Abstract
Ca2+ influx by store-operated Ca2+ channels is a key component of the receptor-evoked Ca2+ signal. In all cells examined, transient receptor potential canonical (TRPC) channels mediate a significant portion of the receptor-stimulated Ca2+ influx. Recent studies have revealed how STIM1 activates TRPC1 in response to store depletion; however, the role of STIM1 in TRPC channel activation by receptor stimulation is not fully understood. Here, we established mutants of TRPC channels that could not be activated by STIM1 but were activated by the “charge-swap” mutant STIM1(K684E,K685E). Significantly, WT but not mutant TRPC channels were inhibited by scavenging STIM1 with Orai1(R91W), indicating the STIM1 dependence and independence of WT and mutant TRPC channels, respectively. Importantly, mutant TRPC channels were robustly activated by receptor stimulation. Moreover, STIM1 and STIM1(K684E,K685E) reciprocally affected receptor-activated WT and mutant TRPC channels. Together, these findings indicate that TRPC channels can function as STIM1-dependent and STIM1-independent channels, which increases the versatility of TRPC channel function and their role in receptor-stimulated Ca2+ influx.
Keywords: Calcium, Calcium Channels, G Protein-coupled Receptors (GPCRs), Signal Transduction, TRP Channels, STIM1, Store-dependent, TRPC Channels
Introduction
A central component of the agonist-evoked Ca2+ signal is Ca2+ influx across the plasma membrane that follows Ca2+ release from the endoplasmic reticulum (ER)3 (1). This Ca2+ influx is mediated by the store-operated Ca2+ influx channels (SOCs) (2). The molecular identity of the channels and their gating mechanism by the ER Ca2+ store have been elucidated with the discovery of STIM1 (3, 4) and the Orai channels (5–7).
STIM1 is an ER-resident multidomain Ca2+-binding protein that clusters at plasma membrane/ER junctions in response to Ca2+ release from the ER (3, 4), together with SOCs (2, 8–11). Several domains in STIM1 have been shown to participate in its function. An N-terminal Ca2+-binding EF hand keeps STIM1 in a non-clustered form when it is bound to Ca2+ (12); a cytoplasmic ERM domain mediates interaction with the transient receptor potential cation (TRPC) channels (10); a SOAR domain within the ERM domain gates the Orai channels (13–15); a stretch of seven negatively charged residues within the ERM domain participates in fast Ca2+-dependent inactivation (16–18); and a polybasic lysine-rich domain (K-domain) is required for the clustering of STIM1 at the plasma membrane (19) and the gating of TRPC channels (11).
Two types of channels, the Orai (7, 20) and TRPC channels (10, 11, 21), interact with and are regulated by STIM1. The Orai channels mediate the calcium release-activated calcium current, which is typified by high Ca2+ selectivity and strong inward rectification (1). STIM1 is obligatory for the Orai channels to function as channels (7, 20, 22). An important finding was that the 98-residue SOAR domain is sufficient to fully activate all Orai channels (13), indicating that no other STIM1 domain is essential for opening of these channels, including the polybasic K-domain. The severe combined immune deficiency-causing Orai1(R91W) mutation results in channel-dead Orai1 (5, 23). However, Orai1(R91W) interacts normally with STIM1 (23) and thus can be used as a STIM1 scavenger (24).
The gating of TRPC channels by STIM1 is more complex. The SOAR domain alone cannot activate TRPC1 (11). Although several TRPC channels are gated by STIM1 (21), TRPC1, TRPC4, and TRPC5 appear to directly interact with STIM1, whereas TRPC3 and TRPC6 do not (10, 21). However, in the absence of TRPC1 and TRPC4, TRPC3 and TRPC6 can be activated by receptor stimulation (21).
A key finding in the regulation of TRPC channels by STIM1 is that the STIM1 K-domain is essential for activation of the TRPC channels (11, 25). Moreover, STIM1 gates TRPC1 by electrostatic interaction between the negatively charged TRPC1 Asp-639 and Asp-640 residues and the positively charged STIM1 Lys-684 and Lys-685 residues (11). Hence, neutralizing or reversing the charges in TRPC1 or STIM1 results in channel-dead TRPC1 and dominant-negative STIM1. Significantly, complementary switching the charges in TRPC1 and STIM1 results in rescue of activation of TRPC1(D639K,D640K) by STIM1(K684E,K685E) (11). The negative charges are conserved in all TRPC channels (see Fig. 2). This allowed us to address several key questions in the regulation of TRPC channels by STIM1: (a) to obtain further evidence for the gating of TRPC channels by STIM1, (b) to determine whether the negative charges in all TRPC channels are required for their gating by STIM1, including the charges in TRPC3 and TRPC6, which do not interact directly with STIM1; (c) to determine whether TRPC channels activated by receptor stimulation function as STIM1-dependent and/or STIM1-independent channels; and (d) to determine whether channels functioning in a STIM1-independent mode recouple with STIM1 by increasing the STIM1/TRPC channel ratio.
