Abstract
Ca2+-dependent activator protein for secretion (CAPS) regulates exocytosis of catecholamine- or neuropeptide-containing dense-core vesicles (DCVs) at secretion sites, such as nerve terminals. However, large amounts of CAPS protein are localized in the cell soma, and the role of somal CAPS protein remains unclear. The present study shows that somal CAPS1 plays an important role in DCV trafficking in the trans-Golgi network. The anti-CAPS1 antibody appeared to pull down membrane fractions, including many Golgi-associated proteins, such as ADP-ribosylation factor (ARF) small GTPases. Biochemical analyses of the protein-protein interaction showed that CAPS1 interacted specifically with the class II ARF4/ARF5, but not with other classes of ARFs, via the pleckstrin homology domain in a GDP-bound ARF form-specific manner. The pleckstrin homology domain of CAPS1 showed high affinity for the Golgi membrane, thereby recruiting ARF4/ARF5 to the Golgi complex. Knockdown of either CAPS1 or ARF4/ARF5 expression caused accumulation of chromogranin, a DCV marker protein, in the Golgi, thereby reducing its DCV secretion. In addition, the overexpression of CAPS1 binding-deficient ARF5 mutants induced aberrant chromogranin accumulation in the Golgi and consequently reduced its DCV secretion. These findings implicate a functional role for CAPS1 protein in the soma, a major subcellular localization site of CAPS1 in many cell types, in regulating DCV trafficking in the trans-Golgi network; this activity occurs via protein-protein interaction with ARF4/ARF5 in a GDP-dependent manner.
Keywords: Exocytosis, Golgi, Neuroscience, Secretion, Trafficking
Introduction
The Ca2+-dependent activator protein for secretion (CAPS)2 family is involved in dense-core vesicle (DCV) exocytosis (1–3) and, in mammals, consists of two family members, CAPS1 (2) and CAPS2 (4, 5). Many previous studies have suggested that CAPS1 plays a role in the secretion of catecholamines (e.g. norepinephrine (2, 3)), neuropeptides (e.g. neuropeptide Y (6)), and peptide hormones (e.g. insulin (7)) by binding to phosphatidylinositol 4,5-bisphosphate at the priming step of DCV exocytosis (8, 9). On the other hand, recent studies using knock-out (KO) or knockdown (KD) approaches have led to fresh debates concerning the involvement of CAPS1 in the priming of synaptic vesicle exocytosis (10) as well as in the vesicular loading of catecholamine or serotonin (11, 12). Although these previous studies focused on the role of CAPS protein in exocytosis at secretion sites, such as nerve terminals or the cell periphery, a large fraction of CAPS protein in many neuronal cell types is actually localized in the soma rather than at these secretion sites (2, 13, 14). Therefore, the role of somal CAPS proteins, which constitute the largest amount of CAPS proteins in the cell, also needs to be elucidated.
In this study, we have investigated the role of somal CAPS1 proteins in the DCV secretory pathway, including biogenesis-trafficking-secretion events. We showed an interaction between CAPS1 and the class II ADP-ribosylation factor (ARF) small GTPase on the Golgi membrane, depending on the ARF-GDP mode. The ARF family consists of three classes and a total of six members: class I, consisting of ARF1 to -3; class II, consisting of ARF4 and -5; and class III, which comprises ARF6 (15). The class I (16–18) and class III ARFs (19, 20) have been implicated as important regulators for membrane trafficking, but little is known about the role of the class II ARFs in membrane trafficking. Knockdown of CAPS1 or of the class II ARFs caused accumulation of a DCV marker protein, chromogranin, in the Golgi, resulting in reduced chromogranin secretion. Overexpression of ARF5 mutants that fail to bind CAPS1 also induced accumulation of chromogranin in the Golgi, resulting in a reduction of chromogranin secretion. These results suggest that CAPS1 has a regulatory role, in concert with GDP/GTP binding state-dependent class II ARFs, in DCV trafficking and/or biogenesis in the trans-Golgi network.
EXPERIMENTAL PROCEDURES
Plasmids
Mouse full-length cDNA clones were purchased from the RIKEN FANTOM cDNA library (21). Mouse cDNAs were subcloned into pEF-BOS (kindly provided by Dr. Shigekazu Nagata, Osaka, Japan), which contains the EF-1α promoter (22).
Antibodies
Rabbit polyclonal anti-ARF5 antibodies were raised against a mouse ARF5 (ERVQESADELQKMLQEDC) peptide-KLH conjugate and were affinity-purified against maltose-binding protein (MBP)-tagged full-length ARF5 proteins covalently coupled to CNBr-activated Sepharose 4B. Rabbit anti-ARF5 antibodies were used for Western blotting (1:1,000 dilution). Rabbit anti-CAPS1 antibody was raised against glutathione S-transferase (GST)-tagged mouse CAPS1 (amino acids 266–366) that had been expressed bacterially, and it was affinity-purified against the MBP-tagged antigenic protein that was covalently coupled to CNBr-activated Sepharose 4B. Rabbit anti-CAPS1 antibody was used for Western blotting (1:2,000 dilution).
The following primary antibodies were also used for Western blotting: rabbit polyclonal anti-CAPS2 (1:1,000 dilution) (5), mouse monoclonal anti-FLAG (1:1,000 dilution; catalog no. F1804, Sigma), rat monoclonal anti-hemagglutinin epitope (HA) (1:1,000 dilution; catalog no. 1867423, Roche Applied Science), mouse monoclonal anti-VAMP2 (1:10,000 dilution; catalog no. 104211, Synaptic Systems), rabbit polyclonal anti-VAMP4 (1:1,000 dilution; catalog no. PA1-768, Affinity BioReagents), mouse monoclonal SNAP25 (1:2,500 dilution; catalog no. SMI-81, Sternberger), mouse monoclonal anti-ARF (clone 1D9) (1:500 dilution; catalog no. ab2806, Abcam), mouse monoclonal anti-GM130 (1:250 dilution; catalog no. 610822, BD Biosciences), mouse monoclonal anti-GS28 (1:1,000 dilution; catalog no. 611184, BD Biosciences), mouse monoclonal anti-p115 (1:1,000 dilution; catalog no. 612260, BD Biosciences), mouse monoclonal anti-Bcl-2 (1:500 dilution; catalog no. 610538, BD Biosciences), mouse monoclonal anti-BiP/GRP78 (1:250 dilution; catalog no. 610978, BD Biosciences), rabbit polyclonal anti-syntaxin 5 (1:1,000 dilution; catalog no. 110053, Synaptic Systems), mouse monoclonal anti-syntaxin 6 (1:2,500 dilution; catalog no. 610635, BD Biosciences), rabbit polyclonal anti-syntaxin 16 (1:1,000 dilution; catalog no. 110162, Synaptic Systems), mouse monoclonal anti-Vti1a (1:2,500 dilution; catalog no. 611220, BD Biosciences), and goat polyclonal anti-GST (1:1,000 dilution; catalog no. 27-4577, Amersham Biosciences). The following primary antibodies were used for immunocyto- or immunohistochemistry: mouse monoclonal anti-chromogranin A (1:5,000 dilution; catalog no. 611844, BD Biosciences), mouse monoclonal anti-syntaxin 6 (1:500 dilution; catalog no. 610635, BD Biosciences), mouse monoclonal anti-FLAG (1:250 dilution; catalog no. F1804, Sigma), and rat monoclonal anti-HA (1:250 dilution; catalog no. 1867423, Roche Applied Science).
