Abstract
Thrombin uses three principal sites, the active site, exosite I, and exosite II, for recognition of its many cofactors and substrates. It is synthesized in the zymogen form, prothrombin, and its activation at the end of the blood coagulation cascade results in the formation of the active site and exosite I and the exposure of exosite II. The physiological inhibitors of thrombin are all serpins, whose mechanism involves significant conformational change in both serpin and protease. It has been shown that the formation of the thrombin-serpin final complex disorders the active site and exosite I of thrombin, but exosite II is thought to remain functional. It has also been hypothesized that thrombin contains a receptor-binding site that is exposed upon final complex formation. The position of this cryptic site may depend on the regions of thrombin unfolded by serpin complexation. Here we investigate the conformation of thrombin in its final complex with serpins and find that in addition to exosite I, exosite II is also disordered, as reflected by a loss of affinity for the γ′-peptide of fibrinogen and for heparin and by susceptibility to limited proteolysis. This disordering of exosite II occurs for all tested natural thrombin-inhibiting serpins. Our data suggest a novel framework for understanding serpin function, especially with respect to thrombin inhibition, where serpins functionally “rezymogenize” proteases to ensure complete loss of activity and cofactor binding.
Keywords: Antithrombin, Protease Inhibitor, Prothrombin, Serpin, Thrombin, Exosites, Zymogenicity
Introduction
Thrombin is formed from its zymogen prothrombin through sequential cleavage at two sites by the prothrombinase complex (1). The first cleavage at Arg320 (prothrombin numbering, and Arg15 in chymotrypsin template numbering (2), used throughout the text) is the classic chymotrypsin (S1)-family activation event, resulting in the formation of a new N terminus at Ile16, which subsequently inserts into the zymogen activation pocket and stabilizes the catalytic site. In most serine proteases, cleavage activation results in a disordered-to-ordered conformational transition involving a distinct fraction of the molecule, known as the “zymogen activation domain,” and the most important event in promoting this conformational change is the formation of a salt bridge between the new N terminus (Ile16) with Asp194 on the catalytic loop (3). This interaction results in the formation of the principal substrate-binding site (the S1 pocket) and orders the oxyanion hole (formed by Gly193 and Ser195). The second cleavage event at Arg271 (prothrombin numbering) liberates thrombin from the Gla and two kringle domains of prothrombin (the F1.2 fragment).
Thrombin relies on three main sites to exert its activity, the active site and two anion-binding exosites, known as exosites I and II (for review, see Ref. 4), none of which is available in prothrombin. The active site is part of the zymogen activation domain and is thus disordered/unfolded in prothrombin, and similarly, exosite I is disordered in prothrombin and only becomes capable of binding to substrates and cofactors after cleavage at Arg320 (5–7). Exosite II, on the other hand, is fully formed but is blocked by a tight interaction with the second kringle domain of the F1.2 fragment (8). Thus, the zymogen form of thrombin is deficient in all three functional sites, allowing prothrombin to circulate at a high reservoir concentration in an inactive state that will not prematurely bind to substrates or cofactors, such as fibrinogen, thrombomodulin (TM),3 glycosaminoglycans, or platelet receptors.
The physiological inhibitors of thrombin are all serpins (antithrombin (AT), heparin cofactor II (HCII), protease nexin 1 (PN1), plasminogen activator inhibitor 1 (PAI1), protein C inhibitor, and the Pittsburgh variant of α1-antitrypsin, α1ATpitts) (9). Serpins utilize a unique, well characterized mechanism of protease inhibition, where the protease attacks the P1-P1′ bond of the serpin reactive center loop as if it were a normal substrate (10, 11). Upon formation of the acyl-enzyme intermediate, the protease is flung to the opposite pole of the serpin due to the rapid incorporation of the reactive center loop into the A sheet. The ester bond between the side chain Oγ of Ser195 of the protease and the main chain carbon of the P1 residue of the serpin is stabilized by the distension of the catalytic loop and the loss of the oxyanion hole. In the original crystal structure, some 40% of the protease could not be resolved in electron density, suggesting that about half of the protease had effectively unfolded (12). However, the extent of protease unfolding by serpins in solution for physiologically relevant pairs is somewhat controversial. A recent crystal structure of another serpin-protease complex showed a distension of the catalytic loop but no protease unfolding (13), and an NMR study by the same group suggested complete protease unfolding into a molten globule-like state (14). Other studies have shown loss of specific ligand binding (15–17) and induced proteolytic susceptibility (18–20), supporting the idea that some structural disordering of proteases is a general feature of complexation by serpins. Although there is no crystal structure of a serpin-thrombin complex, two functional studies have shown a specific loss of exosite I function (16, 17).
Determining the extent of unfolding of thrombin when complexed by serpins is critical for understanding how thrombin is disengaged from its various substrates and cofactors and also for understanding how serpin-thrombin complexes are cleared (21). Here we investigate the effect of serpin complexation on thrombin structure, with particular emphasis on exosite II. We demonstrate a significant loss of affinity for exosite II ligands concomitant with complex formation and an induced and extensive increase in proteolytic susceptibility. The extraction of Asp194 from the activation pocket of the protease by serpins results in the obligate unfolding of the zymogen activation domain, and in thrombin, the consequent disordering of exosite I. The disordering of exosite II is likely the result of clashes between thrombin surface loops with the serpin in the final complex. The loss of ordered structure in the catalytic site and the two principal exosites upon complexation by serpins functionally rezymogenizes thrombin to promote dissociation from substrates and cofactors. The conformational consequences of their mechanism help explain why serpins control blood coagulation and other highly regulated proteolytic cascades critical for life.