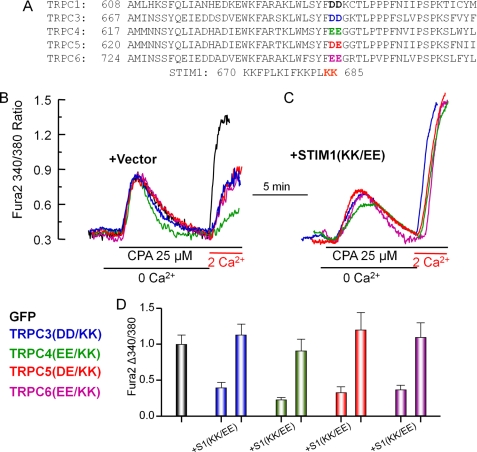
FIGURE 2.
Inhibition of native SOC by the charge-swap TRPC mutants and rescue of SOC by STIM1(K684E,K685E). A, alignment of a C-terminal region of TRPC channels encompassing the conserved negative charges that are highlighted in boldface colored letters. The two negative charges were mutated to the positively charged lysines. B and C, HEK cells were transfected with GFP (black traces) and the following TRPC channel mutants: TRPC3(D697K,D698K) (TRPC3(DD/KK); blue traces and bars), TRPC4(E648K,E649K) (TRPC4(EE/KK); green traces and bars), TRPC5(D651K,E652K) (TRPC5(DE/KK); red traces and bars), and TRPC6(E755K,E756K) (TRPC6(EE/KK); magenta traces and bars). The cells were also transfected with empty vector (B) or STIM1(K684E,K685E) (S1(KK/EE); C). The Fura-2-loaded cells were incubated in Ca2+-free medium, treated with 25 μm cyclopiazonic acid (CPA) for 7.5 min to deplete the stores, and then exposed to medium containing 2 mm Ca2+ to assay Ca2+ influx. D, mean ± S.E. of at least 15 cells from two separate experiments.
We report here that switching the conserved negative charges (DD/E) to positive charges (KK) located distal to the last transmembrane domain in the C terminus of TRPC channels results in channels that inhibit Ca2+ influx activated by passive store depletion. This Ca2+ influx could be rescued by the reverse charge mutant STIM1(K684E,K685E), indicating that gating by electrostatic interaction with STIM1 is common to all TRPC channels. When activated by stimulation of the G protein-coupled M3 receptors, the TRPC channel charge-swap mutants mediated robust Ca2+ influx, which was inhibited by wild-type STIM1 and augmented by the charge-swap mutant STIM1(K684E,K685E). Similarly, receptor stimulation activated wild-type and mutant TRPC channel currents. However, most notably, the wild-type but not mutant TRPC channels current was inhibited by scavenging STIM1 with Orai1(R91W), and the inhibition was rescued by overexpression of STIM1. Moreover, wild-type STIM1 and STIM1(K684E,K685E) reciprocally affected the TRPC3 and TRPC3(D697K,D698K) currents, with inhibition of the TRPC3 current by STIM1(K684E,K685E) but not by STIM1 and with inhibition of the TRPC3(D697K,D698K) current by STIM1 but not by STIM1(K684E,K685E). Similar behavior was observed with TRPC5(D651K,E652K) and TRPC6(E755K,E756K). These findings reveal that TRPC channels can function in STIM1-dependent and STIM1-independent modes. The STIM1/TRPC channel ratio determines the STIM1-dependent mode of the channels to tune their SOC-dependent and SOC-independent function.
MATERIALS AND METHODS
Solutions, Reagents, and Clones
TRPC1, TRPC3, TRPC5, and TRPC6 cDNAs were cloned into the p3×FLAG-CMV7.1 vector using NotI (5′) and SalI (3′). The TRPC4, STIM1, and STIM1(K684E,K685E) clones were described previously (10, 13). The Orai1 cDNA was cloned into the p3×FLAG-CMV expression vector using SalI (5′) and NotI (3′) as described (13). A site-directed mutagenesis kit from Stratagene was used to generate the DD/E-to-KK mutants of all TRPC channels and Orai1(R91W).
