Abstract
Protein-protein interfaces are usually large and complementary surfaces, but specific side chains, representing energetic “hot spots,” often contribute disproportionately to binding free energy. We used a computational method, comprehensive interface design, to identify hot spots in the interface between the stalk regions of the β3 and the complementary αIIb and αv integrin subunits. Using the Rosetta alanine-scanning and design algorithms to predict destabilizing, stabilizing, and neutral mutations in the β3 region extending from residues Lys532 through Gly690, we predicted eight alanine mutations that would destabilize the αIIbβ3 interface as well as nine predicted to destabilize the αvβ3 interface, by at least 0.3 kcal/mol. The mutations were widely and unevenly distributed, with four between residues 552 and 563 and five between 590 and 610, but none between 565 and 589, and 611 and 655. Further, mutations destabilizing the αvβ3 and αIIbβ3 interfaces were not identical. The predictions were then tested by introducing selected mutations into the full-length integrins expressed in Chinese hamster ovary cells. Five mutations predicted to destabilize αIIb and β3 caused fibrinogen binding to αIIbβ3, whereas three of four predicted to be neutral or stabilizing did not. Conversely, a mutation predicted to destabilize αvβ3, but not αIIbβ3 (D552A), caused osteopontin binding to αvβ3, but not fibrinogen binding to αIIbβ3. These results indicate that stability of the distal stalk interface is involved in constraining integrins in stable, inactive conformations. Further, they demonstrate the ability of comprehensive interface design to identify functionally significant integrin mutations.
Keywords: Computation, Glycoprotein Structure, Integrin, Ligand-binding Protein, Platelet
Introduction
Integrins are transmembrane (TM)4 heterodimers that transmit signals bidirectionally across cell membranes and reside on cell surfaces in a finely tuned equilibrium between resting low affinity and active high affinity conformations (1). Thus, binding of ligands to integrin extracellular domains initiates intracellular signaling pathways (“outside-in signaling”), whereas intracellular signaling increases the affinity of integrin extracellular domains for ligands (“inside-out signaling”). Integrin-mediated bidirectional signaling has been particularly well studied for the integrin αIIbβ3 and the homologous integrin αvβ3 because of the role these integrins play in hemostasis, thrombosis, and angiogenesis (2).
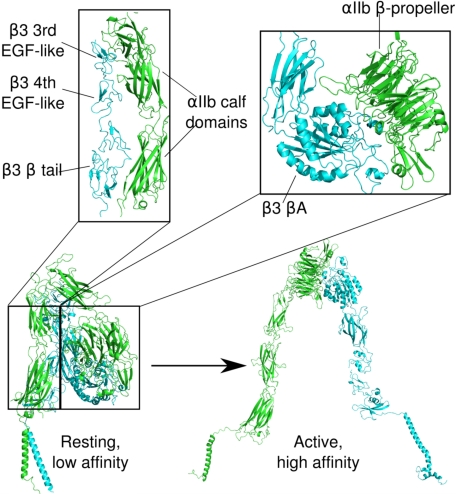
Electron microscope images of rotary-shadowed αIIbβ3 depict an extended heterodimer consisting of an 8 × 12-nm nodular head containing its ligand-binding site and two 18-nm flexible stalks containing its TM and cytoplasmic domains (3). However, crystal structures of the extracellular portions of inactive αIIbβ3 (4) and αvβ3 (5) revealed that the molecules in the crystals were severely bent with their ligand-binding site on the nodular head oriented toward the cell surface. Thus, large conformational changes appear to occur upon αIIbβ3 and αvβ3 activation (6). A preponderance of evidence suggests that these changes result in a shift from the bent to a fully extended conformation (7). Other results, however, suggest that bent integrins are fully capable of binding ligands (8). Regardless, physiologic αIIbβ3 activation is initiated when the cytoplasmic proteins talin and kindlin-3 bind to the β3 cytoplasmic tail (9) and is thought to be followed by cytoplasmic and TM domain separation, as well as separation of the membrane-proximal portions of the α and β subunit extracellular stalks (4, 6) (Fig. 1). This suggests that an extended interface involving portions of the juxtamembrane cytoplasmic, TM, and extracellular stalk domains constrains αIIbβ3 in its inactive conformation.
FIGURE 1.
Models of the transition of αIIbβ3 from a resting, low affinity to an active, high affinity conformation that illustrate the presence of contacts between the distal αIIb and β3 stalks in the resting conformation. The TM domains are the helices located at the bottom of the figure. αIIb is shown in green, β3 in cyan. The models are based on an extensive body of evidence that activation converts αIIbβ3 from a bent to an extended molecule (4, 6), including our own rotary-shadowed electron microscope images of αIIbβ3 bound to fibrinogen in which αIIbβ3 is fully extended with separated stalks (3, 6). Nonetheless, there is also evidence that integrins in the bent conformation are able to interact with ligands (8).