EXPERIMENTAL PROCEDURES
Materials
Human α-thrombin was purchased from Hematologic Technologies Inc. (Essex Junction, VT). The chromogenic substrate S-2238 was purchased from Chromogenix (Milano, Italy). Nα-Fmoc-protected amino acids, solvents, and reagents for peptide synthesis were purchased from Novabiochem (Darmstadt, Germany) or Bachem AG (Bubendorf, Switzerland). All other reagents and solvents were of analytical grade and purchased from Fluka.
Production of Recombinant Thrombins
Thrombin variants S195A, and R93E were created by site-directed mutagenesis following the Stratagene protocol and verified by DNA sequencing. Recombinant thrombins were purified from Escherichia coli and refolded as described previously (22).
Preparation of Fluorescein-labeled Fibrinogen γ′-Peptide
The fibrinogen γ′ 408–427 peptide (408VRPEHPAETEY*DSLY*PEDDL427, where Y* denotes phosphorylated tyrosine), γ′(408–427), was synthesized by the solid-phase method using the Fmoc chemistry (23) on a model PS3 automated synthesizer from Protein Technologies International (Tucson, AZ). The peptide chain was assembled stepwise on a Wang resin (Novabiochem) derivatized with Fmoc-Leu (0.45 milliequivalence/g). The crude peptide was fractionated by reverse phase-HPLC on a Zorbax (Agilent Technologies, Santa Clara, CA) C18 analytical column, eluted with a linear acetonitrile-0.1% TFA gradient from 25 to 45% in 30 min. The N-terminal fluoresceinated derivative was obtained by adding a solution of purified γ′(408–427) peptide (20 μl, 80 nmol) in 0.1 m NaHCO3, pH 9.0, to a solution of fluorescein isothiocyanate (Sigma) in dimethyl sulfoxide (10 μl, 25 mm). The reaction was allowed to proceed for 1 h at room temperature with a final yield >90%. After lyophilization, the reaction mixture was fractionated on a Grace-Vydac (Hesperia, CA) C-18 column (4.6 × 25 cm) eluted with a linear acetonitrile-0.1% TFA gradient from 15 to 30% in 30 min at a flow rate of 0.8 ml/min. The absorbance of the effluent was recorded at 226 nm, and the peptide material was analyzed by mass spectrometry on a Mariner ESI-TOF instrument from PerSeptive Biosystems (Stafford, TX), which yielded mass values in agreement with the theoretical mass within 20 ppm accuracy.
Binding of Fibrinogen γ′-Peptide to Thrombin
All experiments were performed in buffer containing 50 mm Tris-HCl, pH 7.4, 50 mm NaCl with 0.1% PEG8000 on a PerkinElmer Life Sciences LS50B fluorometer at 22 °C. Fluorescence emission spectra were collected in 2-ml cuvettes containing 50 nm fluorescein-γ′, exciting at 475 nm with slits set at 2.5 nm for excitation and 4 nm for emission. An average of three scans was taken for each spectrum. Thrombin was added to a final concentration of 340 nm before the addition of a small excess of inhibitor. Small volume additions were used, and dilution was accounted for. To ensure full inhibition of thrombin by α1ATpitts, complex was preformed at high concentration (50 μm thrombin and 135 μm serpin) and incubated for 1 h. Full inhibition was verified by S-2238 hydrolysis and SDS-PAGE (data not shown). 20 μl of the reaction mixture was added to yield a final concentration of 495 nm complexed thrombin, and spectra were recorded as previously. A spectrum of the R93E thrombin variant was obtained under similar conditions (480 nm). Dissociation constants for thrombin (wild-type and variants) were determined by monitoring change of 50 nm fluorescein-γ′ fluorescence signal at 516 nm with increasing thrombin concentration and fitting the resulting curve to a one-site specific binding equation using the software PRISM. For fluorescein-γ′ displacement experiments, 340 nm thrombin was added to buffer containing 50 nm fluorescein-γ′. Time drive was used to monitor the continuous fluorescence change at 516 nm after the addition of serpins. In the parallel activity assay for monitoring loss of thrombin activity, AT (7.5 μl, 482 nm final concentration) was added to 1 ml of buffer containing 340 nm thrombin (conditions identical to the fluorescein-γ′ displacement experiment). At time intervals, 3 μl of the reaction mixture was withdrawn and added to a microtiter plate well containing 200 μl of 300 μm S-2238, and the residual thrombin activity was monitored by the change of absorbance at 405 nm.
Analytical Heparin-Sepharose Chromatography
The thrombin-α1ATpitts complex was formed by incubating 100 μg of thrombin with a 1:1.5 or 1.5:1 molar ratio of α1ATpitts in 500 μl of 50 mm Tris, 50 mm NaCl, 5 mm EDTA, pH 7.4, for 30 min at room temperature. Heparin-Sepharose chromatography was conducted on a 1-ml HiTrap heparin-Sepharose column (GE Healthcare, Uppsala, Sweden). Proteins were eluted with a gradient from 50 mm to 1 m NaCl in 50 mm Tris, pH 7.4. Plasma thrombin and recombinant variants, alone and inhibited with α1ATpitts or PPACK, were subjected to chromatography under identical conditions, and peak conductivities were recorded (see Table 1). Samples from the selected peaks were subjected to SDS-PAGE.
TABLE 1.