Co-immunoprecipitation and Western Blotting
HEK cells were transfected with TRPC1, TRPC3, TRPC4, TRPC5, and TRPC6 and with or without YFP-STIM1 using Lipofectamine 2000 (Invitrogen). Cells were lysed in 1× PBS (pH 7.4) containing 1% Triton X-100, 10 mm sodium pyrophosphate, 50 mm NaF, 1 mm NaVO3, and protease inhibitor mixture. Native STIM1 or YFP-STIM1 was immunoprecipitated with 1 μg of anti-STIM1 monoclonal antibody (BD Biosciences) or anti-GFP polyclonal antibody (Invitrogen) to determine co-immunoprecipitation of the various TRPC channels. Western blotting was done using anti-HA monoclonal antibody (Covance) for TRPC1/4, anti-HRP-FLAG monoclonal antibody (Sigma) for TRPC3/6, and anti-STIM1 monoclonal antibody (BD Biosciences).
Measurement of [Ca2+]i
[Ca2+]i was measured at ∼24 h post-transfection using a Nikon image acquisition system with MetaFluor software. [Ca2+]i was measured by loading the cells with Fura-2, recording Fura-2 fluorescence at excitation wavelengths of 340 and 380 nm, and collecting the light emitted at wavelengths above 500 nm. [Ca2+]i is expressed as the 340/380 nm ratio.
Current Measurements
TRPC channel current was measured in transiently transfected HEK cells as described previously (11). The pipette solution contained 140 mm CsCl, 2 mm MgCl2, 1 mm ATP, 5 mm EGTA, 1.5 mm CaCl2 (to clamp free Ca2+ at 70 nm), and 10 mm HEPES at pH 7.2 (with CsOH). This solution eliminates the K+ current and prevents inhibition of the TRPC channel by cytoplasmic Ca2+. The bath solution contained 140 mm NaCl or 140 mm N-methyl-d-glucamine (NMDG)-Cl, 5 mm KCl, 0.5 mm EGTA, and 10 mm HEPES at pH 7.4 (with NaOH or NMDG-OH−). Cells were transfected with TRPC channels; with the M3 receptor; and with or without STIM1, STIM1(K684E,K685E), and Orai1(R91W). The current was recorded by 400-ms rapid alterations of membrane potential from −100 to +100 mV from a holding potential of 0 mV. The current recorded at −100 mV was used to calculate current density as pA/picofarads (pF), and the current recorded in multiple experiments was used to obtain the mean ± S.E. and to calculate significance by Student's t test.
RESULTS AND DISCUSSION
STIM1 Gates TRPC Channels by Electrostatic Interaction
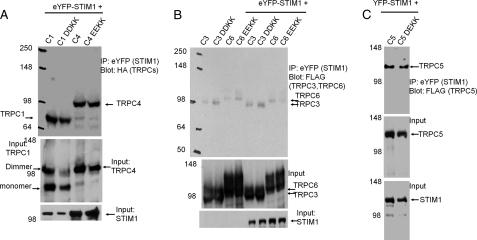
The binding of STIM1 to TRPC1 is mediated by the STIM1 ERM domain, whereas the gating of TRPC1 is mediated by the STIM1 K-domain, which is not required for the binding of STIM1 to the TRPC channels (10, 11). However, the gating of TRPC1 by STIM1 is mediated by electrostatic interaction between the STIM1 Lys-684 and Lys-685 residues and the TRPC1 Asp-639 and Asp-640 residues, and the TRPC1(D639K,D640K) mutant cannot be activated by wild-type STIM1 (11). Because STIM1 also regulates the activity of other TRPC channels (21), and the negative charges required for the electrostatic interaction are conserved in all TRPC channels (see Fig. 2A), we asked whether the conserved negative charges have a role in the regulation of other TRPC channels by STIM1. To ascertain that the effects of disruption of the electrostatic interaction are not due to inhibition of STIM1-TRPC channel interaction, we tested the effect of mutating the TRPC channel conserved negative charges (DD/E) to positive charges (KK) on interaction with STIM1. Fig. 1 shows the similar interaction of wild-type and mutant TRPC channels with STIM1. This is in agreement with our previous findings with TRPC1, showing that disruption of the electrostatic interaction does not affect STIM1-TRPC1 interaction (11) and that the STIM1 K-domain is not required for interaction between STIM1 and TRPC1 (10).
FIGURE 1.