Secretion Assay
Mouse chromogranin A and secretogranin II cDNA was subcloned into pEF-BOS with the C-terminal triple HA tag to create pEF-BOS-ChgA-HA and pEF-BOS-SgII-HA, respectively. Transfections were carried out in 12-well plates using Lipofectamine 2000 reagent (Invitrogen) as described previously (5). In these experiments, we also transfected PC12 cells with Lipofectamine complexed with 0.2 μg of pEF-BOS-granin-HA and 20 pmol of CAPS1 stealth siRNA (catalog no. RSS304417, Invitrogen). After incubation with Lipofectamine for 24 h, cells were washed and grown in DMEM containing 100 ng/ml NGF. 48 h after transfection, two different DMEMs were used as a stimulating medium: standard DMEM (catalog no. 11965, Invitrogen) and high KCl DMEM (based on 11965 but with 50 mm KCl, 65 mm NaCl (specially ordered); Invitrogen). Both DMEMs were fully equilibrated in a 5% CO2 atmosphere at 37 °C before use. Culture media were collected after control and high KCl stimulation assays, and cell lysates were also collected.
Immunocytochemistry
A yellow-shifted GFP derivative, YPet (23), was generated from Venus (24). A (GGGGS)3 linker separated CAPS1 from the C-terminal triple HA-tagged YPet in the pEF-BOS-CAPS1-YPet-HA plasmid. Dissociated PC12 cells were plated onto a glass coverslip (12 mm in diameter; Matsunami Glass, Japan) coated with poly-l-lysine (Sigma) and then cultured in DMEM supplemented with 1% horse serum and 5% fetal bovine serum at 37 °C in a humidified 5% CO2 atmosphere. Transfection was carried out using Lipofectamine 2000 reagent (Invitrogen). Forty-eight hours after transfection, cells were fixed with Zamboni's fixative (2% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4, containing 0.2% picric acid) at room temperature for 15 min. After washing three times with PBS, cells were permeabilized in PBS containing 0.02% Triton X-100 at room temperature for 5 min. After blocking with 5% normal donkey serum in PBS at room temperature for 60 min, cells were incubated with specific primary antibodies at 4 °C overnight, rinsed in PBS, and then incubated with Alexa-conjugated secondary antibodies (1:2,000 dilution; Invitrogen) at room temperature for 1 h and again rinsed in PBS. Immunoreacted cells were mounted with Vectashield (Vector) mounting medium. Images were acquired with a microscope (Olympus BX51) equipped with a CCD camera (VB-7000, Keyence). Digital images were processed using Adobe Photoshop 6.0 software.
Golgi Localization Study
The type II membrane-anchored protein galactosyltransferase, B4galt1, has been used as a marker of the Golgi complex (25). The N-terminal 81 amino acids of mouse B4galt1 were fused to tdTomato (catalog no. 632534, Clontech) to make the Golgi indicator, B4galt1-tdTomato. PC12 cells were transfected with B4galt1-tdTomato together with either control or CAPS1 siRNA. Endogenous ARF5 immunoreactivity merged with immunoreactivities for B4galt1-tdTomato (a marker of the Golgi complex) and whole-cell soma was quantified using Metamorph software (Universal Imaging Corp.).
Immunoaffinity Purification of Microsomal Fractions
To purify the CAPS1-associated subfraction from the mouse cerebellar microsomal fractions using immunoaffinity, anti-CAPS1 antibody-coupled magnetic beads were made as follows. 40 μl of superparamagnetic, polystyrene beads covalently coated with sheep anti-rabbit IgG (Dynabeads M-280; catalog no. DB11203, Invitrogen) were incubated for 1 h at 4 °C in phosphate buffer (PBS, pH 7.4, 1% BSA) containing 2 μg of primary rabbit polyclonal anti-CAPS1 antibody (5). Similarly, rabbit polyclonal anti-VAMP2 antibody (13) and control rabbit IgG (Jackson ImmunoResearch, West Grove, PA) were coupled to magnetic beads. P21 mouse cerebella were dissected and homogenized in homogenization buffer (50 mm HEPES, pH 7.4, 5 mm EDTA, 0.32 m sucrose, and protease inhibitor mixture). Homogenates were centrifuged at 2,000 × g for 10 min at 4 °C, and microsomal fractions were sedimented from the supernatant at 20,000 × g for 15 min as described elsewhere (26). After incubation with 40 μl of the magnetic beads (with no primary antibody) in homogenization buffer for 2 h, the preabsorbed microsomal supernatant was diluted with an equal volume of PBS containing 10% skimmed milk and incubated with the primary antibody-bound beads for 2 h at 4 °C. The microsome-bound beads were collected and washed three times with PBS containing 5% skimmed milk and 2 mm EDTA and then twice with PBS containing 2 mm EDTA.
Immunoprecipitation
Mouse CAPS1 cDNA was subcloned in frame in front of the triple HA epitope tag sequence in pEF-BOS to create the C-terminally HA-tagged CAPS1 construct, pEF-BOS-CAPS1-HA. Similarly, mouse ARF family cDNAs were subcloned in frame in front of the triple FLAG epitope tag sequence to create the constructs, pEF-BOS-ARF-FLAG. Lipofectamine 2000 reagent was used to transiently transfect 5 × 105 cells in 6-well plates with 4 μg of pEF-BOS-CAPS1-HA and 1 μg of pEF-BOS-ARF-FLAG plasmids. 48 h after transfection, transfected COS-7 cells were harvested and lysed in 1.3 ml of lysis buffer (50 mm HEPES, pH 7.4, 10% glycerol, 100 mm NaCl, 1 mm CaCl2, 0.5 mm MgCl2, and 0.3% Triton X-100) containing a mixture of protease inhibitors. When required for the CAPS1-ARF binding assay, 10 μm GDP or 10 μm GTPγS was included in the lysis buffer (see Fig. 3D). After preabsorption with protein A-Sepharose, the supernatants were divided equally into two tubes and incubated with 0.5 μg of anti-HA or anti-FLAG antibody, and the immunocomplexes were then associated with protein A-Sepharose resins. The resins were washed five times with lysis buffer, and the bound proteins were separated on an SDS-polyacrylamide gel and transferred to a nitrocellulose membrane for analysis with anti-HA or anti-FLAG antibodies.
