free enzyme (Fig. 2B). Representative peptides covering the regions of the B′- and C-helices (residues 101–134) and the F-G cassette (residues 193–243) show much lower solvent accessibility in the presence of 4-CPI than in the absence of ligand. In the presence of 4-CPI, the B′- and C-helices showed the greatest difference in deuteration levels at about 1000 s when compared with the ligand-free H-D exchange profile (Fig. 2B, peptide 1). The F-G cassette shows similar behavior, with the greatest difference between the 4-CPI and ligand-free profiles coming at about 10,000 s (Fig. 2B, peptide 2). The remainder of the protein shows remarkably small differences between the solution structural conformation of P450 2B4dH without ligand and with 4-CPI, as exemplified by Fig. 2B, peptides 3 and 4. As a control for the possibility of changes in protein behavior due to the presence of methanol or CYMAL-5, DXMS experiments were completed at one time point, 10,000 s, for protein alone, in the presence of 1% methanol, and in the presence of 1% methanol and 1 mm CYMAL-5. This time point was selected due to the large differences in deuteration level between ligand-free P450 2B4dH and the protein in the presence of 4-CPI. Neither methanol alone nor methanol and CYMAL-5 changed the deuterium exchange profile of the protein (data not shown).
FIGURE 2.
DXMS behavior of P450 2B4dH upon ligand binding. A, stick model structures of 4-CPI and 1-PBI. Volumes for ligands calculated are listed in parentheses. B, time course of deuterium exchange in the B′-C loop region and the F-G cassette, respectively, of peptides 1 (residues 95–115) and 2 (residues 225–243), which show differences in DXMS exchange. Peptides in the D-E helices and I-J helices, respectively, are illustrated as 3 (residues 155–178) and 4 (residues 313–340). The scale of the axis is the maximum number of exchangeable amides.
In contrast to the results with 4-CPI, the solution dynamic behavior of P450 2B4dH in the presence of 1-PBI (Fig. 2A) is more similar to ligand-free enzyme. Accordingly, 1-PBI did not elicit much change in deuteration rate in the B′- and C′-helices or F-G cassette, (Fig. 2B, peptides 1 and 2). The static regions of the protein also show little change in exchange rate upon binding of 1-PBI to the protein (Fig. 2B, peptides 3 and 4). The differences in the behavior of P450 2B4dH when in the presence of 4-CPI or 1-PBI correspond to the major changes in conformation seen in the respective crystal structures of the enzyme complexes (11, 18). Because of the similarity of the DXMS results of protein alone and in the presence of 1-PBI, a DXMS experiment was carried out at 10,000 s with a 10:1 ligand to protein ratio, where the expected percent saturation of the protein is above 99%. Increasing the ligand concentration did not alter the exchange profile compared with a ligand to protein ratio of 1.2:1(data not shown). As in the absence of ligand, 1 mm CYMAL-5 did not alter the DXMS results at 10,000 s in the presence of 10:1 1-PBI:P450.
Structure of Ligand-free P450 2B4dH
Crystallization of P450 2B4dH was carried out concurrently with deuterium exchange experiments in light of recent success in the laboratory utilizing facial amphiphiles as co-crystallization agents. The ligand-free P450 2B4dH protein crystallized in the P31 space group with two molecules in the asymmetric unit. The R-factor of 19.6% and R-free of 22.3% were achieved by continued iterative model building and refinement. The final model of the three-dimensional crystal structure contains protein residues 28–492 in each subunit A and B, with the terminal histidine residue being a part of the larger C-terminal His tag. Numbers correspond to the full-length enzyme, which has 491 residues; in some cases, portions of the C-terminal histidine tag were visible in the electron density maps. Residues 473 and 474 were disordered in each of the two subunits. The model also contains 729 water molecules and 4 molecules of CYMAL-5. Two CYMAL-5 molecules in each protein chain were located in the hydrophobic pocket near residues Phe212 and Phe296, respectively, and were built during the final rounds of refinement. Density for the maltose group of two observed CYMAL-5 molecules was slightly disordered in each of the chains. The presence of 3α,7α,12α-tris[(β-d-maltopyranosyl)ethyloxy]cholane yielded higher quality crystals, which diffracted to a higher resolution than crystals obtained in the absence of the facial amphiphile. This molecule was not observed in the electron density maps. Data collection and refinement statistics are shown in Table 1. The structure was evaluated using MolProbity (62), and 97.8% of the residues fell into the favored regions of the Ramachandran plot (Table 1). The overall geometry ranked into the 98th percentile (MolProbity score of 1.33) in the final model with no bad angles or bonds. The clash score for “all atoms” was 2.75 corresponding to the 99th percentile compared with other structures at similar resolution (100th percentile is best among structures of comparable resolution).