Peak elution conductivities of thrombin and its complexes from an analytical heparin Sepharose column and dissociation constants for fluorescein-γ′
ND, not determined.
| Proteins | Peak conductivity | Dissociation constant for fluorescein-γ′ |
|---|---|---|
| mS/cm | μm | |
| Plasma thrombin | 54.4 | 0.175 ± 0.025 |
| Plasma thrombin-α1ATpitts complex | 33.0 | ND |
| Cleaved plasma thrombin-α1ATpitts complex | 24.8 | ND |
| Plasma thrombin-PPACK | 55.9 | ND |
| Recombinant thrombin | 60.1 | 0.201 ± 0.015 |
| Recombinant R93E thrombin | 39.4 | 14.1 ± 0.6 |
| Recombinant thrombin- α1ATpitts complex | 39.7 | ND |
| Recombinant R93E thrombin- α1ATpitts complex | 35.7 | ND |
Limited Proteolysis
Limited proteolysis assays were carried out by incubating 0.23 nmol of plasma-derived thrombin with 0.13–0.17 nmol of AT, α1ATpitts, or PN1, in the presence or absence of exosite ligands, at 37 °C for 1–2 h to allow the excess thrombin (10-μl reaction volume) to cleave the covalently linked serpin-thrombin complex. Thrombin alone was incubated under the identical condition as control. 12% Bis-Tris non-reducing SDS-PAGE (Invitrogen) was used to analyze the cleavage products. For N-terminal sequencing, bands were transferred onto PVDF membrane after SDS-PAGE, and sequencing was carried out at the Department of Biochemistry, University of Cambridge.
RESULTS
Fibrinogen γ′-Peptide Binding Is Disrupted by Serpins
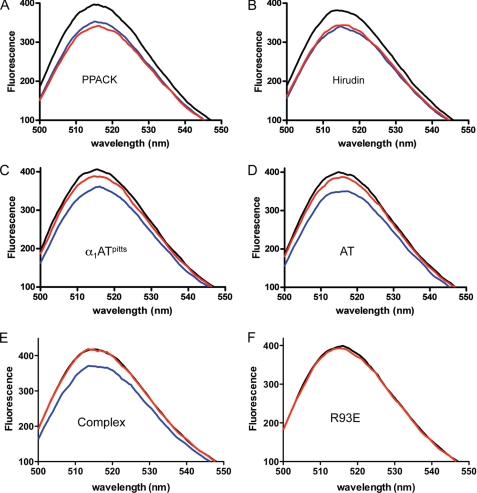
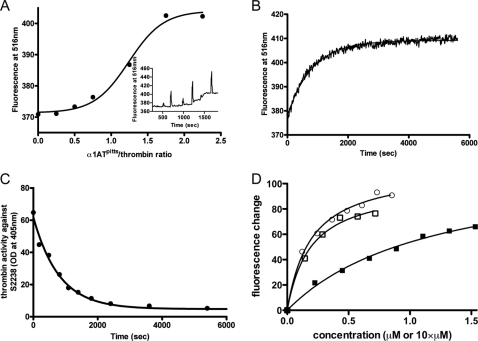
Thrombin binds to several substrates and cofactors via exosite II, including heparin (and other glycosaminoglycans such as heparan sulfate exposed on intact blood vessels and chondroitin sulfate expressed on a fraction of TM), the C-terminal portion of platelet receptor GpIbα, and the γ′-peptide of fibrinogen (4). Crystal structures of these have been published, and although binding modes vary, the ligands generally utilize exosite II residues Arg93, Arg101, Arg126, Arg233, Lys235, Lys236, and Lys240, with Arg93 of principal importance. To test the effect of serpin complexation on exosite II of thrombin, we created a fluorescein-labeled γ′-peptide. Fluorescence spectra of fluorescein-γ′ with thrombin and inhibitors are shown in Fig. 1. The binding of plasma thrombin to the fluorescein-γ′-peptide caused an 11% fluorescence quench at the emission peak of 516 nm. The addition of the covalent active site inhibitor PPACK to the cuvette caused a small (4%) additional quench (Fig. 1A), suggesting that PPACK binding results in slightly higher affinity for the fluorescein-γ′-peptide. The addition of full-length hirudin to thrombin, which binds in both exosite I and the active site, did not result in a change in fluorescence signal (Fig. 1B). In contrast, when serpins α1ATpitts (Fig. 1C) or AT (Fig. 1D) were added to the cuvette, a large loss of the original quench was observed (63 and 78%, respectively), suggesting that the binding of the fluorescein-γ′-peptide to exosite II had been significantly diminished. The residual quench observed in the serpin-inhibited samples was found to be due to incomplete inhibition under the low concentrations used. This was demonstrated by preincubating thrombin and serpin at high concentrations for 1 h before adding a small volume of the complex to a cuvette containing the fluorescently labeled γ′-peptide (Fig. 1E). No fluorescence change was observed despite the high final complex concentration (500 nm). Control experiments showed that inhibitors alone did not bind to fluorescein-γ′ or cause any fluorescence change (data not shown). These data are consistent with the interpretation that forming the final complex with the serpin causes thrombin to lose affinity for the γ′-peptide. To confirm this, we examined how much α1ATpitts is needed to fully expel the fluorescein-γ′-peptide from thrombin. As shown in Fig. 2A, the addition of increasing amounts of α1ATpitts resulted in a saturable loss of quench at ∼1.5-fold molar excess of inhibitor, very close to the stoichiometry of inhibition under similar conditions (stoichiometry of inhibition = 1.2) (24). The apparent sigmoidal nature of Fig. 2A is likely due to the slower rates of inhibition at low serpin concentrations, consistent with an overestimation of the stoichiometry of inhibition using these short (1–2-min) incubations. To confirm the concomitant loss of fluorescence quench with inhibition using excess serpin, we followed fluorescence and thrombin activity over time at a single AT concentration (Fig. 2, B and C) and obtained identical rates (1.25 × 10−3 s−1 and 1.22 × 10−3 s−1, respectively). We conclude that thrombin inhibition by serpins results in a loss of affinity for the fluorescein-γ′-peptide and that expulsion of the γ′-peptide from exosite II occurs concomitant with formation of the final complex.