Site-directed mutagenesis has allowed the identification of interactions that maintain integrins in the resting state. Activating mutations are widely distributed throughout the proteins and result in remodeling or dissociation of subunit-subunit interactions spanning from the extracellular, ligand-binding domain to the cytoplasmic domains. As a result, mutations that disrupt subunit-subunit and interdomain interactions of the resting state appear to activate the protein by destabilizing the default resting state. For example, disruption of intersubunit interactions near the juxtamembrane cytoplasmic domains (10, 11) and the TM (12, 13) domains leads to integrin activation. Similarly, disruption of interactions between the head and stalk domains that appear to stabilize the bent conformation also causes integrin activation (14, 15).
What has been less clear is the extent to which dissociation of the extracellular stalk domains is needed for integrin activation. Experiments in which disulfides and/or coiled coils have been used to hold the C-terminal ends of the extracellular stalks in proximity lead to inactive integrins (6, 16), although it is not entirely clear whether the lack of activity reflects mere proximity effects or constraining the integrin to a nonnatively cross-linked conformation. In this case, a complementary strategy to destabilize the intersubunit stalk interface should lead to activation rather than inactivation.
Point mutations often provide substantial insight into the folding, association, and function of proteins (17, 18). However, it is often not clear in advance which mutations will be most informative. A potential solution for this problem is to use computational methods that predict the energetic effect of mutations (19). If such methods are sufficiently robust, stabilizing or destabilizing mutants could be selected computationally when a structure is known, and experiments could focus on determining the biological effects of mutations, rather than iteratively searching for informative ones. In particular, with a comprehensive set of accurate predictions, not only disruptive, but also stabilizing mutations could be introduced into proteins and the biological effects studied.
To test whether destabilizing the intersubunit stalk interface results in integrin activation, we used the Rosetta energy functions to predict mutations in distinct regions of the β3 stalk that would destabilize, stabilize, or have little effect on its interactions with αIIb or αv (20, 21). Using these computational methods, we identified energetic “hot spots” that contribute disproportionately to the binding free energy between the distal β3 stalk and αIIb and αv. We then tested whether mutating these hot spots would result in integrin activation by measuring the activation state of full-length integrins containing mutated hot spots expressed in CHO cells. The results of these experiments demonstrate that maintaining the interface between the membrane-proximal region of the β3 stalk with αIIb and αv is critical for maintaining the resting, low affinity state of αIIbβ3 and αvβ3. Further, they suggest that small molecules designed to stabilize this extracellular interface could be useful inhibitors of integrin activation.
EXPERIMENTAL PROCEDURES
Computational Design
Predicted disruptive mutations were identified using the interface alanine-scanning Rosetta server (20, 21). For the structure of αIIbβ3 (4), chains A and B were selected for the calculation because these chains are more complete than chains C and D. For the structure of αvβ3, the original structure (5) was used. Residues in the stalk where alanine substitutions were predicted to destabilize both interfaces by greater than 0.3 kcal/mol were selected for mutagenesis.
Other calculations were performed using the Rosetta molecular modeling program (22). The arguments interface and pmut_scan were used for all calculations. Following Sammond et al. (23), for dampened repulsion energies, the soft_rep_design was used (24). Calculations that allowed side chains to repack also included the argument repack_neighbors.
Stable Expression of αIIbβ3 in Chinese Hamster Ovary (CHO) Cells
Full-length cDNAs for human αIIb and β3 were inserted into pcDNA3.1(+) Neo and pcDNA3.1(+) Zeo (Invitrogen), respectively. Single amino acid substitutions were introduced into the β3 cDNA using the QuikChange Site-directed Mutagenesis kit (Stratagene). All mutated sequences were confirmed by DNA sequence analysis.
CHO cells were cultured in Ham's F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone Laboratories). Plasmids containing cDNAs for αIIb and β3 were introduced into CHO cells using FuGENE 6 according to the manufacturer's instructions (Roche Applied Science). Two days after transfection, cells were transferred to selection medium containing 500 μg/ml G418 (Invitrogen) and 300 μg/ml Zeocin (Invitrogen). After 3 weeks of selection, αIIbβ3 expression was assessed by flow cytometry using A2A9, a monoclonal antibody (mAb) that recognizes the human αIIbβ3 heterodimer. Transfected cells were sorted by fluorescence-activated cell sorting (FACS) to obtain cell lines expressing high levels of αIIbβ3. CHO cells also express endogenous hamster αv that can form αvβ3 heterodimers with transfected human β3. The amount of αvβ3 on the transfected cells was assessed by flow cytometry using the αvβ3-specific monoclonal antibody LM609 (Chemicon International) (supplemental Fig. 1).