Interaction of STIM1 with wild-type and mutant TRPC channels. A, HEK cells were transfected with eYFP-STIM1 and HA-TRPC1 (C1), HA-TRPC1(D639K,D640K) (C1 DDKK), HA-TRPC4 (C4), and HA-TRPC4(E645K,E649K) (C1 EEKK) and were used to immunoprecipitate (IP) eYFP (STIM1) and blot for HA (TRPC1 and TRPC4). B, HEK cells were transfected with eYFP-STIM1 and FLAG-TRPC3 (C3), FLAG-TRPC3(D697K,D698K) (C3 DDKK), FLAG-TRPC6 (C6), and FLAG-TRPC6(E755K,E756K) (C6 EEKK) and were used to immunoprecipitate eYFP (STIM1) and blot for FLAG (TRPC3 and TRPC6). C, HEK cells were transfected with eYFP-STIM1 and FLAG-TRPC5 (C5) or FLAG-TRPC5(D651K,E652K) (C5 DEKK) and used to immunoprecipitate eYFP (STIM1) and blot for FLAG (TRPC5). Note that the DD/E-to-KK mutations had no effect on the interaction of STIM1 with the TRPC channels.
Next, we tested the effect of the TRPC channel mutants on store- and receptor-stimulated Ca2+ influx. Fig. 2 (B and D) shows that expression of the TRPC channel mutants resulted in partial inhibition of the native SOC activity triggered by passive store depletion with the sarco/endoplasmic reticulum Ca2+-ATPase pump inhibitor cyclopiazonic acid. Thus, the results in Fig. 2 (B and D) suggest that the mutant TRPC channels cannot mediate Ca2+ influx when Ca2+ influx is activated by passive store depletion. Because the mutants can bind STIM1 (Fig. 1), inhibition of native SOC is likely mediated by scavenging of native STIM1 by the overexpressed channels. This is verified by the findings in Fig. 2 (C and D), in which expression of the STIM1(K684E,K685E) mutant rescued Ca2+ influx by the TRPC channel mutants.
The findings in Fig. 2 have several implications. First, inhibition of native SOC by the mutants and its rescue by STIM1(K684E,K685E) indicate that the TRPC channels functionally interact with STIM1 (see also results with Orai1(R91W) below). Second, gating by electrostatic interaction with STIM1 is common to all TRPC channels.
STIM1-dependent and STIM1-independent Function of TRPC Channels
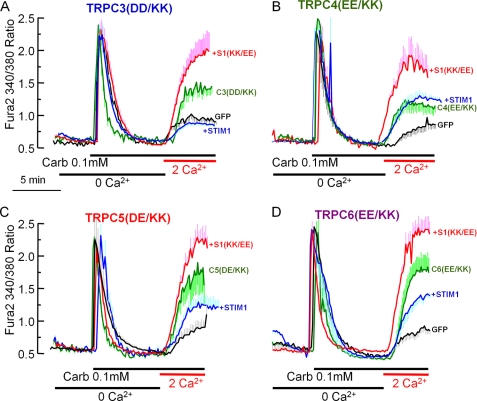
Regulation of TRPC3 and TRPC6 by STIM1 is observed only when these channels are expressed at relatively low levels (21), suggesting that these and perhaps other TRPC channels can function in a STIM1-dependent and STIM1-independent mode. To determine whether this is the case, we analyzed the effect of STIM1 and STIM1(K684E,K685E) and of the STIM1 scavenger Orai1(R91W) mutant on the function of the charge-swap TRPC channel mutants. We also asked if the channels behaved differentially when activated by store depletion and by receptor stimulation. Cells were transfected with the M3 muscarinic receptor, with the TRPC channel mutants, and with and without STIM1 and STIM1(K684E,K685E). Fig. 3 shows the results obtained with TRPC3(D697K,D698K), TRPC4(E648K,E649K), TRPC5(D651K,E652K), and TRPC6(E755K,E756K).
FIGURE 3.
STIM1-dependent and STIM1-independent function of receptor-activated TRPC channels. Black traces are for HEK cells transfected with GFP and M3 receptors only and show the native receptor-stimulated Ca2+ influx. HEK cells were also transfected with M3 receptors, the indicated TRPC channel mutants, and empty vector (green traces) with wild-type STIM1 (blue traces) or with STIM1(K684E,K685E) (S1(KK/EE); red traces). The cells in Ca2+-free medium were stimulated with 0.1 mm carbachol (Carb) and then incubated with medium containing carbachol and 2 mm Ca2+ to assay Ca2+ influx. A, TRPC3(D697K,D698K) (TRPC3(DD/KK), C3(DD/KK)); B, TRPC4(E648K,E649K) (TRPC4(EE/KK), C4(EE/KK)); C, TRPC5(D651K,E652K) (TRPC5(DE/KK), C5(DE/KK)); D, TRPC6(E755K,E756K) (TRPC6(EE/KK), C6(EE/KK)). The traces are the mean ± S.E. of at least five cells and are representative of three more experiments with similar results.