FIGURE 1.
The fluorescence spectra of fluorescein-labeled fibrinogen γ′-peptide in the presence of free and inhibited thrombin. In each case, the black trace is the solution containing fluorescein-γ′ alone, the blue trace is after the addition of thrombin to the solution, and the red trace is after the further addition of a thrombin inhibitor (A, PPACK; B, hirudin; C, α1ATpitts; and, D, AT). E, no fluorescence quench is observed when thrombin inhibition is complete. The black trace is buffer with the fluorescein-γ′ alone; red is after the addition of preformed α1ATpitts-thrombin complex; and blue is fluorescein-γ′ with uninhibited thrombin. F, exosite II variant R93E is deficient in fluorescein-γ′ binding, although a small quench is observed upon the addition of the thrombin variant (red).
FIGURE 2.
The loss of fibrinogen γ′-peptide binding is concomitant with thrombin inhibition by serpins. A, the stoichiometry of α1ATpitts required to fully expel fluorescein-γ′ from thrombin was determined by following fluorescein-γ′-thrombin fluorescence while titrating in α1ATpitts. The inset is a time drive showing fluorescence change upon each addition (spikes reflect mixing following additions of α1ATpitts). B, time course of fluorescence change upon the addition of excess AT to a cuvette containing 50 nm fluorescein-γ′-peptide and thrombin and its corresponding fit. C, time course of loss of thrombin activity with the identical solution as in B, monitored by absorbance of chromogenic substrate S-2238 (with single exponential fit). OD, optical density. D, titration of plasma (circles), recombinant S195A (open squares), and R93E (closed squares) thrombins into solutions containing the fluorescein-γ′-peptide, with non-linear fits as lines. The concentration of plasma and S195A thrombins is μm units, whereas the concentration of R93E is 10 times higher than indicated.
Heparin Binding Affinity Is Reduced by Serpins
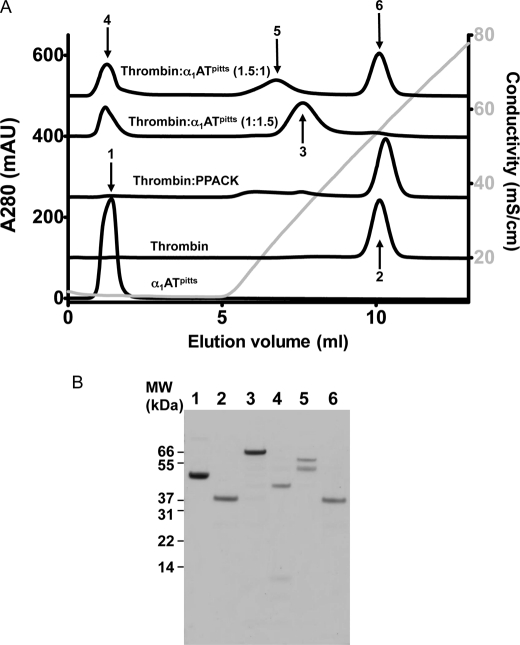
The binding of the fibrinogen γ′-peptide to exosite II of thrombin is highly specific and of moderate affinity (25, 26). To test whether the less avid and nonspecific binding of heparin to thrombin (27, 28) is also affected by serpin complexation, we assessed heparin affinity by elution from an analytical heparin-Sepharose column. For these studies, it was critical to use a serpin incapable of binding to heparin, and therefore limited our studies to complexes with α1ATpitts. Elution profiles from heparin-Sepharose are shown in Fig. 3A, and peak conductivities are given in Table 1. As expected, α1ATpitts eluted in the void volume, confirming that it does not bind to heparin. Uninhibited thrombin eluted during the NaCl gradient at 54 mS/cm (equivalent to ∼590 mm NaCl), and inhibition by PPACK resulted in a small shift to the right (56 mS/cm), indicative of a small increase in affinity, consistent with what was observed for the fluorescein-γ′-peptide. The thrombin-α1ATpitts complex eluted in a position intermediate between thrombin and α1ATpitts, indicative of a reduction in the affinity of complexed thrombin for heparin. When α1ATpitts is in excess, the intact final complex elutes around 33 mS/cm (310 mm NaCl). However, when thrombin is in excess over α1ATpitts, and the complex and free thrombin are incubated at 37 °C to allow the final complex to be nicked before loading on the column, the complex elutes even earlier (25 mS/cm). SDS-PAGE was used to confirm the identities of the peak fractions (Fig. 3B). It is clear that complexation of thrombin by a serpin results in a substantial decrease in heparin affinity.
FIGURE 3.
Analytical heparin-Sepharose column elution profiles for thrombin, α1ATpitts, and their complex. A, elution profile of proteins and complexes, as indicated. It can be seen that α1ATpitts, native or cleaved (peaks 1 and 4), does not bind to heparin-Sepharose and elutes in the void volume. Thrombin and thrombin bound to PPACK bind to the heparin column with high affinity and elute late in the NaCl gradient (gray line). Thrombin-α1ATpitts complex, whether intact (peak 3) or nicked by excess thrombin (peak 5), has weaker affinity for heparin-Sepharose than native thrombin. mAU, milliabsorbance units. B, non-reducing SDS-PAGE analysis of the peaks labeled in A. MW, molecular weight.