Fibrinogen Binding to CHO Cells Expressing αIIbβ3
Fibrinogen binding to αIIbβ3 expressed on the CHO cell surface was measured as described previously (25). Briefly, CHO cells (2 × 106 cells/ml) were incubated with the β3-specific mAb SSA6 on ice for 30 min. Labeled cells were washed and incubated for 30 min at 37 °C with phycoerythrin-conjugated anti-mouse IgG (Molecular Probes), 200 μg/ml fibrinogen conjugated with Alexa Fluor 488 (Molecular Probes), and freshly made 5 mm dithiothreitol (DTT) ± 5 mm EDTA. The cells were washed, fixed with 0.37% formalin in phosphate-buffered saline (PBS), and examined by two-color FACS analysis. Fibrinogen specifically bound to αIIbβ3 was defined as fibrinogen binding inhibited by EDTA.
Specific fibrinogen binding data were used to calculate a fibrinogen binding index (Fi), using the equation
where FBcmut represents constitutive fibrinogen binding to αIIbβ3; FBDTTmut, fibrinogen binding to mutant αIIbβ3 induced by 5 mm DTT; FBcWT, constitutive fibrinogen binding to wild-type (WT) αIIbβ3; and FBDTTWT, fibrinogen binding to WT αIIbβ3 induced by 5 mm DTT.
Rupture Force Spectroscopy (Laser Tweezers)
A custom-built laser tweezers apparatus assembled from a Nikon Diaphot 300 inverted microscope, 100 × 1.3 NA Fluor lens, and a Spectra Physics FCBar Nd:YAG laser was used to measure the strength of osteopontin (OPN) binding to CHO cells, as described previously for fibrinogen binding to platelets and CHO cells and OPN binding to platelets (26–28). Briefly, recombinant human OPN (R & D Systems) was covalently bound to 1.87-μm carboxylate-modified latex beads. Transfected CHO cells were permitted to settle and attach spontaneously to the polylysine-coated bottom of the chamber. An OPN-coated bead, trapped by the laser light, was brought into proximity of an immobilized cell, oscillated at 10 Hz with a 0.8-μm peak-to-peak amplitude, and then brought into repeated intermittent contact with the cell by micromanipulation. Data collection was initiated at the first contact. Rupture forces following repeated contacts were collected into 5-pN-wide bins. The percentage of events in a particular bin (frequency, %) represents the probability of rupture events at that tension. Optical artifacts observed with or without trapped latex beads produced signals that appear as forces below 10 pN; rupture forces in this range were not considered when data were analyzed.
RESULTS
Comprehensive Interface Design
Although protein-protein interfaces are usually composed of relatively large and complementary surfaces, specific side chains in the interface often contribute disproportionately to the binding free energy (18, 29). Thus, despite the presence of crystal and solution structures for these interfaces, the residues responsible for the binding affinity may not be apparent, and extensive mutagenesis is commonly required to identify these energetic hot spots. However, computational algorithms have reached the point where destabilizing (20, 21) and stabilizing mutations (23) can be successfully predicted, in many cases obviating the need for time-consuming and expensive experimental effort. Here, we have employed a combined strategy, which we term “comprehensive interface design,” to select a set of destabilizing, stabilizing, and neutral mutations that characterize the extracellular stalk interfaces of the integrins αIIbβ3 and αvβ3.
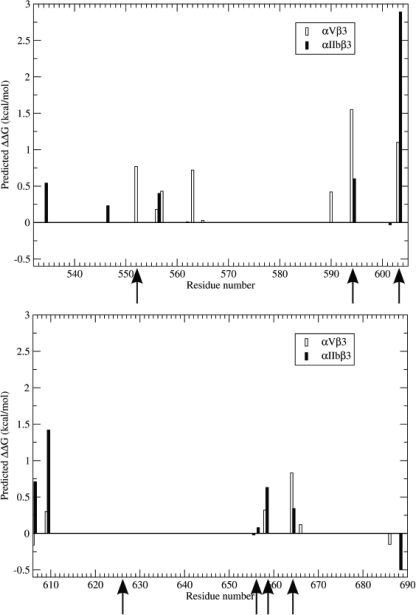
Structures for the αIIbβ3 and αvβ3 stalk interfaces are available (4, 5). Consequently, we inputted these structures, extending from β3 residues Lys532 through Gly690, into the Rosetta alanine-scanning algorithm to identify alanine substitutions that would be predicted to destabilize the αIIbβ3 and αvβ3 extracellular stalk interfaces (20, 21). This algorithm mutates every residue in a protein-protein interface to alanine and reports a predicted change in binding free energy, optimized to reproduce experimental changes in free energy of binding observed for alanine mutants. Using this algorithm, we identified eight alanine replacements predicted to destabilize the αIIbβ3 interface, as well as nine replacements predicted to destabilize the αvβ3 interface, by at least 0.3 kcal/mol (Table 1; Figs. 2 and 3). It is noteworthy that these replacements were widely but unevenly distributed along the β3 sequence examined. Thus, there were four destabilizing replacements between residues 552 and 563 and five between residues 590 and 609, but none between residues 565 and 589 and residues 610 and 655. Moreover, there were often notable differences in the effects on the free energy of β3 binding to αv and αIIb. For example, although Asp552 is present in the vicinity of the interface of β3 with the αIIb and αv β propeller domains, D552A was predicted to destabilize the interaction of β3 with αv by 0.77 kcal/mol, but have no effect on its interaction with αIIb. Conversely, a T609A replacement was predicted to destabilize β3 binding to αIIb by 1.4 kcal/mol, but only destabilize its binding to αv by 0.3 kcal/mol.