The first notable finding is that all TRPC channel mutants were activated by receptor stimulation (green traces in Fig. 3). Note that none of these mutants was activated by passive store depletion (Fig. 2). This suggests that receptor stimulation can activate TRPC channels in a STIM1-independent mechanism. The second finding is that wild-type STIM1 either slightly (TRPC5(D651K,E652K)) or strongly (TRPC3(D697K,D698K) and TRPC6(E755K,E756K)) inhibited the receptor-stimulated Ca2+ influx by the TRPC channel mutants (blue traces). By contrast, STIM1(K684E,K685E) increased the receptor-stimulated Ca2+ influx by the TRPC channel mutants (red traces). These findings indicate that (a) TRPC channels are regulated by STIM1 because STIM1 inhibits and STIM1(K684E,K685E) increases the activity of the TRPC channel mutants; (b) the activity of TRPC channels can be modulated by STIM1 (if the activating STIM1 is limiting, they can be further activated by STIM1); and (c) most importantly, when activated by receptor stimulation, TRPC channels can function in a STIM1-independent manner, as evident from Ca2+ influx by the charge-swap TRPC channel mutants in the absence of STIM1(K684E,K685E). The channels can be recoupled with STIM1 and made to function in a STIM1-dependent manner when STIM1 is expressed together with the channel at a favorable STIM1/TRPC channel ratio. This is concluded from the increased Ca2+ influx by the TRPC channel mutants when expressed with STIM1(K684E,K685E) and from their inhibition by wild-type STIM1.
STIM1 and TRPC Channel Currents
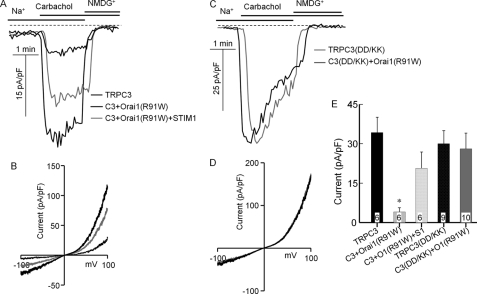
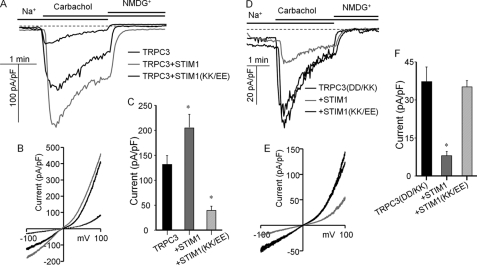
To obtain further independent evidence for the STIM1-dependent and STIM1-independent function of the TRPC channels, we tested the effect of scavenging STIM1 with Orai1(R91W) on channel function of wild-type and mutant TRPC channels. To better isolate the function of the TRPC channels, we measured the Na+ current carried by these channels. Fig. 4 shows the current activated by receptor stimulation of cells expressing TRPC3 or TRPC3(D697K,D698K) in the presence and absence of Orai1(R91W). Orai1(R91W) does not mediate any current, but it can bind STIM1, similar to wild-type Orai1 (23). Hence, Orai1(R91W) can be used as a scavenger of STIM1 to determine the dependence of channel function on STIM1. Fig. 4 (A, B, and E) shows that when expressed with wild-type TRPC3, Orai1(R91W) strongly inhibited the TRPC3 current. That this is likely due to scavenging of STIM1 is shown by the reverse of the inhibition of the TRPC3 current by STIM1 when coexpressed with Orai1(R91W). The TRPC3(D697K,D698K) mutant showed a large receptor-stimulated Na+ current, with I/V typical of TRPC3 (Fig. 4D). Significantly, Orai1(R91W) did not inhibit the current measured with TRPC3(D697K,D698K) (Fig. 4, C–E), indicating that TRPC3(D697K,D698K) functions in a STIM1-independent mode.
FIGURE 4.
Scavenging STIM1 with Orai1(R91W) inhibits the TRPC3 but not TRPC3(D697K,D698K) current. HEK cells were transfected with M3 receptors; wild-type TRPC3 (A and B) or TRPC3(D697K,D698K) (TRPC3(DD/KK), C3(DD/KK); C and D); and empty vector (thin black traces), Orai1(R91W) (thick black traces), or Orai1(R91W) + STIM1 (gray traces). A and C, examples of the time dependence of the current; B and D, I/V at peak current; E, mean ± S.E. of current density at −80 mV in pA/pF from the indicated number of experiments. *, p < 0.01 of TRPC3 + Orai1(R91W) (O1(R91W)) relative to wild-type TRPC3. All others were not different from wild-type TRPC3.