TABLE 1.
Data collection and refinement statistics
Values for the highest resolution shell are shown in parentheses.
| Construct | 2B4dH |
| Crystal space group | P31 |
| Crystal unit cell parameters | |
| a = b | 91.47 Å |
| c | 150.40 Å |
| α = β | 90 |
| γ | 120 |
| Data collection statistics | |
| Beamline | SSRL 7-1 |
| Wavelength | 0.97 Å |
| Resolution range | 150.0 to 1.76 Å |
| Completeness | 97.9% (96.8%) |
| Redundancy | 4.7 (4.5) |
| Rmerge | 6 (34%) |
| I/s | 7.9 (2.2) |
| No. of observations | 643,945 |
| No. of unique reflections | 135,654 |
| Refinement statistics | |
| R-factor | 19.6% |
| R-free | 22.3% |
| r.m.s.d. | |
| Bond lengths | 0.013 Å |
| Bond angles | 1.443° |
| Average B-factor | 26.276 Å2 |
| Ramachandran plot | |
| Preferred | 97.8% |
| Allowed | 100% |
Overall, the structure of the ligand-free P450 2B4dH (Fig. 3) with a predominant α-helical domain was observed to be strikingly similar to that of the P450 2B4dH-4-CPI and P450 2B4dH-1-CPI complexes (10, 11), implying a closed conformation not previously observed without ligand. Electron density for partially occupied water molecules was observed near the heme in the ligand-free P450 2B4dH structure. UV-visible spectroscopy of the heme absorbance in a single crystal and in solution showed no evidence of any ligand besides water in the active site (supplemental Fig. S1). Crystals of P450 2B4dH without the internal H226Y mutation could be obtained under similar conditions; however, they diffracted to a much lower resolution.
FIGURE 3.
Representation of ligand-free cytochrome P450 2B4dH in a closed conformation. β-Sheets, α-helices, and loops are shown in purple, orange, and green, respectively. Heme is shown in red and CYMAL-5 in yellow.
Additionally, to compare the active site of the ligand-free closed conformation of P450 2B4dH with the 4-CPI bound structure, VOIDOO (63) measurements were made of the volume occupied by the cavity in each of these structures. Calculations were performed without ligands (if present) other than heme, using a probe-occupied radius of 1.4 Å. The cavity volumes in the ligand-free closed structure and 4-CPI complex were 358 and 266 Å3, respectively, predominantly as a result of Glu301 flipping away from the active site as opposed to coordinating 4-CPI. Moreover, Phe297 protrudes further into the active site to interact with 4-CPI, thereby reducing the cavity volume of the P450 2B4dH-4-CPI structure.