TABLE 1.
β3 Alanine replacements predicted to destabilize β3 binding to αIIb and αv by ≥0.3 kcal/mol
| αIIb | αv |
|---|---|
| E534 | Q552 |
| Y556 | Y557 |
| Y594 | R563 |
| T603 | Q590 |
| D606 | Y594 |
| T609 | T603 |
| K658 | T609 |
| V664 | K658 |
| V664 |
FIGURE 2.
Calculated effects of replacing β3 residues Lys532–Gly690 individually with alanine on the binding free energy (ΔG) of the β3 stalk with the stalks of αIIb (filled bars) and αv (open bars). Arrows indicate the locations of β3 mutations whose effect on αIIbβ3 and αvβ3 function was subsequently studied.
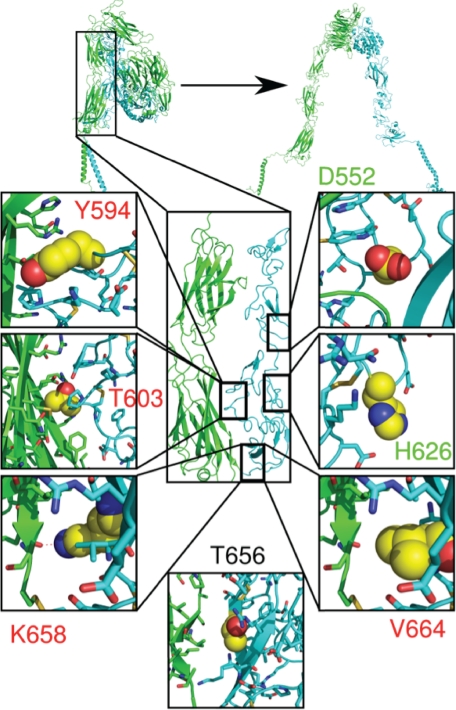
FIGURE 3.
Location of the designed β3 mutations in the interface of the αIIb and β3 stalks. The seven mutated β3 residues are shown in the crystal structure reported by Zhu et al. (4). The αIIb stalk is shown in green and the β3 stalk in cyan. The boxes in the figure show the side chains of the mutated WT residues as yellow spheres. Alanine mutations predicted to be destabilizing and neutral are lettered in red and green. A residue predicted to be stabilizing when mutated to tryptophan is indicated by black lettering.
We then employed three strategies to select negative control mutations. First, we selected D552A as an alanine substitution predicted to destabilize the αvβ3, but not the αIIbβ3, interface. Second, we sought stabilizing mutations using a protocol previously shown to be successful in detecting such mutations (23) and searched for a residue predicted to have no change on the binding free energy. H626A met this criterion as it was predicted to have no effect on the binding interface when the protein side chains were repacked.
In the third strategy, we used the Rosetta design program to predict additional stabilizing mutations. The protocol of Sammond et al. (23) uses dampening repulsive energies to identify mutations that increase hydrophobicity without requiring repacking of nearby side chains. Thus, we sought hydrophobic residues that could be mutated to larger hydrophobic residues or polar residues not involved in hydrogen bonds that could be mutated to hydrophobic residues. The most stabilizing mutation of a residue fitting these criteria was V664L. We also tried a modified protocol that allowed nearby protein side chains to repack, potentially identifying larger residues that could further stabilize the interface. Thus, the same Rosetta energy function was used, but repulsive forces were not dampened, and side chains were allowed to repack. The most stabilizing prediction changed hydrophobic Thr656 to Trp, a much larger residue that provides a greater buried hydrophobic surface area.