Function in a STIM1-dependent (TRPC3) and STIM1-independent (TRPC3(D697K,D698K)) mode raised the question as to whether the STIM1 dependence of the channel can be modulated by varying the expression of STIM1. In Fig. 5, this was tested by determining the effects of STIM1 and STIM1(K684E,K685E) on channel activity. Fig. 5 (A–C) shows that STIM1 significantly increased channel activity when coexpressed with TRPC3. Most notably, the receptor-stimulated TRPC3 current was markedly inhibited by STIM1(K684E,K685E). The reciprocal experiment is shown in Fig. 5 (D–F). The current mediated by TRPC3(D697K,D698K) was not influenced by STIM1(K684E,K685E). However, the activity of TRPC3(D697K,D698K) was markedly inhibited by wild-type STIM1.
FIGURE 5.
Effect of STIM1 and STIM1(K684E,K685E) on TRPC3 and TRPC3(D697K,D698K) currents. HEK cells were transfected with M3 receptors; TRPC3 (A–C) or TRPC3(D697K,D698K) (TRPC3(DD/KK); D–F); and empty vector (thin black traces), wild-type STIM1 (gray traces), or STIM1(K684E,K685E) (STIM1(KK/EE); thick black traces). The Na+ current was measured as described under “Materials and Methods.” A and D, examples of the time dependence of the current; B and E, I/V at peak current; C and F, mean ± S.E. of current density in pA/pF from seven experiments. *, p < 0.05 or better relative to the respective controls.
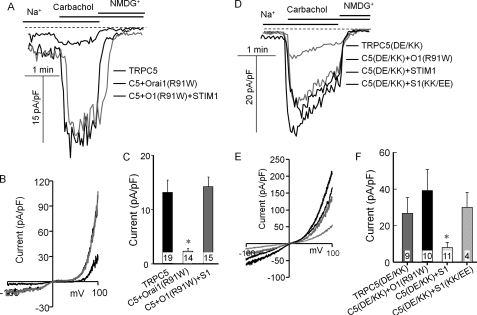
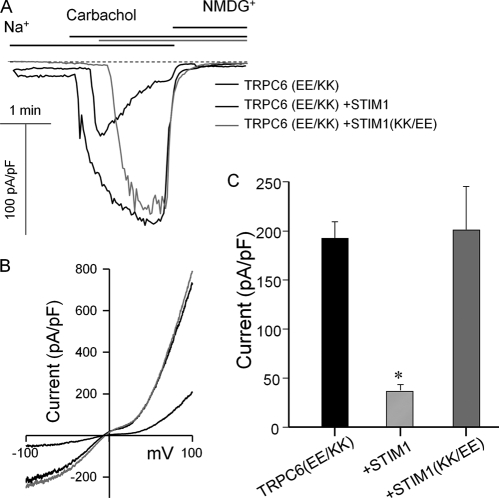
The effects of Orai1(R91W) and the STIM1 mutants on wild-type and mutant TRPC5 are shown in Fig. 6. Fig. 6 (A–C) shows that Orai1(R91W) markedly inhibited the TRPC5 current and that the inhibition could be reversed by overexpression of STIM1 with Orai1(R91W). By contrast, the TRPC5(D651K,E652K)-mediated current was not inhibited by Orai1(R91W). Moreover, wild-type STIM1 inhibited the TRPC5(D651K,E652K) current, whereas STIM1(K684E,K685E) did not. Finally, Fig. 7 shows similar experiments with TRPC6(E755K,E756K). This mutant was fully activated by receptor stimulation. Although the activity of TRPC6(E755K,E756K) was not affected by coexpressed STIM1(K684E,K685E), it was markedly inhibited by coexpressed wild-type STIM1. Hence, TRPC6 can also function in a STIM1-independent mode and be made to couple to STIM1.
FIGURE 6.
Effect of Orai1(R91W), STIM1, and STIM1(K684E,K685E) on TRPC5 and TRPC5(D651K,E652K) currents. A–C, HEK cells were transfected with M3 receptors; TRPC5 (C5); and empty vector (thin black traces), Orai1(R91W) (thick black traces), or Orai1(R91W) + STIM1 (O1(R91W)+S1; gray traces). D–F, HEK cells were transfected with M3 receptors; TRPC5(D651K,E652K) (TRPC5(DE/KK), C5(DE/KK)); and empty vector (thin black traces), Orai1(R91W) (thin gray traces), STIM1 (thick gray traces), or STIM1(K684E,K685E) (S1(KK/EE); thick black traces). The Na+ current was measured as described under “Materials and Methods.” A and D, examples of the time dependence of the current; B and E, I/V at peak current; C and F, mean ± S.E. of current density in pA/pF from the indicated number of experiments. *, p < 0.01 relative to the respective controls.