Exosite II Defect Is Similar to Charge Reversal at Arg93
The small fluorescence quench observed when adding serpins to cuvettes containing thrombin and the fluorescein-γ′-peptide (Fig. 1, C and D) suggested either residual binding or an alteration in the quantum yield of the bound peptide. However, when complex was preformed to ensure that all thrombin had been inhibited, the fluorescence spectrum of the fluorescein-γ′-peptide returned to what was observed in the absence of thrombin (Fig. 1E). Although this could still be due to a change in quantum yield of bound peptide, this explanation would require the quantum yield of bound peptide to be identical to that of free peptide. A more reasonable explanation is that thrombin in complex with serpins does not appreciably bind to the fluorescein-γ′-peptide under the conditions used in the experiment (50 nm peptide and 500 nm complex). Attempts to quantify the reduction in affinity for the fluorescein-γ′-peptide by fluorescence were unsuccessful due principally to our inability to concentrate complex sufficiently to obtain a signal. However, it is possible to estimate the magnitude of loss of affinity by comparison with a well characterized exosite II variant, R93E. This variant (and R93A) has been shown to be deficient in exosite II binding by various methods, including heparin-Sepharose elution and rate of inhibition by AT in the presence of heparin (29, 30). We produced the variant in our E. coli system and compared its elution from heparin-Sepharose with recombinant wild-type when inhibited by α1ATpitts (Table 1). R93E thrombin eluted at 39.4 mS/cm, comparable with the thrombin-serpin complex, which eluted at 39.7 mS/cm. Formation of the complex between R93E thrombin with α1ATpitts resulted in a small additional reduction in the conductivity of the elution peak (35.7 mS/cm). These data suggest that the defect in exosite II binding to thrombin caused by complexation with serpins is similar to that caused by charge reversal at a critical exosite II residue.
The ability of the R93E thrombin variant to bind to fluorescein-γ′ was tested by taking a fluorescence spectrum, as before, with the variant at 480 nm (Fig. 1F). The resulting spectra revealed a small but appreciable quench of fluorescein-γ′ fluorescence upon the addition of the variant. We concentrated the R93E variant to 16 mg/ml, performed a titration following fluorescence quench, and obtained a Kd value of 14.1 μm (Fig. 2D). Titrations using plasma-derived thrombin or recombinant thrombin gave Kd values of 175 and 200 nm, respectively (Fig. 2D). Thus, the R93E mutation results in a 70–80-fold loss in affinity for the fluorescein-γ′-peptide.
Exosite II Is Susceptible to Proteolysis When Complexed by AT
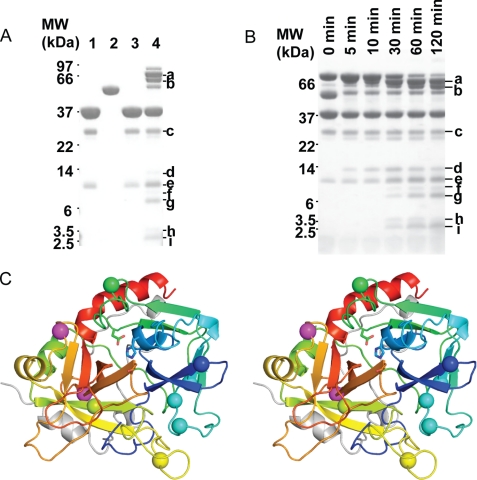
The preceding sections demonstrate that exosite II is compromised when thrombin is complexed by serpins, but the loss of exosite II interactions/affinity might be due either to the serpin blocking exosite II or to serpin-induced conformational change/disordering of exosite II. To address this, we probed thrombin structure/disorder by determining the sites of autolysis in the absence and presence of AT. We incubated thrombin with a 2/3rd equivalence of AT at 37 °C for 2 h and observed digestion of the final complex by SDS-PAGE. Under these conditions, the majority of the final complex is digested into two major large molecular weight fragments and a number of smaller fragments. N-terminal sequencing was carried out to identify the cleavage sites (Table 2 and Fig. 4). Most of the autolysis sites were found to be in exosite I (Arg35, Arg67, Lys69, Arg73, Fig. 4A, bands f, h, and i); however, the most important exosite II residue, Arg93 (Fig. 4A, bands a, f, and g), is also cleaved when thrombin is in complex with AT. These results confirm that exosite I is disordered upon complexation by serpins, as predicted from previous studies that showed a loss of exosite I binding affinity (16, 17), and suggest that exosite II is similarly disordered. To determine whether Arg93 cleavage was the result of a general destabilization due to cleavage elsewhere in thrombin, we conducted a time course of thrombin digestion. As shown in Fig. 4B, the major Arg93 containing band a was the first to appear, indicating that it is likely to be the preferred autolysis site when thrombin is complexed by AT.
TABLE 2.
N-terminal sequencing of thrombin cleavage products for the thrombin-AT complex
Sequences marked with * are unique sites that only occur in thrombin when in complex with AT. Multiple N termini are generally observed for each band due to disulfide bonds and the covalent bond between Ser195 of thrombin and Arg393 of AT. Cleavage at the very C terminus of thrombin (e.g. exosite II residues Arg233, Lys235, Lys236, and Lys240) would produce bands too small to detect by SDS-PAGE and may therefore have been missed.