Molecular Dynamics Simulation of the P450 2B4dH Crystal Structure
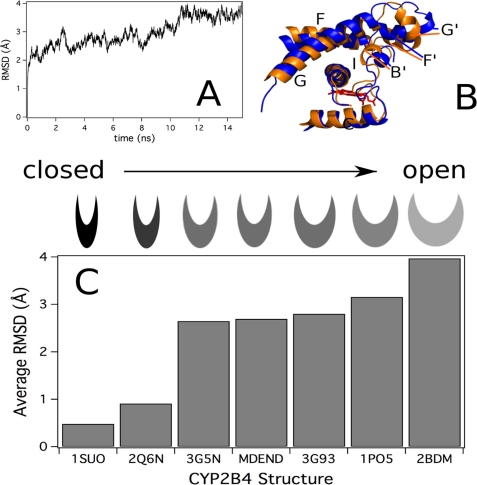
To investigate the energetically accessible and preferred conformations of P450 2B4dH, an MD simulation was performed of the enzyme for 15 ns. The r.m.s.d. of the Cα backbone during the MD simulation versus the starting closed ligand-free structure (PDB code 3MVR with residues 20–27, 473, 474, and 492–495 added with MODELLER) rapidly equilibrated in ∼100 ps to an r.m.s.d. of 2 Å, as reported previously (64). This was followed by a slower equilibrium representing larger molecular motions of P450 2B4dH that equilibrated in ∼10 ns with an r.m.s.d. of ∼4 Å and persisted for at least 5 ns (Fig. 4A). Conformational differences among structures in the MD simulation became apparent when the starting structure (PDB code 3MVR with residues 20–27, 473, 474, and 492–495 added with MODELLER) was aligned to the structure of the enzyme at 15 ns (Fig. 4B). Most notably, the F-G and B′-C loop regions were significantly shifted away from their initial positions. The r.m.s.d. was calculated to qualitatively compare the enzyme during the MD simulation with the crystal structures of P450 2B4dH (Fig. 4C). The 4-CPI (PDB code 1SUO) and 1-CPI (PDB code 2Q6N) complexes had the lowest r.m.s.d. from the starting structure and were thus most similar to the closed ligand-free P450 2B4dH structure at the beginning of the simulation. In contrast, the average r.m.s.d. of the equilibrated ligand-free P450 2B4dH structure during the MD simulation (MDEND) was similar to the r.m.s.d. of P450 2B4dH complexes with 1-PBI (2.70 Å, PDB codes 3G5N and 3G93). Even larger r.m.s.d. values were observed for the open ligand-free enzyme (3.16 Å, PDB code 1PO5) and the bifonazole-bound P450 2B4 structures (3.97 Å, PDB code 2BDM). Although the preferred orientation from MD resembles that of the 1-PBI complex, the MD simulation shows that a wide range of conformations are energetically accessible to P450 2B4.
FIGURE 4.
MD simulation and analysis of “closed” ligand-free P450 2B4dH. A, r.m.s.d. of the Cα backbone during the MD simulation from the MD starting structure (PDB code 3MVR with residues 20–27, 473, 474, and 492–495 added using MODELLER). B, ligand-free P450 2B4dH structure at 15 ns of the MD simulation (blue) aligned with the starting structure (green) with the F-G loop region and I-helix colored and labeled. The heme is shown in red. Residues 80–140 and 274–284 are hidden from the figure for clarity. C, r.m.s.d. of the Cα backbone of the P450 2B4dH crystal structures and MD simulation from the closed ligand-free P450 2B4dH structure. The category axis is labeled with the PDB code of the crystal structure or with MDEND for the average r.m.s.d. between 12 and 15 ns of the MD simulation (PDB code 1SUO, P450 2B4dH with 4-CPI; PDB code 2Q6N, P450 2B4dH with 1-CPI; PDB code 3G5N, P450 2B4dH with three 1-PBI molecules; PDB code 3G93, P450 2B4dH with one 1-PBI molecule; PDB code 1PO5, P450 2B4dH in the open conformation; and PDB code 2BDM, P450 2B4dH with bifonazole).
DISCUSSION
Despite the intense focus in recent years on the ligand recognition and binding step in the P450 catalytic cycle, there is still uncertainty as to how a single enzyme handles a diverse assortment of compounds (65–67). Data from a host of complementary techniques, including NMR, molecular dynamics simulations, and crystallography, indicate an integral role of protein dynamics in ligand binding and catalysis for many enzymes, including P450s (38, 68–71). The results presented here describe for the first time the use of DXMS to analyze the solution dynamic behavior of a mammalian drug-metabolizing cytochrome P450 in the absence and presence of ligands. These results indicate that the snapshots from the various crystal structures of P450 2B4dH provide a good representation of the dynamic solution behavior of this enzyme in the absence or presence of ligands (10, 11, 15, 18, 19). Additionally, a new ligand-free crystal structure of this protein in an alternate, closed conformation combined with a molecular dynamics simulation show that both the open and closed conformations of ligand-free P450 2B4dH are energetically accessible.