FIGURE 7.
Effect of wild-type STIM1 and STIM1(K684E,K685E) on the TRPC6(E755K,E756K) current. HEK cells were transfected with M3 receptors; TRPC6(E755K,E756K) (TRPC6(EE/KK)); and empty vector (thin black traces), STIM1 (thick black traces), or STIM1(K684E,K685E) (STIM1(KK/EE); gray traces). The Na+ current was measured as described under “Materials and Methods.” A, examples of the time dependence of the current; B, I/V at peak current; C, mean ± S.E. of current density in pA/pF from 15–19 experiments. *, p < 0.01.
The results of (1) inhibition of the wild-type TRPC channel current by Orai1(R91W), (2) the lack of effect of Orai1(R91W) on the mutant TRPC channel current, and (3) the reciprocal effect of STIM1 and STIM1(K684E,K685E) on the wild-type and mutant channel currents together indicate that TRPC channels have two functional modes: STIM1-dependent and STIM1-independent.
Conclusions
In this work, we sought to determine the role of STIM1 in receptor-stimulated TRPC channel activity. To study the role of STIM1 in channel activity stimulated by G protein-coupled receptors, we used the strategy of mutating the conserved negative charges in the C-terminal domain of the TRPC channels (Fig. 2) (11) that are required for activation of the channels by STIM1. The advantage of this strategy is that the mutant channels cannot be activated by wild-type STIM1, so their activity and regulation by STIM1 can be clearly isolated. The STIM1 independence of the mutant TRPC channels is best illustrated by their resistance to scavenging STIM1 with the Orai1(R91W) mutant. That the main function of Orai1(R91W) is to scavenge STIM1 is shown by overcoming the inhibition by overexpression of STIM1 (Figs. 4E and 6C).
The Ca2+ influx and Na+ currents of the mutant TRPC channels are inhibited by wild-type STIM1, and Ca2+ influx is activated by the charge-swap STIM1(K684E,K685E) mutant. This indicates that, like TRPC1 (11), all TRPC channels are gated by electrostatic interaction with STIM1 that requires matching of the two negative charges in the C-terminal domain of the TRPC channels with the last two positively charged lysines of STIM1. We note that, unlike Ca2+ influx, the Na+ current of the mutant TRPC channels is not further increased by STIM1(K864E,K865E). At present, we do not have a clear explanation for this finding. One possibility is that since the TRPC channels can function in a STIM1-independent mode, apparently the Na+ current of the mutants in the absence of STIM1 is already maximal, and further interaction with STIM1 does not appear to increase the Na+ current. Another possibility is that the modes of Na+ and Ca2+ transport by the TRPC channels are not identical, as has been shown for many transient receptor potential channels (26). Nevertheless, both modes are regulated by STIM1, as evident from inhibition of the wild-type TRPC channel Na+ current by Orai1(R91W) and the mutant TRPC channel Na+ current by STIM1.
The most important finding of this work is that TRPC channels can function as STIM1-dependent and STIM1-independent channels. This is concluded from the findings that the wild-type channels are inhibited by the STIM1 scavenger Orai1(R91W), whereas the charge-swap mutant channels are not. Moreover, when expressed alone, the mutant TRPC channels inhibit the native SOC activity (Fig. 2), yet robust Ca2+ influx by all the charge-swap channel mutants is activated by stimulation of the M3 muscarinic receptor (Fig. 3). Notably, the activity of these channels is inhibited by wild-type STIM1. This is most prominent with TRPC3(D697K,D698K), TRPC5(D651K,E652K), and TRPC6(E755K,E756K). The current mediated by these mutants is markedly inhibited by wild-type STIM1 (Figs. 5–7). Because the charge-swap TRPC channel mutants are not inhibited by Orai1(R91W), they cannot be activated by STIM1, and, thus, suppress the native SOC activity. The Ca2+ influx and current of these channels, when activated by receptor simulation, must be STIM1-independent. At the same time, the activity of wild-type TRPC3 and TRPC5 (and TRPC1; see Ref. 11) requires functional STIM1 because the activity of the channels is inhibited by Orai1(R91W) and STIM1(K684E,K685E). Thus, together, our findings suggest that TRPC channels can function in a STIM1-dependent and STIM1-independent mode.