| Band ID | Sequence | Origin of sequence |
|---|---|---|
| a | YNWREN* | Thrombin, H chain, Arg93 cut |
| a | TFGSGE | Thrombin, L chain, N terminus |
| a | HGSPVD | AT, N terminus |
| a | SLNPNR | Cleaved AT, C-terminal fragment |
| b | SLNPNR | Cleaved AT, C-terminal fragment |
| b | HGSPVD | AT, N terminus |
| b | GQPSV | Thrombin, H chain, Lys149 cut |
| b | VTGWGN* | Thrombin, H chain, Arg137 cut |
| c | IVEGSD | Thrombin, H chain, N terminus |
| c | TFGSGE | Thrombin, L chain, N terminus |
| d | IVEGSD | Thrombin, H chain, N terminus |
| e | IVEGSD | Thrombin, H chain, N terminus |
| e | GQPSV | Thrombin, H chain, Lys149 cut |
| f | YNWREN* | Thrombin, H chain, Arg93 cut |
| f | TFGSG | Thrombin, L chain, N terminus |
| f | KSPQEL* | Thrombin, H chain, Arg35 cut |
| g | YNWREN* | Thrombin, H chain, Arg93 cut |
| g | TFGSGE | Thrombin, L chain, N terminus |
| h | IGKHSR* | Thrombin, H chain, Arg67 cut |
| h | TRYER* | Thrombin, H chain, Arg73 cut |
| h | HSRTR* | Thrombin, H chain, Lys69 cut |
| i | TRYER* | Thrombin, H chain, Arg73 cut |
| i | HSRTRY* | Thrombin, H chain, Lys69 cut |
| i | IGKHSR* | Thrombin, H chain, Arg67 cut |
FIGURE 4.
Thrombin exosites I and II are susceptible to autolysis when in complex with AT. A, incubation of the thrombin-AT complex with excess thrombin leads to several degradation products (denoted a–i), as observed by SDS-PAGE. Lanes 1 and 2 are thrombin and AT alone without incubation; lanes 3 and 4 are thrombin alone and thrombin-AT complex (0.23 nmol of thrombin, 0.13 nmol of AT) incubated at 37 °C for 2 h. Bands a–i were subjected to N-terminal sequencing. MW, molecular weight. B, time course of thrombin digestion of the thrombin-AT complex (same ratio as in A) at 37 °C. C, stereo view of the heavy chain of thrombin in standard orientation, colored from N to C termini (blue-to-red), with identified cleavage sites indicated by balls. Magenta balls are the additional sites cleaved in the α1ATpitts-thrombin and PN1-thrombin complexes.
Exosite Ligands Protect Thrombin from Autolysis in Serpin Complexes
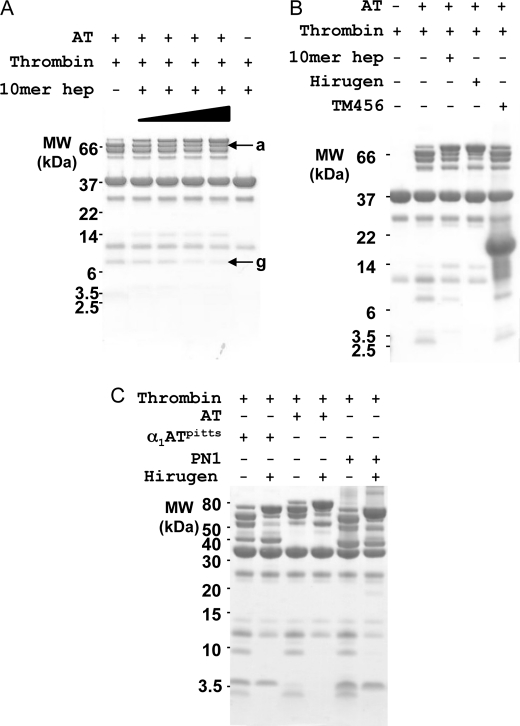
Because thrombin has reduced affinity for heparin due to the disordering of exosite II in complex with serpins, we hypothesized that a large excess of heparin would stabilize the exosite and protect it from autolysis. Indeed, as shown in Fig. 5A, increasing the concentration of 10-mer heparin (final, 1 mm) reduced the exosite II cleavage products (bands a and g) and consequently increased the amount of intact complex. We then tested two exosite I ligands of thrombin, the hirugen peptide and TM456 (EGF domains 4–6 of TM at final concentration of 0.5 mm, Fig. 5B), to see whether they could similarly protect thrombin from autolysis at exosite I. The hirugen peptide protected thrombin even more effectively than heparin, but TM456 had no discernable effect. This is unlikely to be due to steric hindrance from the serpin, because thrombin can cleave at exosite I, and is probably due to the greater sensitivity of TM binding to the conformation of exosite I. Similarly, hirugen binds to the partially unfolded pro-exosite I of prothrombin, whereas TM (despite its 1000-fold higher affinity for thrombin) is unable to bind to the pro-exosite (6, 7).
FIGURE 5.
Exosite ligands protect thrombin-serpin complexes from autolysis. A, cleavage at exosite II (bands a and g) of thrombin in complex with AT is decreased with increasing concentration of 10-mer heparin (10mer hep, from 0.4 to 1 mm, indicated by ramp). MW, molecular weight. B, heparin and hirugen confer protection from proteolysis, but TM456 does not. C, all tested thrombin-serpin complexes (α1ATpitts, AT, and PN1) result in similar cleavage patterns on SDS-PAGE and are similarly protected from limited proteolysis by hirugen.
Finally, to determine whether the degree of proteolytic susceptibility depends on the serpin used, we tested other thrombin-specific serpins. As for AT, thrombin complexation by α1ATpitts, PN1, or protein C inhibitor (not shown) in the presence of excess thrombin resulted in similar cleavage patterns on SDS-PAGE (Fig. 5C). In addition, cleavage of the final complex was reduced by adding hirugen, as for AT. N-terminal sequencing of the thrombin-α1ATpitts and thrombin-PN1 complex cleavage products confirmed cleavage at all of the sites found with AT (listed in Table 2) and two additional sites, Arg175 and Lys202 (Fig. 4C, magenta balls).