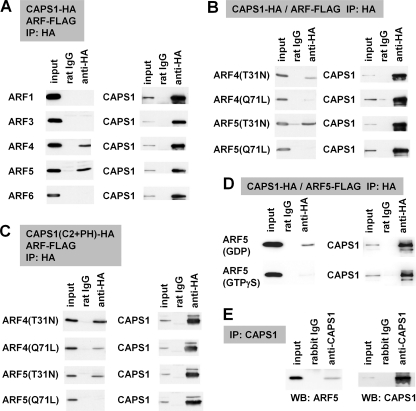
FIGURE 3.
CAPS1 interacts with GDP-locked class II ARF4/5. A–D, protein-protein interaction between CAPS1-HA constructs and ARF-FLAG constructs coexpressed in COS-7 cells was analyzed by coimmunoprecipitation (IP) with anti-HA antibody followed by immunoblotting (WB) with anti-FLAG and anti-HA antibodies. A, coimmunoprecipitation of CAPS1-HA with FLAG-tagged ARF1, ARF3, ARF4, ARF5, and ARF6; B, coimmunoprecipitation of CAPS1-HA with the GDP-locked form (T31N) and the GTP-locked form (Q71L) of ARF4 and ARF5; C, coimmunoprecipitation of the HA-tagged C2 and PH domain of CAPS1 (CAPS1(C2+PH)-HA) with the GDP-locked form (T31N) and GTP-locked form (Q71L) of ARF4-FLAG and ARF5-FLAG; D, coimmunoprecipitation of CAPS1-HA with ARF5-FLAG in the presence of GDP or GTPγS in lysis and assay buffers. E, protein-protein interaction between CAPS1 and ARF5 in mouse cerebellum in vivo. Endogenous ARF5 was coimmunoprecipitated with endogenous CAPS1 by anti-CAPS1 antibody from cerebellar lysates of P21 mice. The blots were immunostained for ARF5 (left) and CAPS1 (right).
GST Pull-down Assay
The purified GST-fused CAPS1 derivatives and MBP-fused ARF5 protein were incubated with glutathione-Sepharose 4B beads (GE Healthcare) in binding buffer (50 mm HEPES-KOH, pH 7.4, 100 mm NaCl, 0.5 mm MgCl2, 2 mm EGTA, 10% glycerol, 0.3% Triton X-100, and 10 μm GDP) for 1 h at room temperature. After washing five times with 1 ml of binding buffer, proteins trapped within the beads were analyzed by Western blotting.
RESULTS
CAPS1 Is Associated with the Golgi Membrane via the PH Domain
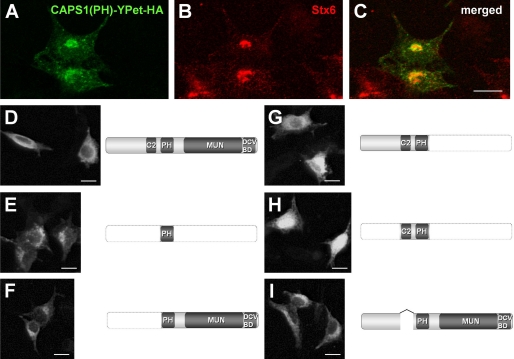
CAPS1 consists of four major protein domains: a C2 domain, a PH domain, a Munc13-1 homology domain, and a DCV binding domain (2). An exogenously expressed PH domain (a fusion construct with fluorescent protein YPet) (Fig. 1A) was colocalized with Stx6 (syntaxin 6) (Fig. 1B) in PC12 cells (Fig. 1C) (14). This suggested involvement of the PH domain in the association of CAPS1 with the trans-Golgi. To elucidate subcellular function of somal CAPS1, we analyzed the relationship between these protein domains and subcellular localization by expressing a series of domain-specific truncation mutants fused with YPet and HA tag in the PC12 neuroendocrine cell line. Full-length CAPS1 exogenously expressed in PC12 cells was distributed diffusely throughout the cytoplasm, but its large fraction was concentrated around the Golgi apparatus (Fig. 1D). The PH domain alone was localized predominantly in Golgi-like structures (Fig. 1E). The N-terminally truncated protein spanning from the PH domain to the C terminus (ΔN-Ter) showed a Golgi-like accumulation pattern (Fig. 1F), similar to that of the PH domain alone (Fig. 1E). However, both the C-terminally truncated protein spanning from the N terminus to the PH domain (ΔC-Ter) (Fig. 1G) and the N/C-terminal truncated protein containing the C2 and PH domains (ΔN/C-Ter) (Fig. 1H) were distributed diffusely throughout the cytoplasm. By contrast, the C2 domain-deleted protein (ΔC2) exhibited a Golgi-like accumulation pattern (Fig. 1I). There were slight differences in the expression levels of these constructs in PC12 cells (supplemental Fig. S1). These results suggest that CAPS1 associates with the Golgi complex via the PH domain and that the CAPS1-Golgi association is controlled by the C2 domain.
FIGURE 1.
CAPS1 is associated with the Golgi membrane via the PH domain. A–C, accumulated expression of CAPS1(PH)-YPet-HA (the PH domain of CAPS1 C-terminally fused to the fluorescent protein YPet and the HA epitope tag) around Stx6-immunopositive Golgi structures of PC12 cells. A, CAPS1(PH)-YPet-HA (green); B, Stx6 (red); C, merged image. Scale bars, 20 μm. D–I, subcellular distribution of six deletion derivatives of CAPS1-YPet-HA expressed in PC12 cells; immunostaining for HA. D, wild-type CAPS1-YPet-HA; E, PH domain alone (PH)-YPet-HA; F, PH domain and Munc13-1-homologous domain (ΔN-Ter)-YPet-HA; G, the region from the first amino acid to the PH domain (ΔC-Ter)-YPet-HA; H, C2 and PH domain (ΔN/C-Ter)-YPet-HA; I, C2 domain-skipped CAPS1 (ΔC2)-YPet-HA. Scale bars, 20 μm.
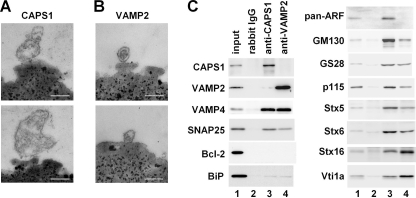
To investigate whether endogenous CAPS1 associates with the Golgi membrane, we tried purifying the CAPS1-associated membrane fraction from the microsomal fraction of mouse cerebella (postnatal day 21) using CAPS1 antibody-coated magnetic beads. Electron microscopic examination of the affinity-purified fraction revealed the presence of membranous structures bound to CAPS1 affinity beads (Fig. 2A) but not to normal rabbit IgG-coated magnetic beads (data not shown). It was noteworthy that the membranous structures bound to the CAPS1 affinity beads tended to be larger than the vesicles bound to VAMP2 (a synaptic vesicle marker) affinity beads (Fig. 2B). Western blot analysis showed that various Golgi marker proteins (GM130, GS28, p115, Stx5, Stx6, Stx16, Vti1a, and VAMP4) were included in the membranous fractions purified with CAPS1 affinity beads (Fig. 2C). Moreover, the vesicle trafficking-related proteins SNAP25 and ARFs were detected in the CAPS1 affinity fractions. The mitochondrial protein Bcl-2 was hardly detected in the CAPS1 affinity fraction, and only a trace amount of BiP/GRP78, a marker of the endoplasmic reticulum, was present in this crude fraction (Fig. 2C), indicating that the CAPS1 affinity method concentrated the Golgi membranes. These results indicated that some of the endogenous CAPS1 indeed associates with the Golgi membrane.