Results of DXMS experiments indicate that the amide hydrogens of P450 2B4dH are provided differing solvent protection depending upon the presence or absence of ligand and the identity of that ligand. Peptides covering large portions of the previously identified plastic regions of the protein (15), the B′- and C-helices, residues Ile101–Asp134, and the F-G cassette, residues Pro183–Thr255, showed significant slowing in the H-D exchange rate in the presence of 4-CPI compared with the exchange rate of ligand-free P450 2B4dH. These differences are in agreement with changes observed in these plastic regions in the previously reported crystal structures of open ligand-free and 4-CPI-bound P450 2B4dH (supplemental Fig. S2) (11, 19). Conversely, P450 2B4dH with and without 1-PBI shows little difference in H-D exchange rates across the entire protein. These results indicate that binding of 1-PBI does not increase protection from amide hydrogen exchange for the plastic regions of the protein in the B′-C loop and the F-G cassette but do not contradict the differences in conformation between the crystal structures of P450 2B4dH complexed with 1-PBI and the open ligand-free structure (Fig. 2B and supplemental Fig. S2). Although DXMS demonstrates that the large majority of P450 2B4dH adopts a more open conformation in the absence of ligand, averaging across the entire population of protein means that closed conformers representing minor populations would not be observed with this method.
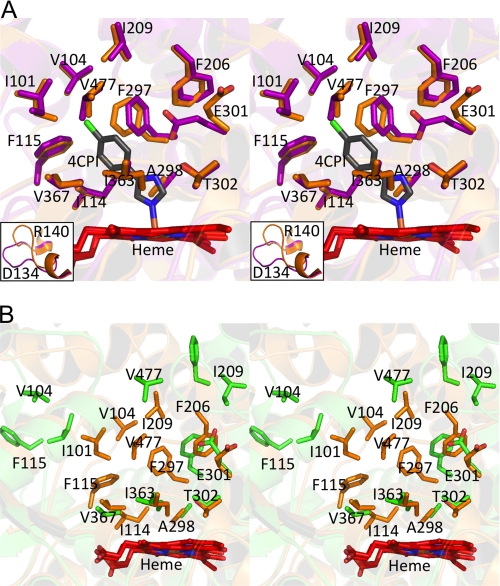
In contrast to the DXMS results, the new closed crystal structure of ligand-free P450 2B4dH was essentially super-imposable onto the 4-CPI complex, with an overall root mean square deviation (r.m.s.d.) of 0.49 Å in a Cα overlay. The prosthetic heme group bound to Cys436 and the residues previously found within a 5-Å radius of 4-CPI in 2B4dH show minimal changes in orientation between the 4-CPI-bound and closed ligand-free conformations (Fig. 5A). Ile101, Phe206, Phe297, and Val477 show small rearrangements, presumably because of ligand binding, whereas Glu301 flips away from the active site, as seen in the P450 2B6dH-4-CPI (72) and P450 2B4dH-1-CPI (10) structures. Most of these residues show major differences in orientation between the closed and open ligand-free structures (Fig. 5B). Transitioning between such open and closed states appears to be facilitated by reorientation of the F- and G-helices and the B-C loop. Moreover, the open ligand-free structure forms a dimer via coordination of His226 of each monomer with the heme iron of the other monomer. Such dimer formation was prevented in the subsequent structures and the DXMS studies by mutation of His226 to Tyr. The DXMS studies indicate a predominantly open conformation in the absence of ligand in contrast to the new closed ligand-free crystal structure. The only major difference observed between the closed ligand-free structure and the previously reported 4-CPI-bound complex was in the positioning of the C-D loop, residues Asp134–Arg140 (Fig. 5A, insets). Interestingly, the C-D loop of P450 2B4dH appears to have high solvent accessibility in both the absence and presence of ligands as seen by both DXMS and the difference in arrangement of the loop in the various crystal structures of the enzyme. This finding is interesting in light of recent work investigating interactions between cytochrome P450 2B6 and cytochrome P450 reductase (73). Chemical cross-linking experiments indicate that the C-D loop interacts with cytochrome P450 reductase.
FIGURE 5.