Effect of β3 Stalk Mutations on the αIIbβ3 Activity State
In most current models of integrin regulation, the resting low affinity state of αIIbβ3 has a large protein-protein interface, including an interface involving the extracellular stalks of αIIb and β3, whereas in the active high affinity state, the stalks are separated (6) (Fig. 1). If these models are accurate, then destabilizing the resting stalk interface should shift αIIbβ3 toward its high affinity state, which can be accessed experimentally by its ability to bind fibrinogen constitutively (13). To test this hypothesis, we selected nine computationally identified mutations: five predicted to be destabilizing, two predicted to be neutral, and two predicted to be stabilizing (Table 2) and introduced them into WT β3 by site-directed mutagenesis. The mutants were co-expressed with WT αIIb in CHO cells, after which unstimulated and DTT-stimulated fibrinogen binding to the recombinant αIIbβ3 was measured by flow cytometry. The resulting fibrinogen-binding measurements were then expressed as a fibrinogen binding index which compares the magnitude of constitutive fibrinogen binding to the mutant αIIbβ3 with that to WT αIIbβ3.
TABLE 2.
Predicted consequences of β3 stalk mutations
βTD, β-tail domain.
| β3 mutant | Location | Predicted ΔΔG | Classification |
|---|---|---|---|
| kcal/mol | |||
| Y594A | EGF-4 | 0.60 | Destabilizing |
| T603A | βTD | 2.89 | Destabilizing |
| K658A | βTD | 0.63 | Destabilizing |
| V664A | βTD | 0.34 | Destabilizing |
| V664K | βTD | Destabilizing | |
| D552A | EGF-3 | 0 | Neutral |
| H626A | βTD | 0 | Neutral |
| V664L | βTD | −0.7 | Stabilizing |
| T656W | βTD | −1.5 | Stabilizing |
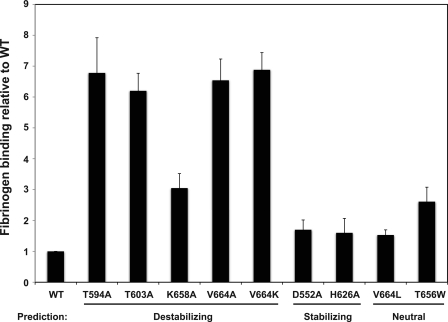
The four alanine mutations in β3 predicted to destabilize the stalk interface caused αIIbβ3 activation when introduced into the full-length integrin, although the alanine-scanning algorithm did not correctly rank order the mutation effects (Fig. 4). Thus, although the mutation predicted to be most destabilizing, T603A with a predicted ΔΔG of 2.89 kcal/mol, caused substantial αIIbβ3 activation, two other alanine substitutions, Y594A and V664A, induced nearly identical fibrinogen binding, but with ΔΔG values of 0.60 and 0.34 kcal/mol, respectively (Table 1).
FIGURE 4.
Fibrinogen binding to CHO cells expressing αIIbβ3 containing β3 stalk mutations. Alexa Fluor 488 fibrinogen binding to αIIbβ3 was measured by flow cytometry in the absence (constitutive binding) or presence (stimulated binding) of 5 mm DTT. Fibrinogen-binding measurements were used to calculate a fibrinogen-binding index relative to WT for each mutation, as described under “Experimental Procedures.” Data shown are the mean ± S.E. (error bars) of three to six measurements.
Val664 is of particular interest because although V664A was correctly predicted to destabilize the stalk interface (Fig. 2), a V664L mutation was predicted to be stabilizing. To clarify these results, we replaced Val664 with positively charged Lys and found that the mutation caused constitutive fibrinogen binding to an extent that was similar to V664A, implying that this mutation disrupted the stalk interface as well. Taken together, the results indicate that residue 664 is a hot spot at which the large hydrophobic side chains of Val or Leu contribute significantly to the binding free energy.
The alanine-scanning algorithm also accurately predicted neutral substitutions (D552A and H626A) that have no effect on αIIbβ3 activity. D552A is noteworthy because although it was predicted to have no effect on the free energy of β3 binding to αIIb, its ΔΔG for β3 binding to αv was 0.8 kcal/mol. However, the Rosetta design program was less successful in predicting stabilizing mutations. Although it accurately predicted that V664L would not activate αIIbβ3 (whereas both V664A and V664K are activating), it also predicted that T656W would be stabilizing. However, T656W induced fibrinogen binding to αIIbβ3 that was not statistically different from K658A (t test, p = 0.73), a mildly activation mutation.