DISCUSSION
Using specific exosite II probes and limited proteolysis, we have demonstrated that exosite II of thrombin partially unfolds in response to complexation with serpins. However, as with previous studies that showed a loss of exosite I binding (16, 17), quantification of the magnitude of loss of affinity proved problematic. It is possible to interpret the abrogation in quench of the fluorescein-γ′-peptide when thrombin is complexed by serpins either as reflecting a lack of binding (under the conditions used) or as a change in the fluorescence properties of the probe when bound to complexed thrombin. However, only the former interpretation is consistent with the other experiments that demonstrate a reduction in binding to heparin-Sepharose and the proteolytic susceptibility of the principal exosite II residue Arg93. Although we were not successful in quantifying the affinity of serpin-complexed thrombin for the γ′-peptide by direct measurement, we were able to concentrate the R93E variant sufficiently to conduct titrations and determine a Kd. Charge reversal at Arg93 resulted in an ∼80-fold reduction in affinity for the γ′-peptide. This corresponded to a reduction in peak conductivity for elution from heparin-Sepharose from 60.1 to 39.4 mS/cm, identical to the elution conductivity for the recombinant thrombin-α1ATpitts complex (39.7 mS/cm). We are thus able to conclude that the reduction in affinity for exosite II ligands when thrombin is complexed by serpins is approximately 2 orders of magnitude.
Two previous studies showed a loss of exosite I binding for thrombin-serpin complexes, using various probes, including fluorescently labeled hirugen (16, 17). The integrity of exosite II was probed in one of the studies (17), and the authors concluded that exosite II remained intact based on modest reductions in apparent binding to an aptamer and to heparin-Sepharose. In light of the current data, it is necessary to review the previous work in some detail. Binding of thrombin to a DNA aptamer was monitored by fluorescence. Briefly, they reacted free amines (Lys residues) on prothrombin with fluorescein isothiocyanate, converted the labeled material to thrombin, and followed fluorescence change upon titration of the DNA aptamer HD-22. The thrombin used in this study was likely a mixture of species with a distribution of Lys residues labeled with the negatively charged probe fluorescein. When HD-22 was titrated into the fluorescein-thrombin solution, a fluorescence quench of ∼7% was registered, providing a Kd of 230 nm. This value is about 500 times the reported Kd for unlabeled thrombin, indicating that labeling of the surface Lys residues affected the interaction significantly. When the aptamer was titrated into a solution containing fluorescein-thrombin-serpin complexes, they recorded 2–4% fluorescence quenches but similar Kd values. On this basis, they concluded that exosite II binding was unaffected. However, complexes were formed by incubating fluorescein-thrombin with serpins at a 2.5–10-fold molar excess for 15 min. It is likely that the observed signal reflects the fraction of thrombin that was not inhibited by the serpins. Assuming unaltered rates of inhibition and a 10-fold excess for all serpins, HCII would only have inhibited half of the thrombin under the conditions used, and AT would have inhibited 80%. However, we know that altering the surface properties of thrombin by modifying Lys residues is likely to slow the rate of inhibition. This is particularly relevant for HCII because its inhibition of thrombin requires a functional exosite I. In addition, the contention that HD-22 is an exosite II-directed aptamer (31) is at best overstated because it fully inhibits the ability of thrombin to clot fibrinogen. In addition, HD-22 is a derivative of a 15-mer aptamer that similarly blocked fibrinogen cleavage and whose crystal structure with thrombin showed principal exosite I binding and a crystal contact with exosite II (32). Furthermore, the heparin-Sepharose elution profiles reported for thrombin, alone and in complexes with AT, HCII, and α1ATpitts (17), are quite similar to what we observe in this study and suggest a large reduction in heparin binding affinity. They reported that thrombin eluted at 450 mm NaCl, and the complexes with AT, HCII, and α1ATpitts eluted at 340, 340, and 250 mm. The high values with AT and HCII are caused by the ability of these serpins to bind to heparin, and the 1.8-fold decrease in NaCl concentration reported for the complex with α1ATpitts is remarkably similar to the 1.9-fold decrease found here (590–310 mm NaCl).
Thus, it is clear from this and previous studies that thrombin is globally disordered when complexed by serpins and that the loss of structure is manifested by a marked reduction in ability to bind to substrates and cofactors via exosites I and II. This explains to a large extent the reason why the serpin mechanism is uniquely suited to physiological thrombin inhibition and can be understood as a functional reversal of the formation/exposure of exosites upon activation of prothrombin. The active site and exosites on prothrombin must be inaccessible to prevent prothrombin from swamping the exposed TM and heparan sulfate on the intact vascular wall and preloading fibrinogen and platelet receptors. Freely mobile prothrombin must remain at a high concentration in the circulation to be delivered rapidly and in large amounts to the site of clot formation, and exposure of the exosites at the site of action ensures that the activity of thrombin will be directed by competition for those sites (33). It is similarly critical that the active sites of thrombin and exosites are compromised when inhibited by serpins to allow extraction of the inhibited thrombin from its bound substrates and cofactors and to permit free circulation of the complex to the liver for clearance. As a necessary consequence of the serpin mechanism, the zymogen activation contact between Ile16 and Asp194 is reversed, resulting in the disordering of the oxyanion hole, and in thrombin, the loss of functional exosite I. In the crystal structure of the α1AT-trypsin complex, the region corresponding to exosite I of thrombin was entirely missing from electron density due to flexibility. Here we show that exosite II is also severely compromised when thrombin forms a complex with serpins. This is not predicted from the structure of the serpin-protease complex but makes good biological sense; it permits the unobstructed transport of the complexes to the liver, where the unfolded portion of the protease may contribute to the binding of clearance receptors.
This work was supported, in whole or in part, by National Institutes of Health Grant HL68629 (J. A. H.) and by grants from the British Heart Foundation (to J. A. H.) and the Medical Research Council (J. A. H.).