Stereo representations of an overlay using the ligand-free closed conformation structure of P450 2B4dH. A, superposition of the active site residues of the P450 2B4dH 4-CPI complex (purple) onto the closed ligand-free P450 2B4dH (orange) illustrates the striking similarities between the two structures. Also included is the 4-CPI ligand colored gray. The orientation of residues Ile101, Glu301, Val477, Phe206, and Phe297 is the only considerable difference in the active site of two structures. The inset shows residues 134–140, which are in significantly different orientations in the two structures. B, superposition of the closed conformation structure (orange) of ligand-free P450 2B4dH onto the previously determined open structure (green). Residues defined as part of the active site in the 4-CPI structure are shown.
The results reported here enabled analysis of structural plasticity of P450 2B4dH in terms of existing models of protein structural dynamics. Koshland's induced fit mechanism postulates changes in enzyme conformation after the binding of a ligand (74). In contrast, Monod-Wyman-Changeaux allostery theory proposes heterogeneity of conformations in solution whereby the ligand induces a change by binding to a specific conformer and affecting the equilibrium of conformers (75). These two models are represented visually in Scheme 1. Previous reports of ligand interactions with P450 2B4 inferred the induced fit model (18, 20, 76), but recent studies suggest that bacterial P450s exist in multiple conformers in solution, which more closely resembles the Monod-Wyman-Changeaux theory. For example, two ligand-free crystal structures of P450 EryK were reported in markedly different conformations influenced by salt concentration (71), and NMR and MD studies of CYP119 suggest multiple conformers in solution (77). Most recently, a crystal structure of P450cam was solved in an open ligand-free conformation (78). However, no evidence has been presented that distinguishes between these two models of ligand binding in mammalian P450s. In the case of P450 EryK, changing salt concentration altered the population distribution of states with high salt shifting the equilibrium to the closed state. The substrate erythromycin D bound more tightly to the closed conformation of the enzyme, indicating involvement of population shift in protein-ligand interactions (71). However, in the case of P450 2B4dH, protein-ligand interactions are most readily explained by induced fit (Scheme 1, path 1). Results from DXMS and MD point to the majority of the protein existing in an open orientation in solution (Figs. 2 and 4). Binding of 1-PBI to the open conformation would require little rearrangement, because this is close to the final bound structure (Fig. 4 and supplemental Fig. S2). Conversely, 4-CPI could bind either to an open or closed conformer but most likely interacts with the open conformer, which is the predominant solution form of the enzyme. Although induced fit protein interactions are the most straightforward explanation of these results, there is a possibility that 4-CPI could interact with P450 2B4dH by binding preferentially to a more closed form of the enzyme (Scheme 1, path 2).
SCHEME 1.
Protein ligand interaction models. Path 1, Koshland induced fit model. Path 2, the model based on Monod-Wyman-Changeux allostery theory. Green indicates PDB code 1PO5; orange indicates PDB code 3MVR, and purple indicates PDB code 1SUO).
In conclusion, the solution structural behavior of P450 2B4dH revealed by DXMS is very consistent with the predicted behavior based upon previously reported crystal structures. Interestingly, a new crystal structure of ligand-free P450 2B4dH was found in a closed conformation that is strikingly different from the previously reported open ligand-free structure. The molecular dynamics simulation allowed for conversion from this closed ligand-free structure to a conformation similar to the 1-PBI-bound and open ligand-free structures. On the whole, utilization of DXMS in structural studies of mammalian drug-metabolizing cytochromes P450 demonstrates that movements in the plastic regions of P450 2B4dH in solution are in close accord with the differences seen in available crystal structures of the protein. This finding is supported by the molecular dynamics simulation, thus providing valuable insight into both ligand-independent as well as ligand-dependent mechanisms of protein conformational changes in solution.
Supplementary Material
Acknowledgments
We are grateful to Dr. Sean Gay for critical reading of the manuscript and also to the staff at the Stanford Synchrotron Radiation Lightsource, operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences for assistance with data collection. The Stanford Synchrotron Radiation Lightsource is supported by the National Institutes of Health, NCRR, the Biomedical Technology Program, and the United States Department of Energy of Biological and Environmental Research.
This work was supported, in whole or in part, by National Institutes of Health Grants ES003619 (to J. R. H.), CA099835, CA118595, AI076961, AI081982, AI2008031, GM020501, GM066170, and RR029388 (to V. L. W.), and GM073197 (to Q. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (code 3MVR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Previous research involving P450 2B4, including the truncation of the protein and the open ligand-free structure, was completed using protein with the native His226. However, because of the formation of a dimer involving coordination of His226 of each monomer with the heme iron of the other monomer, subsequent solution biophysical work and crystallography efforts utilized the internal mutant H226Y. In this study, P450 2B4dH will refer to P450 2B4dH(H226Y) unless otherwise indicated.