FIGURE 2.
Endogenous CAPS1 is associated with the Golgi membrane. A and B, electron micrograms of submicrosomal fractions affinity-purified using immune magnetic beads. Cerebellar microsomal fractions of P21 mice were incubated with magnetic beads coated with anti-CAPS1 antibody (A) or anti-VAMP2 antibody (B), and immunoaffinity-purified submicrosomal fractions were subjected to electron microscopic observation. Scale bars, 200 nm. C, Western blot analysis of submicrosomal fractions affinity-purified using normal rabbit IgG, anti-CAPS1 antibody, or anti-VAMP2 antibody immunomagnetic beads. Each immunopurified fraction was immunoblotted with anti-CAPS1, anti-VAMP2, anti-VAMP4, anti-SNAP25, anti-Bcl-2, anti-BiP/GRP78, anti-pan-ARF (1D9), anti-GM130, anti-GS28, anti-p115, anti-Stx5, anti-Stx6, anti-Stx16, and anti-Vti1a antibodies. Anti-pan-ARF (1D9) antibody recognizes all of the ARF family proteins (supplemental Fig. S3).
CAPS1 Interacts with the GDP-bound Form of Class II ARFs
Most PH domains of various PH-containing proteins have been shown to localize to the plasma membrane. However, the PH domains of several proteins, including oxysterol-binding protein, Goodpasture antigen-binding protein, and phosphatidylinositol 4-phosphate adaptor protein, have the ability to target the Golgi membrane (27–29). An interaction between the PH domain of these three proteins and phosphatidylinositol 4-phosphate or the ARF1 small GTPase, a regulator of membrane trafficking, is the common mechanism underlying their targeting to the Golgi membrane (28, 29). Membrane fractions that were affinity-purified using anti-CAPS1 antibody contained proteins of the ARF family (Fig. 2C). Therefore, we investigated the possibility that CAPS1 binds ARFs. All members of the ARF family of proteins (class I ARF1 to -3, class II ARF4 and ARF5, and class III ARF6) were expressed ubiquitously in the mouse brain (supplemental Fig. S2). Notably, among all of these ARFs, only the class II ARFs (ARF4 and ARF5) were coimmunoprecipitated with CAPS1 (Fig. 3A).
Similar to other small GTPases, such as Ras, the T31N (Thr31 → Asn) and Q71L (Gln71 → Leu) substitution mutants of ARF4/5 have been shown to give rise to a GDP-bound conformation (30) and a GTP-bound conformation (31), respectively. Interestingly, CAPS1 preferentially coimmunoprecipitated with the GDP-locked form (T31N) of ARF4/5 (Fig. 3B), and the region encompassing the C2 and PH domains alone (C2 + PH) continued to retain this coimmunoprecipitation ability (Fig. 3C). Moreover, the preference of CAPS1 to bind to the GDP-ARF5 form was revealed by coimmunoprecipitation of CAPS1 and ARF5 from cell lysates in GDP-containing lysis buffer but not in GTPγS-containing lysis buffer (Fig. 3D). To confirm the interaction between CAPS1 and the class II ARFs in vivo, we generated an ARF5-specific antibody (supplemental Fig. S3) and carried out coimmunoprecipitation of endogenous CAPS1 from cell lysates of mouse cerebellum. The results showed that endogenous ARF5 was coimmunoprecipitated with CAPS1 (Fig. 3E). These findings indicate that CAPS1 interacts with class II ARF4/5 via the region containing the C2 and PH domains, depending on the GDP-binding state of ARF4/5.
Protein-Protein Interaction between the PH Domain of CAPS1 and the N-terminal Region of ARF5
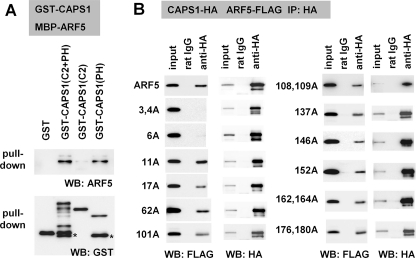
We also examined the structural and functional properties of the CAPS1-ARF5 interaction. Pull-down experiments using the bacterially expressed fusion proteins GST-CAPS1 and MBP-ARF5 indicated that CAPS1 interacts directly with ARF5 via the PH domain but not the C2 domain (Fig. 4A). We then analyzed the ability of CAPS1 to bind to each of 12 GDP-locked ARF5(T31N) mutants that had single or double amino acid substitutions of 16 residues that were identical only in class II ARF4/5 and not in the other classes of ARFs (supplemental Fig. S4). Among the mutants with Ala residue substitutions of class II-specific residues, those with substitutions of the N-terminal Leu, Thr, and Ser, located at the third, fourth, and sixth positions, respectively (L3A, T4A, and S6A), lacked CAPS1 binding activity in coimmunoprecipitation tests (Fig. 4B). These results suggest that the N-terminal region of ARF5 is critical for CAPS1 binding (supplemental Fig. S4).
FIGURE 4.
The PH domain of CAPS1 binds to the N-terminal region of ARF5. A, in vitro binding assay using bacterially expressed recombinant proteins reveals the involvement of the PH domain of CAPS1 in binding to ARF5. MBP-tagged ARF5 protein was pulled down by glutathione-Sepharose beads on which GST, GST-tagged C2 and PH domain (C2 + PH), C2 domain (C2), and PH domain (PH) proteins were immobilized, followed by Western blot analysis (IB) with anti-ARF5 (top) and anti-GST (bottom) antibodies. B, ARF5 binds CAPS1 via the N-terminal region. The region of ARF5 that binds to CAPS1 was screened by generating a series of Ala substitutions in ARF5(T31N)-FLAG at 16 amino acid positions, 3, 4, 6, 11, 17, 62, 101, 108, 109, 137, 146, 152, 162, 164, 176, and 180, all of which are unique to class II ARF4/5 among the ARF family of proteins. CAPS1-HA and Ala-substituted ARF5(T31N)-FLAG constructs were coexpressed in COS-7 cells, and cell lysates were subjected to coimmunoprecipitation (IP) with anti-HA antibody followed by Western blot analysis with anti-FLAG (top) and anti-HA (bottom) antibodies.