- TM
- thrombomodulin
- TM456
- thrombomodulin epidermal growth factor domains 4, 5, and 6
- AT
- antithrombin
- α1ATpitts
- α1-antitrypsin Pittsburgh mutant
- PN1
- protease nexin 1
- HCII
- heparin cofactor II
- PPACK
- d-Phe-Pro-Arg-chloromethyl ketone
- Fmoc
- fluorenylmethyloxycarbonyl
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- mS
- millisiemens.
REFERENCES
- 1.Bianchini E. P., Orcutt S. J., Panizzi P., Bock P. E., Krishnaswamy S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10099–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode W., Mayr I., Baumann U., Huber R., Stone S. R., Hofsteenge J. (1989) EMBO J. 8, 3467–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehlhammer H., Bode W., Huber R. (1977) J. Mol. Biol. 111, 415–438 [DOI] [PubMed] [Google Scholar]
- 4.Huntington J. A. (2005) J. Thromb. Haemost. 3, 1861–1872 [DOI] [PubMed] [Google Scholar]
- 5.Kroh H. K., Tans G., Nicolaes G. A., Rosing J., Bock P. E. (2007) J. Biol. Chem. 282, 16095–16104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L. W., Ye J., Johnson A. E., Esmon C. T. (1991) J. Biol. Chem. 266, 23633–23636 [PubMed] [Google Scholar]
- 7.Wu Q., Picard V., Aiach M., Sadler J. E. (1994) J. Biol. Chem. 269, 3725–3730 [PubMed] [Google Scholar]
- 8.Arni R. K., Padmanabhan K., Padmanabhan K. P., Wu T. P., Tulinsky A. (1993) Biochemistry 32, 4727–4737 [DOI] [PubMed] [Google Scholar]
- 9.Rau J. C., Beaulieu L. M., Huntington J. A., Church F. C. (2007) J. Thromb. Haemost. 5, Suppl. 1, 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huntington J. A. (2006) Trends Biochem. Sci 31, 427–435 [DOI] [PubMed] [Google Scholar]
- 11.Gettins P. G. (2002) Chem. Rev. 102, 4751–4804 [DOI] [PubMed] [Google Scholar]
- 12.Huntington J. A., Read R. J., Carrell R. W. (2000) Nature 407, 923–926 [DOI] [PubMed] [Google Scholar]
- 13.Dementiev A., Dobó J., Gettins P. G. (2006) J. Biol. Chem. 281, 3452–3457 [DOI] [PubMed] [Google Scholar]
- 14.Peterson F. C., Gettins P. G. (2001) Biochemistry 40, 6284–6292 [DOI] [PubMed] [Google Scholar]
- 15.Calugaru S. V., Swanson R., Olson S. T. (2001) J. Biol. Chem. 276, 32446–32455 [DOI] [PubMed] [Google Scholar]
- 16.Bock P. E., Olson S. T., Björk I. (1997) J. Biol. Chem. 272, 19837–19845 [DOI] [PubMed] [Google Scholar]
- 17.Fredenburgh J. C., Stafford A. R., Weitz J. I. (2001) J. Biol. Chem. 276, 44828–44834 [DOI] [PubMed] [Google Scholar]
- 18.Stavridi E. S., O'Malley K., Lukacs C. M., Moore W. T., Lambris J. D., Christianson D. W., Rubin H., Cooperman B. S. (1996) Biochemistry 35, 10608–10615 [DOI] [PubMed] [Google Scholar]
- 19.Kaslik G., Patthy A., Bálint M., Gráf L. (1995) FEBS Lett. 370, 179–183 [DOI] [PubMed] [Google Scholar]
- 20.Egelund R., Petersen T. E., Andreasen P. A. (2001) Eur. J. Biochem. 268, 673–685 [DOI] [PubMed] [Google Scholar]
- 21.Strickland D. K., Ranganathan S. (2003) J. Thromb. Haemost. 1, 1663–1670 [DOI] [PubMed] [Google Scholar]
- 22.Johnson D. J., Adams T. E., Li W., Huntington J. A. (2005) Biochem. J. 392, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields G. B., Noble R. L. (1990) Int. J. Pept. Protein Res. 35, 161–214 [DOI] [PubMed] [Google Scholar]
- 24.Zhou A., Carrell R. W., Huntington J. A. (2001) J. Biol. Chem. 276, 27541–27547 [DOI] [PubMed] [Google Scholar]
- 25.Lovely R. S., Moaddel M., Farrell D. H. (2003) J. Thromb. Haemost. 1, 124–131 [DOI] [PubMed] [Google Scholar]
- 26.Pineda A. O., Chen Z. W., Marino F., Mathews F. S., Mosesson M. W., Di Cera E. (2007) Biophys. Chem. 125, 556–559 [DOI] [PubMed] [Google Scholar]
- 27.Olson S. T., Halvorson H. R., Björk I. (1991) J. Biol. Chem. 266, 6342–6352 [PubMed] [Google Scholar]
- 28.Carter W. J., Cama E., Huntington J. A. (2005) J. Biol. Chem. 280, 2745–2749 [DOI] [PubMed] [Google Scholar]
- 29.He X., Ye J., Esmon C. T., Rezaie A. R. (1997) Biochemistry 36, 8969–8976 [DOI] [PubMed] [Google Scholar]
- 30.Sheehan J. P., Sadler J. E. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5518–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tasset D. M., Kubik M. F., Steiner W. (1997) J. Mol. Biol. 272, 688–698 [DOI] [PubMed] [Google Scholar]
- 32.Padmanabhan K., Padmanabhan K. P., Ferrara J. D., Sadler J. E., Tulinsky A. (1993) J. Biol. Chem. 268, 17651–17654 [DOI] [PubMed] [Google Scholar]
- 33.Lane D. A., Philippou H., Huntington J. A. (2005) Blood 106, 2605–2612 [DOI] [PubMed] [Google Scholar]