- P450
- cytochrome P450
- BME
- 2-mercaptoethanol
- CYMAL-5
- 5-cyclohexyl-1-pentyl-β-d-maltoside
- 4-CPI
- 4-(4-chlorophenyl)imidazole
- 1-PBI
- 1-biphenyl-4-methyl-1H-imidazole
- 1-CPI
- 1-(4-chlorophenyl)imidazole
- r.m.s.d.
- root mean square deviation
- DXMS
- hydrogen-deuterium exchange mass spectrometry
- H-D
- hydrogen-deuterium
- ESI
- electrospray ionization
- MALDI
- matrix-assisted laser desorption ionization
- GdnHCl
- guanidine hydrochloride
- MD
- molecular dynamics
- PDB
- Protein Data Bank.
REFERENCES
- 1.Johnson E. F., Stout C. D. (2005) Biochem. Biophys Res. Commun. 338, 331–336 [DOI] [PubMed] [Google Scholar]
- 2.Al Omari A., Murry D. J. (2007) J. Pharm. Pract. 20, 206–218 [Google Scholar]
- 3.Guengerich F. P. (2001) Chem. Res. Toxicol. 14, 611–650 [DOI] [PubMed] [Google Scholar]
- 4.Ingelman-Sundberg M. (2004) Naunyn-Schmiedebergs Arch. Pharmacol. 369, 89–104 [DOI] [PubMed] [Google Scholar]
- 5.Anzenbacher P., Anzenbacherova E., Lange R., Skopalik J., Otyepka M. (2008) Acta Chim. Slov. 55, 63–66 [Google Scholar]
- 6.Mestres J. (2005) Proteins 58, 596–609 [DOI] [PubMed] [Google Scholar]
- 7.Otyepka M., Skopalík J., Anzenbacherová E., Anzenbacher P. (2007) Biochim. Biophys. Acta 1770, 376–389 [DOI] [PubMed] [Google Scholar]
- 8.Poulos T. L. (2005) Drug Metab. Dispos. 33, 10–18 [DOI] [PubMed] [Google Scholar]
- 9.Poulos T. L. (2005) Biochem. Biophys. Res. Commun. 338, 337–345 [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y., Sun L., Muralidhara B. K., Kumar S., White M. A., Stout C. D., Halpert J. R. (2007) Biochemistry 46, 11559–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott E. E., White M. A., He Y. A., Johnson E. F., Stout C. D., Halpert J. R. (2004) J. Biol. Chem. 279, 27294–27301 [DOI] [PubMed] [Google Scholar]
- 12.Williams P. A., Cosme J., Vinkovic D. M., Ward A., Angove H. C., Day P. J., Vonrhein C., Tickle I. J., Jhoti H. (2004) Science 305, 683–686 [DOI] [PubMed] [Google Scholar]
- 13.Wester M. R., Yano J. K., Schoch G. A., Yang C., Griffin K. J., Stout C. D., Johnson E. F. (2004) J. Biol. Chem. 279, 35630–35637 [DOI] [PubMed] [Google Scholar]
- 14.Williams P. A., Cosme J., Sridhar V., Johnson E. F., McRee D. E. (2000) Mol. Cell 5, 121–131 [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y., White M. A., Muralidhara B. K., Sun L., Halpert J. R., Stout C. D. (2006) J. Biol. Chem. 281, 5973–5981 [DOI] [PubMed] [Google Scholar]
- 16.Ekroos M., Sjögren T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domanski T. L., Halpert J. R. (2001) Curr. Drug Metab. 2, 117–137 [DOI] [PubMed] [Google Scholar]
- 18.Gay S. C., Sun L., Maekawa K., Halpert J. R., Stout C. D. (2009) Biochemistry 48, 4762–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott E. E., He Y. A., Wester M. R., White M. A., Chin C. C., Halpert J. R., Johnson E. F., Stout C. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y., Halpert J. R. (2007) Biochim. Biophys. Acta 1770, 402–412 [DOI] [PubMed] [Google Scholar]
- 21.Busenlehner L. S., Armstrong R. N. (2005) Arch. Biochem. Biophys. 433, 34–46 [DOI] [PubMed] [Google Scholar]
- 22.Engen J. R. (2003) Analyst 128, 623–628 [DOI] [PubMed] [Google Scholar]
- 23.Hoofnagle A. N., Resing K. A., Ahn N. G. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 1–25 [DOI] [PubMed] [Google Scholar]
- 24.Tsutsui Y., Wintrode P. L. (2007) Curr. Med. Chem. 14, 2344–2358 [DOI] [PubMed] [Google Scholar]