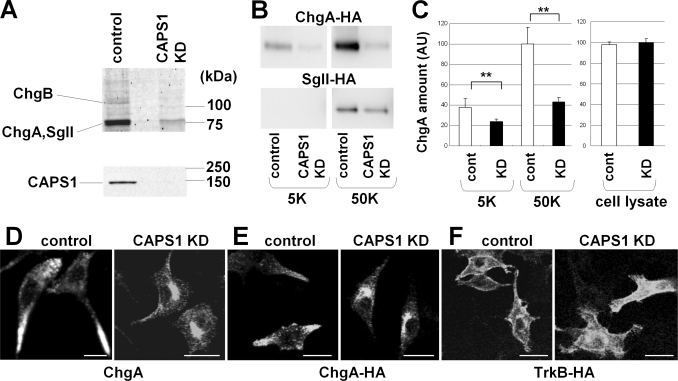
CAPS1 KD Induces Chromogranin Accumulation in the Golgi Complex
To investigate the role of somal CAPS1 in the Golgi function, we examined the KD effect of CAPS1 expression by transfecting specific CAPS1 siRNAs. CAPS1 KD in PC12 cells, in which CAPS1 is expressed and CAPS2 is not (5), resulted in the levels of CAPS1 becoming almost undetectable by Western blot analysis (Fig. 5A, bottom). We then compared the protein components of culture media from control and CAPS1 KD cultures using SDS-PAGE analysis, followed by tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS). As a result, three granin family proteins, chromogranin A, chromogranin B, and secretogranin II, all of which are DCV marker proteins released by DCV secretion, were identified to be dramatically decreased in the culture media of the CAPS1 KD cells (Fig. 5A, top). To confirm the reduction of granin protein secretion by CAPS1 KD cells, HA-tagged chromogranin A (ChgA-HA) or HA-tagged secretogranin II (SgII-HA) was co-transfected with CAPS1 siRNA into PC12 cells. High KCl (50 mm) treatment significantly enhanced the amount of ChgA-HA released into the media of control cultures, compared with treatment with a normal KCl (5 mm) concentration (Fig. 5, B and C). However, CAPS1 KD decreased both the spontaneous release and the high KCl-induced release of ChgA-HA (Fig. 5, B and C). SgII-HA release was also reduced in CAPS1 KD cultures (Fig. 5B). To determine whether CAPS1 KD impairs DCV exocytosis, DCV trafficking, or DCV biogenesis from the Golgi complex, we analyzed subcellular localization of ChgA in KD cells. CAPS1 KD altered the subcellular distribution of endogenous ChgA in PC12 cells; ChgA was localized largely in the processes of the control cells, whereas it accumulated in Golgi-like structures in the CAPS1 KD cells (Fig. 5D). Abnormal accumulation of exogenously expressed ChgA-HA was also observed in Golgi-like structures in the CAPS1 KD cells, in marked contrast to its predominant distribution in processes in the control cells (Fig. 5E). By contrast, transmembrane proteins, including neurotrophin receptors TrkA (data not shown) and TrkB (Fig. 5F), which are not transported by DCVs, were normally targeted to the cell surface of the CAPS1 KD cells. These results implicate somal CAPS1 in DCV trafficking in the Golgi complex, and thus CAPS1 KD has a consequential effect on DCV secretion of granins into the culture media.
FIGURE 5.
CAPS1 knockdown induces chromogranin accumulation in the Golgi complex. A, KD of CAPS1 expression in PC12 cells by siRNA. siRNA specific for CAPS1 was electroporated into PC12 cells, and the culture medium was concentrated and electrophoresed. Top, three bands were greatly reduced in the culture medium of the CAPS1 KD cells and were identified as chromogranin A (ChgA), chromogranin B (ChgB), and secretogranin II (SgII) by LC-MS/MS. Bottom, reduction of CAPS1 expression in PC12 cells by siRNA KD was assayed by immunoblotting of cell lysates with anti-CAPS1 antibody. B, high KCl-induced chromogranin A and secretogranin II release from CAPS1 KD PC12 cells. PC12 cells were transfected with either control siRNA or CAPS1 siRNA together with a ChgA-HA or SgII-HA expression plasmid. The culture media, treated with 5 mm KCl (5K) or 50 mm KCl (50K) (see “Experimental Procedures”), were collected and analyzed by immunoblotting with anti-HA antibody. C, statistical analyses of the effect of CAPS1 KD on ChgA-HA release from PC12 cells. PC12 cells were transfected with ChgA-HA together with the control (white bar) or CAPS1 siRNA (black bar). The amounts of ChgA-HA released into the culture media with 5 or 50 mm KCl stimulation were analyzed by Western blotting, followed by densitometric analysis (n = 6). There was no significant difference in the amount of ChgA-HA in cell lysates between the control and CAPS1 KD groups (n = 6). The signal intensities of the extracellular ChgA-HA bands of the culture media were normalized against those of the intracellular ChgA-HA bands of the cell lysates. AU, arbitrary unit. Error bars, S.E. **, p < 0.01 by Student's t test. D, subcellular localization of endogenous chromogranin A in NGF-differentiated PC12 cells without (left) and with (right) CAPS1 KD; immunostaining for chromogranin A. Scale bars, 20 μm. E, subcellular localization of ChgA-HA expressed in NGF-differentiated PC12 cells without (left) and with (right) CAPS1 KD; immunostaining for HA. Scale bars, 20 μm. F, subcellular localization of C-terminal HA-tagged TrkB expressed in NGF-differentiated PC12 cells without (left) and with (right) CAPS1 KD; immunostaining for HA. Scale bars, 20 μm.
The Interaction between CAPS1 and Class II ARFs Is Indispensable to DCV Trafficking
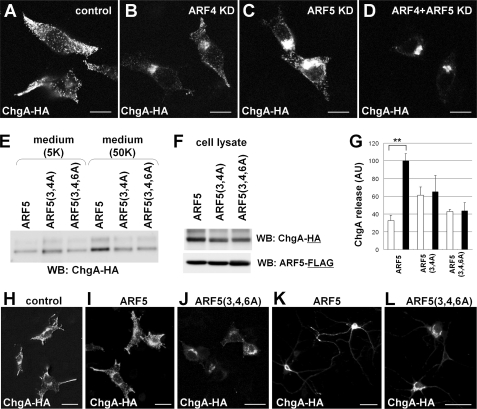
Class I ARFs are known to regulate membrane trafficking (19, 32). To examine whether ARF4/5 plays a role in DCV-Golgi trafficking, we analyzed the KD effects of ARF4/5 expression on the subcellular distribution pattern of ChgA-HA in PC12 cells (Fig. 6, A–D). Exogenously expressed ChgA-HA was densely localized in the tips of processes of control PC12 cells, whereas KD of either ARF4 or ARF5 resulted in stacking of ChgA-HA in Golgi-like structures (Fig. 6, B and C), which was also observed in CAPS1 KD cells (Fig. 5, D and E). Furthermore, when ARF4 and ARF5 were knocked down simultaneously (rather than separately), a greater quantity of ChgA-HA accumulated in Golgi structures (Fig. 6D). These results suggest that ARF4/5 plays a role in DCV-Golgi trafficking.