The reciprocal effect of wild-type STIM1 and STIM1(K684E,K685E) on TRPC3 and TRPC3(D697K,D698K) activity (Fig. 5) has the important implication that the STIM1/TRPC channel ratio determines the STIM1-dependent and STIM1-independent function of the channels and thus tunes the SOC function of TRPC channels. Therefore, access of STIM1 to the channels determines their operational mode. At a low STIM1/TRPC channel ratio, the channels function mostly in a STIM1-independent mode. How receptor stimulation opens the channels when functioning in a STIM1-independent mode remains to be elucidated. On the other hand, at a high STIM1/TRPC channel ratio, the channels are gated by STIM1 and are opened by interaction of the positively charged STIM1 Lys-684 and Lys-685 residues with the conserved two negative charges in the C-terminal domain of the TRPC channels. Receptor stimulation can determine the mode of TRPC channel gating by determining access of STIM1 to the channels. The different modes of operation of the TRPC channels increase their versatility and the precision with which receptor stimulation controls Ca2+ influx.
This work was supported, in whole or in part, by National Institutes of Health Grants DE12302 and DK38938 and by NIDCR/DIR.
- ER
- endoplasmic reticulum
- SOC
- store-operated Ca2+ influx channel
- TRPC
- transient receptor potential cation
- pF
- picofarads
- eYFP
- enhanced YFP
- NMDG
- N-methyl-d-glucamine.
REFERENCES
- 1.Parekh A. B., Putney J. W., Jr. (2005) Physiol. Rev. 85, 757–810 [DOI] [PubMed] [Google Scholar]
- 2.Lee K. P., Yuan J. P., Hong J. H., So I., Worley P. F., Muallem S. (2010) FEBS Lett. 584, 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S. H., Tanasa B., Hogan P. G., Lewis R. S., Daly M., Rao A. (2006) Nature 441, 179–185 [DOI] [PubMed] [Google Scholar]
- 6.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalioti M. D., Zhang J., Volkman H. M., Kahle K. T., Hoffmann K. E., Toka H. R., Nelson-Williams C., Ellison D. H., Flavell R., Booth C. J., Lu Y., Geller D. S., Lifton R. P. (2006) Nat. Genet. 38, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 8.Wu M. M., Buchanan J., Luik R. M., Lewis R. S. (2006) J. Cell Biol. 174, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeHaven W. I., Smyth J. T., Boyles R. R., Bird G. S., Putney J. W., Jr. (2008) J. Biol. Chem. 283, 19265–19273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang G. N., Zeng W., Kim J. Y., Yuan J. P., Han L., Muallem S., Worley P. F. (2006) Nat. Cell Biol. 8, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 11.Zeng W., Yuan J. P., Kim M. S., Choi Y. J., Huang G. N., Worley P. F., Muallem S. (2008) Mol. Cell 32, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stathopulos P. B., Li G. Y., Plevin M. J., Ames J. B., Ikura M. (2006) J. Biol. Chem. 281, 35855–35862 [DOI] [PubMed] [Google Scholar]
- 13.Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawasaki T., Lange I., Feske S. (2009) Biochem. Biophys. Res. Commun. 385, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K. P., Yuan J. P., Zeng W., So I., Worley P. F., Muallem S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullins F. M., Park C. Y., Dolmetsch R. E., Lewis R. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15495–15500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derler I., Fahrner M., Muik M., Lackner B., Schindl R., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 24933–24938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liou J., Fivaz M., Inoue T., Meyer T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. (2006) Nat. Cell Biol. 8, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan J. P., Zeng W., Huang G. N., Worley P. F., Muallem S. (2007) Nat. Cell Biol. 9, 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercer J. C., Dehaven W. I., Smyth J. T., Wedel B., Boyles R. R., Bird G. S., Putney J. W., Jr. (2006) J. Biol. Chem. 281, 24979–24990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., Romanin C. (2008) J. Biol. Chem. 283, 8014–8022 [DOI] [PubMed] [Google Scholar]
- 24.Liao Y., Plummer N. W., George M. D., Abramowitz J., Zhu M. X., Birnbaumer L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3202–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M. S., Zeng W., Yuan J. P., Shin D. M., Worley P. F, Muallem S. (2009) J. Biol. Chem. 284, 9733–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilius B., Owsianik G., Voets T., Peters J. A. (2007) Physiol. Rev. 87, 165–217 [DOI] [PubMed] [Google Scholar]