Effect of β3 Stalk Mutations on OPN Binding to αvβ3
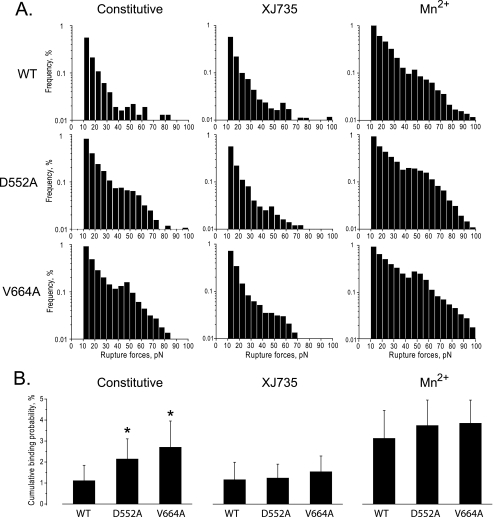
To verify that D552A, as well as V664A, destabilize αvβ3 (Fig. 2), we used laser tweezers-based force spectroscopy to detect the interaction of WT or mutant αvβ3 expressed by CHO cells (supplemental Fig. 1) with OPN, an αv-specific ligand. Previously, we used laser tweezers to measure rupture forces between OPN and single αvβ3 molecules expressed on the surface of human platelets (27). In that case, we found that the rupture forces could be segregated into two populations: a larger population with a rupture force range of 10–30 pN that resulted predominantly from the nonspecific interactions of the OPN-coated beads with the cell surface, and a smaller population with a rupture force range of 30–60 pN that resulted from specific interaction of OPN with active αvβ3 (27). As shown in Fig. 5A, the vast majority of the interactions between cells expressing WT αvβ3 and OPN had exponentially declining rupture forces <30 pN that were not inhibited by the αvβ3-specific inhibitory peptide XJ735, indicating that they were nonspecific. However, preincubating the cells with 1 mm MnCl2 resulted in a new broad peak of rupture forces with a maximum at ≈50 pN and that overlapped to a small extent with the lower rupture force range. This confirms that the WT αvβ3 expressed by the CHO cells was able to bind OPN when activated. By contrast, CHO cells expressing β3 D552A or β3 V664A exhibited the 50-pN rupture force peak in the absence of MnCl2. Further, these rupture forces were inhibited by XJ735, indicating that they resulted from the constitutive binding of OPN to active αvβ3. To confirm these results, we calculated the cumulative probability of β3 D552A and β3 V664A causing rupture forces >10 pN. As shown in Fig. 5B, both the D552A and V664A mutations significantly increased the probability of specific (i.e. XJ735-inhibited) interactions between OPN and αvβ3, although to a lesser extent than exposing the cells to MnCl2.
FIGURE 5.
Effect of β3 stalk mutations on OPN binding to αvβ3 as measured by laser tweezers-based rupture force spectroscopy. Rupture forces between latex beads coated with OPN and CHO cells expressing WT αvβ3 or αvβ3 containing the β3 mutations D552A and V664A were measured as described under “Experimental Procedures.” A, histograms showing the distribution of rupture forces between the OPN-coated beads and αvβ3-expressing CHO cells in the absence of an αvβ3 activator (Constitutive), in the presence of the αvβ3-specific antagonist 100 μm XJ735, and in the presence of the αvβ3 activator 1 mm Mn2+. Measured rupture forces were sorted into histograms and normalized by the total number of bead-cell contacts. The percentage of events in a particular force range (bin) represents the percent frequency of a bond rupture in that range. B, cumulative probabilities of detecting rupture forces >10 pN, normalized by the level of αvβ3 expression, between OPN-coated beads and αvβ3-expressing CHO cells under the conditions described in A. Data shown are the mean ± S.D. (error bars) of 15–40 individual experimental files. *, t tests for statistical significance: D552A versus WT, p = 0.02043; V664A versus WT, p = 0.0076.
DISCUSSION
Crystal structures of the bent extracellular domains of inactive αvβ3 and αIIbβ3 revealed contacts between the distal portion of the β3 stalk and the β-propeller, thigh, calf-1, and calf-2 domains of αv and αIIb (4, 5) (Fig. 1). To circumvent the expensive and time-consuming site-directed mutagenesis required to determine the physiologic significance of these contacts, we used computational protein design methods to identify interacting hot spots whose role in regulating β3 integrin function could be tested (20, 21). Recently, Luo et al. used a similar approach to successfully predict mutations that destabilize the closed conformation of the resting β3 headpiece and enhance αIIbβ3 ligand binding affinity (30). Using the Rosetta alanine-scanning program, we predicted four alanine replacements in the β3 stalk that would destabilize the αIIbβ3 interface and subsequently found that these replacements caused αIIbβ3 activation. Conversely, we identified two alanine replacements that were predicted to have no effect on β3 binding to αIIb and found that neither replacement altered the αIIbβ3 activation state. Thus, these results confirm the utility of the Rosetta alanine-scanning method for identifying functionally significant regions in integrins. Further, they indicate that stability of the distal stalk interface is involved in constraining integrins in stable, inactive conformations.