- 25.Wales T. E., Engen J. R. (2006) Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]
- 26.Wani A. H., Udgaonkar J. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20711–20716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frimpong A. K., Abzalimov R. R., Uversky V. N., Kaltashov I. A. (2010) Proteins 78, 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konermann L., Tong X., Pan Y. (2008) J. Mass Spectrom. 43, 1021–1036 [DOI] [PubMed] [Google Scholar]
- 29.Eyles S. J., Kaltashov I. A. (2004) Methods 34, 88–99 [DOI] [PubMed] [Google Scholar]
- 30.Selevsek N., Rival S., Tholey A., Heinzle E., Heinz U., Hemmingsen L., Adolph H. W. (2009) J. Biol. Chem. 284, 16419–16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutkowska-Wlodarczyk I., Stepinski J., Dadlez M., Darzynkiewicz E., Stolarski R., Niedzwiecka A. (2008) Biochemistry 47, 2710–2720 [DOI] [PubMed] [Google Scholar]
- 32.Ferguson P. L., Kuprowski M. C., Boys B. L., Wilson D. J., Pan J. X., Konermann L. (2009) Curr. Anal. Chem. 5, 186–204 [Google Scholar]
- 33.Bo W., Chu Y. Q., Dai Z. Y., Ding C. F. (2008) Chin. J. Chem. Phys. 21, 217–220 [Google Scholar]
- 34.Liao W. L., Dodder N. G., Mast N., Pikuleva I. A., Turko I. V. (2009) Biochemistry 48, 4150–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaltashov I. A., Zhang M., Eyles S. J., Abzalimov R. R. (2006) Anal. Bioanal. Chem. 386, 472–481 [DOI] [PubMed] [Google Scholar]
- 36.Kaltashov I. A., Bobst C. E., Abzalimov R. R. (2009) Anal. Chem. 81, 7892–7899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamuro Y., Molnar K. S., Coales S. J., OuYang B., Simorellis A. K., Pochapsky T. C. (2008) J. Inorg. Biochem. 102, 364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pochapsky S. S., Dang M., OuYang B., Simorellis A. K., Pochapsky T. C. (2009) Biochemistry 48, 4254–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott E. E., Spatzenegger M., Halpert J. R. (2001) Arch. Biochem. Biophys. 395, 57–68 [DOI] [PubMed] [Google Scholar]
- 40.Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 41.Omura T., Sato R., Cooper D. Y., Rosenthal O., Estabrook R. W. (1965) Fed. Proc. 24, 1181–1189 [PubMed] [Google Scholar]
- 42.Hamuro Y., Anand G. S., Kim J. S., Juliano C., Stranz D. D., Taylor S. S., Woods V. L., Jr. (2004) J. Mol. Biol. 340, 1185–1196 [DOI] [PubMed] [Google Scholar]
- 43.Hamuro Y., Zawadzki K. M., Kim J. S., Stranz D. D., Taylor S. S., Woods V. L., Jr. (2003) J. Mol. Biol. 327, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z., Smith D. L. (1993) Protein Sci. 2, 522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q., Ma X., Ward A., Hong W. X., Jaakola V. P., Stevens R. C., Finn M. G., Chang G. (2007) Angew. Chem. Int. Ed. Engl. 46, 7023–7025 [DOI] [PubMed] [Google Scholar]
- 46.Soltis S. M., Cohen A. E., Deacon A., Eriksson T., González A., McPhillips S., Chui H., Dunten P., Hollenbeck M., Mathews I., Miller M., Moorhead P., Phizackerley R. P., Smith C., Song J., van dem Bedem H., Ellis P., Kuhn P., McPhillips T., Sauter N., Sharp K., Tsyba I., Wolf G. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1210–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leslie A. G. W. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 48.Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 49.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 52.Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 53.Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 54.Oda A., Yamaotsu N., Hirono S. (2005) J. Comput. Chem. 26, 818–826 [DOI] [PubMed] [Google Scholar]