FIGURE 6.
Subcellular distribution and release of chromogranin is disrupted by either ARF4/5 siRNA knockdown or expression of CAPS1-binding-deficient ARF5. A–D, HA-tagged chromogranin (ChgA-HA) expressed in PC12 cells showed a Golgi-accumulated pattern following KD of ARF4/5 expression. Shown are immunocytochemical staining patterns of control (A), ARF4 KD (B), ARF5 KD (C), and ARF4/5 double KD (D) cells with anti-HA antibody. ARF4 and ARF5 siRNAs efficiently reduced the expression levels of ARF4 and ARF5 proteins, respectively (supplemental Fig. S3). Scale bar, 10 μm. E–G, ARF5(3,4A) and ARF5(3,4,6A) had a disrupted N-terminal CAPS1 binding site and showed a decrease in regulated release of ChgA-HA from PC12 cells. ChgA-HA and one of the ARF5 constructs (wild-type ARF5, ARF5(3,4A), or ARF5(3,4,5A)) were transfected into PC12 cells. Culture medium after treatment with 5 mm KCl (5K) or 50 mm KCl (50K) was examined by Western blot analysis with anti-HA antibody (E), and the cell lysates were analyzed by Western blotting (WB) with anti-HA antibody or anti-FLAG antibody (F). G, statistical analyses of the effect of ARF5 and CAPS1 binding-deficient ARF5 on ChgA-HA release from PC12 cells. PC12 cells were transfected with ChgA-HA together with wild-type ARF5, CAPS1 binding-deficient ARF5(3,4A), or ARF5(3,4,6A). The amount of ChgA-HA released into the culture media with 5 mm KCl (white bar) or 50 mm KCl (black bar) stimulation was analyzed by Western blotting followed by densitometric analysis (n = 4). The signal intensities of the extracellular ChgA-HA bands from the culture media were normalized against those of the intracellular ChgA-HA bands from the cell lysates. AU, arbitrary unit. Error bars, S.E. **, p < 0.01 by Student's t test. H–J, CAPS1 binding-deficient ARF5(3,4,6A) coexpressed in PC12 cells induces accumulation of ChgA-HA in the Golgi. Immunocytochemical staining with anti-HA antibody indicates that ChgA-HA is localized in the tips of processes and around the nuclei in control cells (H) and cells coexpressing wild-type ARF5 (I) but is accumulated in the Golgi in cells coexpressing ARF5(3,4,6A) (J). Scale bar, 25 μm. K and L, CAPS1 binding-deficient ARF5(3,4,6A) coexpressed in primary cultured hippocampal neurons (DIV8) induces accumulation of ChgA-HA in the Golgi. Immunocytochemistry with anti-HA antibody shows that expressed ChgA-HA is localized in neurites and in soma of cells coexpressing wild-type ARF5 (K), whereas it is accumulated in the Golgi in cells coexpressing ARF5(3,4,6A) (L). Scale bar, 50 μm.
To investigate whether the association between CAPS1 and class II ARFs has a role in DCV secretion, we compared high KCl-induced ChgA-HA release into the culture media between PC12 cells expressing wild-type ARF5 and PC12 cells with CAPS1 binding-deficient ARF5(3,4A) or ARF5(3,4,6A) (Fig. 6, E–G). The amount of ChgA-HA released into the culture medium by cells expressing wild-type ARF5 was increased about 3-fold in 50 mm KCl as compared with 5 mm KCl, whereas the release of ChgA-HA by cells expressing ARF5(3,4A) or ARF5(3,4,6A) was almost indistinguishable between those treated with 5 and 50 mm KCl (Fig. 6, E and G). The amount of intracellular ChgA-HA detected in cell lysates was slightly decreased by the expression of ARF5(3,4A) or ARF5(3,4,6A) compared with wild-type ARF5 (Fig. 6F). These results suggest that CAPS1-ARF5 association is required for efficient DCV secretion.
We then analyzed whether the CAPS1-ARF5 association affects the subcellular localization of exogenous ChgA-HA in PC12 cells. Coexpression of wild-type ARF5 induced no obvious change in the subcellular distribution of ChgA-HA (Fig. 6I). When coexpressed with ARF5(3,4,6A), ChgA-HA accumulated in the Golgi complex (Fig. 6J), as was observed in CAPS1 KD cells (Fig. 5, D and E) and ARF4/5 KD cells (Fig. 6D). Similarly, coexpression of ARF5(3,4,6A) in primary cultured hippocampal cells altered the somal distribution pattern of ChgA-HA from a diffuse distribution throughout the cytoplasm (Fig. 6K) to a Golgi-like accumulation (Fig. 6L). Taken together, these results suggest that the association between CAPS1 and ARF5 plays a role in trafficking chromogranin-containing DCVs in the Golgi network.
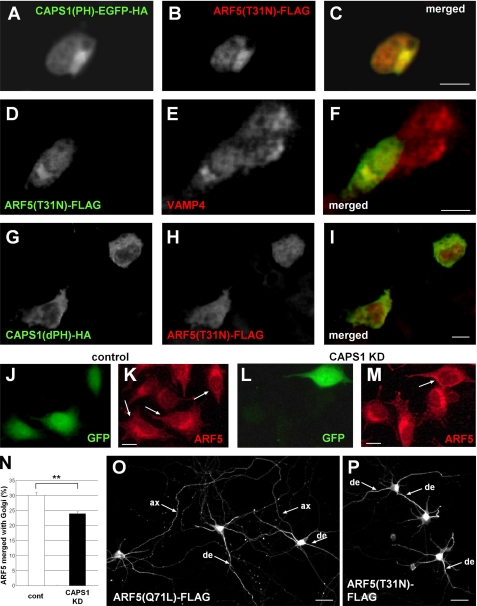
The GDP-bound Form of ARF5 Localizes to the Golgi Complex, Depending on the Interaction with the PH Domain of CAPS1, and Is Not Transported to Axons of Hippocampal Neurons
A previous study showed that GDP-locked class II ARFs have a cytoplasmic, diffuse distribution pattern, whereas GTP-locked class II ARFs associate with the Golgi membrane (33). We investigated whether these guanine nucleotide-dependent distribution patterns of GDP-locked ARF5(T31N) were altered by coexpression of CAPS1 constructs in PC12 cells (Fig. 7, A–I). Coexpression of the CAPS1 PH domain alone (CAPS1(PH)) changed the distribution of ARF5(T31N) from a diffuse cytosolic pattern to a localized pattern (Fig. 7B), which was merged with the Golgi marker protein VAMP4 (Fig. 7F). On the other hand, coexpression of CAPS1 lacking the PH domain (CAPS1(dPH)) resulted in a diffuse cytoplasmic distribution pattern of ARF5(T31N) (Fig. 7H). We next examined whether CAPS1 KD affected the ARF5 localization in the Golgi membrane (Fig. 7, J–M). Endogenous ARF5 was localized in the Golgi in cells transfected with control siRNA (Fig. 7K). However, endogenous ARF5 showed a diffuse cytosolic pattern in KD cells transfected with CAPS1 siRNA (Fig. 7M). These data suggest that GDP-locked ARF5 is recruited to the Golgi complex by binding to CAPS1 via the PH domain. To quantify Golgi localization, PC12 cells were transfected with the fluorescent Golgi marker construct (Golgi marker B4galt1 fused to tdTomato) together with either control or CAPS1 siRNA. ARF5 immunoreactivity merged with immunoreactivities for the Golgi complex and whole-cell soma images was quantified (Fig. 7N). The results showed a modest effect and suggested that CAPS1 induces accumulation of ARF4/5 in the Golgi complex probably by interaction between CAPS1 and ARF4/5.