Although there is an extended interface between the distal extracellular portion of β3 and the α subunits of resting αvβ3 and αIIbβ3, we were able to pinpoint residues in predicted hot spots in the β3 βTD domain that contribute significantly to the interaction energy (Fig. 3). Previous reports, when taken together, corroborate our findings. Takagi et al. reported that acidic and basic coiled peptides, fused to the C termini of the α5β1 extracellular domain, maintained close apposition of the distal α5 and β1 stalks and prevented binding of a fibronectin fragment to the α5β1 headpiece, indicating that apposition of the distal stalks can regulate integrin activity (16). Concurrently, Zang and Springer found that β2 residues Gln525 and Val526, located in the third β2 EGF-like repeat and corresponding to β3 residues Glu534 and Met535, constrained human αXβ2 in an inactive state (31). It is noteworthy in this regard that replacing Glu534 with alanine is predicted to destabilize the αIIbβ3 interface. Kashiwagi et al. also reported an αIIbβ3 variant, T562N, that produced constitutively active αIIbβ3 in CHO cells (32). Nonetheless, as we have predicted, T562A was inactive, as was T562Q. Thus, the mechanism by which T562N activates αIIbβ3 is unclear. Lastly, Arnaout and co-workers have suggested that a loop extending from the βTD domain of β3 (the CD loop) prevents β3 integrin activation by interacting with and preventing downward movement of the α7 helix of the β-I domain (33). However, Zhu et al. either deleted the CD loop in αIIbβ3 (β3 residues 672–676) or replaced residues 672–674 individually with alanine and found, as we predicted, that none of the mutations had an effect on αIIbβ3 or αvβ3 activity (34).
Despite our ability to identify destabilizing and neutral mutations, we had less success using the Rosetta design program to predict stabilizing mutations. Although the program accurately predicted that V664L would not destabilize the αIIbβ3 interface, it predicted that T656W would be stabilizing. However, T656W caused fibrinogen binding to αIIbβ3 that was comparable with a mildly activating mutation, K658A. This finding is consistent with observations that Rosetta is only ∼50% successful at recovering known stabilizing mutations in protein/protein interfaces (23). Moreover, because it is likely more difficult to predict strongly stabilizing mutations than it is to predict destabilizing or neutral ones, our overall agreement between prediction and experiment is larger than might be expected, perhaps reflecting the fact that most of the mutations tested were predicted to be destabilizing rather than stabilizing. In the development of their method, Sammond et al. tested whether increased flexibility, such as repacking side chains, would increase their ability to find stabilizing mutations (23). Unexpectedly, they observed the opposite behavior: repacking side chains reduced their ability to predict stabilizing mutations. Our data support this conclusion as T656W, predicted to stabilize the interface if nearby side chains are allowed to repack, appears to mildly destabilize the interface instead.
Previous computational approaches to identify interface hot spots used a change in binding free energy of 1 kcal/mol to differentiate between neutral and destabilizing alanine substitutions (20, 21). However, we found that a change in predicted binding free energy of 0.3 kcal/mol was sufficient to identify alanine substitutions causing αIIbβ3 and/or αvβ3 activation. The substantially weaker affinity of the predicted β3 hot spots is likely a reflection of the physiologic function of β3 integrins. These integrins, αIIbβ3 in particular, are poised for activation because they must shift from their resting to their active conformations during the exceedingly short time they are exposed to damaged vasculature in flowing blood. Previously, using laser tweezers, we found that αIIbβ3 activation is an all-or-none phenomenon such that individual αIIbβ3 molecules are either in a resting or fully active conformation (35). Thus, any sufficient destabilizing event, regardless of its magnitude, should enable αIIbβ3 to bind fibrinogen. The similar levels of αIIbβ3 activation caused by Y594A, T603A, and V664A imply that these mutations are sufficiently destabilizing, whereas K658A is not. However, unlike mutations that remove hydrophobic interactions at the αIIbβ3 interface, Lys658 removes a side chain to a backbone hydrogen bond that may be alternately satisfied by water molecules. Thus, careful balancing of hydrophobic and hydrogen bonding energy terms will be an important consideration for improving predictive methods. It is also noteworthy that the amounts of fibrinogen binding caused by mutations disrupting the interaction of the β3 TM domain with the αIIb TM domain were comparable to the fibrinogen binding caused by Y594A, T603A, and V664A (25). This provides additional support for the all-or-none nature of αIIbβ3 activation.
In summary, our results confirm the utility of an integrated interface design strategy in identifying functionally significant integrin mutations. Further, they indicate that the stability of the distal stalk interface, like the interfaces of integrin TM and membrane-proximal cytoplasmic domains, is critical for maintaining integrins in stable, inactive conformations. Moreover, because the stalk interfaces of αIIbβ3 and αvβ3 are present in an extracellular location, they could be apt targets for the development of αIIbβ3 and αvβ3 activation inhibitors.
Supplementary Material
Acknowledgments
We thank Dr. Paul C. Billings for a review of the manuscript and Andrey Mekler for assistance with the laser tweezers experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants HL40387 and HL81012.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- TM
- transmembrane
- OPN
- osteopontin
- pN
- piconewtons.