- 55.Froelich J. W., Hearshen D. O., Halpert R. D., Patel S. (1985) Henry Ford Hosp. Med. J. 33, 122–127 [PubMed] [Google Scholar]
- 56.Berendsen H. J., Postma J. P., Vangunsteren W. F., Dinola A., Haak J. R. (1984) J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 57.Oostenbrink C., Soares T. A., van der Vegt N. F., van Gunsteren W. F. (2005) Eur. Biophys. J. 34, 273–284 [DOI] [PubMed] [Google Scholar]
- 58.Darden T., York D., Pedersen L. (1993) J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 59.DeLano W. (2009) The PyMOL Molecular Graphics System, MacPyMOL ed., DeLano Scientific, Palo Alto, CA [Google Scholar]
- 60.Muralidhara B. K., Negi S., Chin C. C., Braun W., Halpert J. R. (2006) J. Biol. Chem. 281, 8051–8061 [DOI] [PubMed] [Google Scholar]
- 61.Morrison J. F. (1969) Biochim. Biophys. Acta 185, 269–286 [DOI] [PubMed] [Google Scholar]
- 62.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 64.Muralidhara B. K., Halpert J. R. (2007) Drug Metab. Rev. 39, 539–556 [DOI] [PubMed] [Google Scholar]
- 65.Davydov D. R., Halpert J. R. (2008) Expert Opin. Drug Metab. Toxicol. 4, 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isin E. M., Guengerich F. P. (2008) Anal. Bioanal. Chem. 392, 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wade R. C., Motiejunas D., Schleinkofer K., Sudarko, Winn P. J., Banerjee A., Kariakin A., Jung C. (2005) Biochim. Biophys. Acta 1754, 239–244 [DOI] [PubMed] [Google Scholar]
- 68.Bakan A., Bahar I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hays A. M., Dunn A. R., Chiu R., Gray H. B., Stout C. D., Goodin D. B. (2004) J. Mol. Biol. 344, 455–469 [DOI] [PubMed] [Google Scholar]
- 70.Henzler-Wildman K. A., Lei M., Thai V., Kerns S. J., Karplus M., Kern D. (2007) Nature 450, 913–916 [DOI] [PubMed] [Google Scholar]
- 71.Savino C., Montemiglio L. C., Sciara G., Miele A. E., Kendrew S. G., Jemth P., Gianni S., Vallone B. (2009) J. Biol. Chem. 284, 29170–29179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gay S. C., Shah M. B., Talakad J. C., Maekawa K., Roberts A. G., Wilderman P. R., Sun L., Yang J. Y., Huelga S. C., Hong W. X., Zhang Q., Stout C. D., Halpert J. R. (2010) Mol. Pharmacol. 77, 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bumpus N. N., Hollenberg P. F. (2010) J. Inorg. Biochem. 104, 485–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koshland D. E., Jr., Némethy G., Filmer D. (1966) Biochemistry 5, 365–385 [DOI] [PubMed] [Google Scholar]
- 75.Monod J., Wyman J., Changeux J. P. (1965) J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 76.Muralidhara B. K., Sun L., Negi S., Halpert J. R. (2008) J. Mol. Biol. 377, 232–245 [DOI] [PubMed] [Google Scholar]
- 77.Lampe J. N., Brandman R., Sivaramakrishnan S., de Montellano P. R. (2010) J. Biol. Chem. 285, 9594–9603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee Y. T., Wilson R. F., Rupniewski I., Goodin D. B. (2010) Biochemistry 49, 3412–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Molday R. S., Englander S. W., Kallen R. G. (1972) Biochemistry 11, 150–158 [DOI] [PubMed] [Google Scholar]
- 81.Oyeyemi O. A., Sours K. M., Lee T., Resing K. A., Ahn N. G., Klinman J. P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10074–10079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.