FIGURE 7.
The binding between the PH domain of CAPS1 and ARF5 induces the accumulation of GDP-locked ARF5 in the Golgi membrane. A–F, subcellular localization of the C-terminal EGFP-HA-tagged CAPS1(PH), the C-terminal FLAG-tagged GDP-locked ARF5(T31N), and endogenous VAMP4 in PC12 cells. A–C, immunostaining of CAPS1(PH)-EGFP-HA and ARF(T31N)-FLAG with anti-HA (A) and anti-FLAG (B) antibodies, respectively, and the merged image (C). D–F, immunostaining of ARF5(T31N)-FLAG and endogenous VAMP4 with anti-FLAG (D) and anti-VAMP4 (E) antibodies, respectively, and the merged image (F). Scale bars (C and F), 10 μm. G–I, subcellular localization of the C-terminal HA-tagged CAPS1(dPH) and the C-terminal FLAG-tagged ARF5(T31N) in PC12 cells. Immunostaining of CAPS1(dPH)-HA and ARF5(T31N)-FLAG with anti-HA (G) and anti-FLAG (H) antibodies, respectively, and the merged image (I). Scale bars, 10 μm. J–M, subcellular localization of endogenous ARF5 in PC12 cells transfected with EGFP together with either control (J and K) or CAPS1 siRNA (L and M). The arrows indicate EGFP-expressing siRNA-transfected cells. Shown is immunostaining of endogenous ARF5 with anti-ARF5 antibody (K and L). Scale bars (K and M), 10 μm. N, ARF5 immunoreactivity levels merged with immunoreactivity for the Golgi complex. PC12 cells were transfected with the Golgi marker B4galt1-tdTomato together with either control (white bar) or CAPS1 siRNA (black bar). The ratio of ARF5 immunoreactivity in the Golgi complex (merged with B4galt1-tdTomato fluorescence) and that in whole cell soma was qualified. **, p < 0.01 by Student's t test. Error bars, S.E. O and P, subcellular distribution of GTP-locked ARF5(Q71L)-FLAG (O) and GDP-locked ARF5(T31N)-FLAG (P) expressed in primary cultured mouse hippocampal neurons. Cultures were transfected at 6 days in vitro, fixed at 8 days in vitro, and immunostained with anti-FLAG antibody. ax, axon; de, dendrite. Scale bar, 50 μm.
Finally, we compared the subcellular localization of GDP-locked and GTP-locked ARF5 exogenously expressed in primary cultured hippocampal neurons. Both ARF5 forms were highly expressed in the neuronal soma, where the Golgi complex is located. Interestingly, the GTP-locked ARF5 was distributed in axons and dendrites, in addition to the soma (Fig. 7O), whereas the GDP-locked ARF5 was transported to dendrites but not to axons (Fig. 7P). Although the biological significance of this differential localization between GDP-locked and GTP-locked ARF5 in hippocampal neurons remains unclear, the function of CAPS1 may be regulated differently for these subcellular compartments of neurons (soma-dendrites versus axons), depending on the localization of GDP-bound class II ARFs.
DISCUSSION
Our findings indicate that CAPS1 interacts with class II ARF4/5 in a GDP/GTP state-dependent manner and that CAPS1 is involved in DCV trafficking in or from the trans-Golgi network. We showed that CAPS1 has the ability to bind GDP-bound, but not GTP-bound, ARF4/5 and the Golgi membrane. ARF4/5 in the GDP-bound state is recruited to the Golgi complex by interaction of its N-terminal domain with the PH domain of CAPS1. This recruitment is indispensable to DCV trafficking for the following reasons: 1) overexpression of CAPS1 binding-deficient ARF5 mutants induced accumulation of ChgA in the Golgi, with a consequent reduction in ChgA release; 2) CAPS1 KD resulted in the accumulation of ChgA in the Golgi network and, thereby, a reduction in ChgA release; and 3) ARF4/5 KD caused abnormal ChgA accumulation in the Golgi network. Collectively, these findings suggest that CAPS1 is required for DCV trafficking in the trans-Golgi network by recruiting class II ARF small GTPases, in a GDP-bound dependent manner, to the Golgi membrane. Because ARF1 reportedly induces vesicle budding in vitro by regulating membrane curvature (34, 35), the class II ARFs may also have a role, in concert with CAPS1, in regulating DCV budding in the trans-Golgi network.
The PH domain of CAPS1 is essential for its accumulation in the Golgi. On the other hand, the C2 domain of CAPS1 appears to have a regulatory role in this subcellular localization, because the N/C-terminal truncated protein containing the C2 and PH domains was distributed throughout the cell soma. We also defined the structural determinants of the CAPS1-ARF4/5 interaction; the PH domain of CAPS1 and the N-terminal region of ARF4/5 interact in a GDP-dependent manner. In addition, we confirmed by substitution of the myristoylation consensus site Gly2 by Ala that the N-terminal myristoylation state of ARF4/5 does not affect CAPS1 interaction (data no shown). These results suggest that the N-terminal region of class II ARFs binds CAPS1 independently of the Golgi membrane anchoring via myristoylation.
The members of the ARF family of proteins are thought to be regulators of membrane trafficking. Various regulators and effectors of the class I and III ARFs have already been identified (16, 19, 36, 37). On the other hand, class II ARF-specific regulators or effectors have not been reported, although previous studies have shown that ARF4 bound Munc18-interacting proteins, which contributed to vesicle trafficking of Alzheimer precursor protein β-APP (38, 39), and that ARF4 bound the epidermal growth factor receptor, which in turn activated PLD2, leading to transcriptional regulation (38, 39). The present study demonstrates that CAPS1 is a novel type of class II ARF target. All ARF guanine nucleotide exchange factors identified to date possess a Sec7 domain, a module of ∼200 amino acids that is sufficient to catalyze the exchange of GDP for GTP on ARF in vitro (16). However, CAPS1 contains no regions that are homologous with the Sec7 domain, and it has no detectable level of guanine nucleotide exchange factor activity (data not shown). Neither CAPS nor class II ARF orthologs are present in Saccharomyces cerevisiae, but both CAPS and class II ARF orthologs are found in Drosophila melanogaster and Caenorhabditis elegans, as well as in vertebrates (37). Thus, the interaction between CAPS and class II ARFs presumably is required for fine regulation of DCV trafficking in multicellular organisms.