REFERENCES
- 1.Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2.Shattil S. J., Newman P. J. (2004) Blood 104, 1606–1615 [DOI] [PubMed] [Google Scholar]
- 3.Weisel J. W., Nagaswami C., Vilaire G., Bennett J. S. (1992) J. Biol. Chem. 267, 16637–16643 [PubMed] [Google Scholar]
- 4.Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., Springer T. A. (2008) Mol. Cell 32, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., Arnaout M. A. (2001) Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 7.Luo B. H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnaout M. A., Mahalingam B., Xiong J. P. (2005) Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]
- 9.Moser M., Legate K. R., Zent R., Fässler R. (2009) Science 324, 895–899 [DOI] [PubMed] [Google Scholar]
- 10.Vinogradova O., Vaynberg J., Kong X., Haas T. A., Plow E. F., Qin J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4094–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinogradova O., Velyvis A., Velyviene A., Hu B., Haas T., Plow E., Qin J. (2002) Cell 110, 587–597 [DOI] [PubMed] [Google Scholar]
- 12.Li R., Mitra N., Gratkowski H., Vilaire G., Litvinov R., Nagasami C., Weisel J. W., Lear J. D., DeGrado W. F., Bennett J. S. (2003) Science 300, 795–798 [DOI] [PubMed] [Google Scholar]
- 13.Li W., Metcalf D. G., Gorelik R., Li R., Mitra N., Nanda V., Law P. B., Lear J. D., Degrado W. F., Bennett J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo B. H., Springer T. A., Takagi J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo B. H., Takagi J., Springer T. A. (2004) J. Biol. Chem. 279, 10215–10221 [DOI] [PubMed] [Google Scholar]
- 16.Takagi J., Erickson H. P., Springer T. A. (2001) Nat. Struct. Biol. 8, 412–416 [DOI] [PubMed] [Google Scholar]
- 17.DeLano W. L. (2002) Curr. Opin. Struct. Biol. 12, 14–20 [DOI] [PubMed] [Google Scholar]
- 18.Clackson T., Wells J. A. (1995) Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 19.Mandell D. J., Kortemme T. (2009) Nat. Chem. Biol. 5, 797–807 [DOI] [PubMed] [Google Scholar]
- 20.Kortemme T., Baker D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortemme T., Kim D. E., Baker D. (2004) Sci. STKE 2004, pl2. [DOI] [PubMed] [Google Scholar]
- 22.Rohl C. A., Strauss C. E., Misura K. M., Baker D. (2004) Methods Enzymol. 383, 66–93 [DOI] [PubMed] [Google Scholar]
- 23.Sammond D. W., Eletr Z. M., Purbeck C., Kimple R. J., Siderovski D. P., Kuhlman B. (2007) J. Mol. Biol. 371, 1392–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dantas G., Corrent C., Reichow S. L., Havranek J. J., Eletr Z. M., Isern N. G., Kuhlman B., Varani G., Merritt E. A., Baker D. (2007) J. Mol. Biol. 366, 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu H., Metcalf D. G., Streu C. N., Billings P. C., Degrado W. F., Bennett J. S. (2010) J. Mol. Biol. 401, 882–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litvinov R. I., Bennett J. S., Weisel J. W., Shuman H. (2005) Biophys. J. 89, 2824–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litvinov R. I., Vilaire G., Shuman H., Bennett J. S., Weisel J. W. (2003) J. Biol. Chem. 278, 51285–51290 [DOI] [PubMed] [Google Scholar]
- 28.Litvinov R. I., Vilaire G., Li W., DeGrado W. F., Weisel J. W., Bennett J. S. (2006) Biochemistry 45, 4957–4964 [DOI] [PubMed] [Google Scholar]
- 29.Bogan A. A., Thorn K. S. (1998) J. Mol. Biol. 280, 1–9 [DOI] [PubMed] [Google Scholar]
- 30.Luo B. H., Karanicolas J., Harmacek L. D., Baker D., Springer T. A. (2009) J. Biol. Chem. 284, 3917–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zang Q., Springer T. A. (2001) J. Biol. Chem. 276, 6922–6929 [DOI] [PubMed] [Google Scholar]
- 32.Kashiwagi H., Tomiyama Y., Tadokoro S., Honda S., Shiraga M., Mizutani H., Handa M., Kurata Y., Matsuzawa Y., Shattil S. J. (1999) Blood 93, 2559–2568 [PubMed] [Google Scholar]
- 33.Xiong J. P., Stehle T., Goodman S. L., Arnaout M. A. (2003) Blood 102, 1155–1159 [DOI] [PubMed] [Google Scholar]
- 34.Zhu J., Boylan B., Luo B. H., Newman P. J., Springer T. A. (2007) J. Biol. Chem. 282, 11914–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Litvinov R. I., Shuman H., Bennett J. S., Weisel J. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7426–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.