Previous studies have shown that CAPS1 is involved in the priming step of DCV exocytosis (8, 9). However, in most cell types (but not hippocampal DG granule cells) (13), the largest amount of endogenous CAPS1 protein is localized in the cytoplasm of the soma rather than in the plasma membrane region near secretion sites. The major role of the cytosolic CAPS proteins present in the cell soma has yet to be determined. The results of this study shed light on another important function of CAPS1 protein in the cell soma, in addition to its regulatory function at DCV secretion sites; CAPS1, together with the class II ARFs, is involved in DCV trafficking in the trans-Golgi network in the cell soma. We therefore suggest that CAPS1 has two functions in the DCV secretion pathway in neurons and neuroendocrine cells: a somal function that regulates DCV-Golgi trafficking and a synaptic (or secretion site-specific) function that regulates DCV exocytosis. In this respect, it seems noteworthy that the GDP-bound ARF5, which is capable of binding to CAPS1, is localized in the soma of cultured hippocampal neurons but is not transported into axons or terminals, where synaptic DCV exocytosis occurs, suggesting that the ARF4/5-CAPS1 interaction is required for the somal function of CAPS1 but not for its synaptic function. Thus, we suggest that the two functions of CAPS1 depend on the presence of its binding partner, GDP-bound class II ARFs, although the basic properties of CAPS1 underlie both functions.
Supplementary Material
Acknowledgments
We are grateful to Dr. Hiroki Inoue (Tokyo University of Pharmacy and Life Sciences) for technical advice and helpful suggestions. We are also grateful to Dr. Akihiko Nakano (University of Tokyo) for fruitful discussions and comments. We thank Dr. Shigekazu Nagata for graciously providing the pEF-BOS expression plasmid. We also thank the Support Unit for Bio-material Analysis, RIKEN BSI Research Resources Center, for help with the MS spectrometry analysis.
This work was supported by grants-in-aid for Scientific Research from the Takeda Science Foundation; the Naito Foundation; the Mochida Memorial Foundation for Medical and Pharmaceutical Research; the Kao Foundation for Arts and Sciences; the Life Science Foundation of Japan; the Japanese Ministry of Education, Culture, Sports, Science, and Technology (Grant 21700421), the Japan Science and Technology Agency; the Japan Society for the Promotion of Science; and the Institute of Physical and Chemical Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- CAPS
- Ca2+-dependent activator protein for secretion
- DCV
- dense-core vesicle
- KD
- knockdown
- ARF
- ADP-ribosylation factor
- MBP
- maltose-binding protein
- PH
- pleckstrin homology
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- EGFP
- enhanced green fluorescent protein
- ARF5(3,4A)
- ARF5(L3A,T4A)
- ARF5(3,4,6A)
- ARF5(L3A,T4A,S6A).
REFERENCES
- 1.Renden R., Berwin B., Davis W., Ann K., Chin C. T., Kreber R., Ganetzky B., Martin T. F., Broadie K. (2001) Neuron 31, 421–437 [DOI] [PubMed] [Google Scholar]
- 2.Berwin B., Floor E., Martin T. F. (1998) Neuron 21, 137–145 [DOI] [PubMed] [Google Scholar]
- 3.Tandon A., Bannykh S., Kowalchyk J. A., Banerjee A., Martin T. F., Balch W. E. (1998) Neuron 21, 147–154 [DOI] [PubMed] [Google Scholar]
- 4.Speidel D., Varoqueaux F., Enk C., Nojiri M., Grishanin R. N., Martin T. F., Hofmann K., Brose N., Reim K. (2003) J. Biol. Chem. 278, 52802–52809 [DOI] [PubMed] [Google Scholar]
- 5.Sadakata T., Mizoguchi A., Sato Y., Katoh-Semba R., Fukuda M., Mikoshiba K., Furuichi T. (2004) J. Neurosci. 24, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita Y., Xu A., Xie L., Arunachalam L., Chou T. C., Jiang T., Chiew S. K., Kourtesis J., Wang L., Gaisano H. Y., Sugita S. (2007) J. Biol. Chem. 282, 21392–21403 [DOI] [PubMed] [Google Scholar]
- 7.Speidel D., Salehi A., Obermueller S., Lundquist I., Brose N., Renström E., Rorsman P. (2008) Cell Metab. 7, 57–67 [DOI] [PubMed] [Google Scholar]
- 8.Grishanin R. N., Kowalchyk J. A., Klenchin V. A., Ann K., Earles C. A., Chapman E. R., Gerona R. R., Martin T. F. (2004) Neuron 43, 551–562 [DOI] [PubMed] [Google Scholar]
- 9.Liu Y., Schirra C., Stevens D. R., Matti U., Speidel D., Hof D., Bruns D., Brose N., Rettig J. (2008) J. Neurosci. 28, 5594–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jockusch W. J., Speidel D., Sigler A., Sørensen J. B., Varoqueaux F., Rhee J. S., Brose N. (2007) Cell 131, 796–808 [DOI] [PubMed] [Google Scholar]
- 11.Brunk I., Blex C., Speidel D., Brose N., Ahnert-Hilger G. (2009) J. Biol. Chem. 284, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 12.Speidel D., Bruederle C. E., Enk C., Voets T., Varoqueaux F., Reim K., Becherer U., Fornai F., Ruggieri S., Holighaus Y., Weihe E., Bruns D., Brose N., Rettig J. (2005) Neuron 46, 75–88 [DOI] [PubMed] [Google Scholar]
- 13.Sadakata T., Itakura M., Kozaki S., Sekine Y., Takahashi M., Furuichi T. (2006) J. Comp. Neurol. 495, 735–753 [DOI] [PubMed] [Google Scholar]
- 14.Grishanin R. N., Klenchin V. A., Loyet K. M., Kowalchyk J. A., Ann K., Martin T. F. (2002) J. Biol. Chem. 277, 22025–22034 [DOI] [PubMed] [Google Scholar]
- 15.Moss J., Vaughan M. (1998) J. Biol. Chem. 273, 21431–21434 [DOI] [PubMed] [Google Scholar]
- 16.Donaldson J. G., Jackson C. L. (2000) Curr. Opin. Cell Biol. 12, 475–482 [DOI] [PubMed] [Google Scholar]
- 17.Jackson C. L., Casanova J. E. (2000) Trends Cell Biol. 10, 60–67 [DOI] [PubMed] [Google Scholar]
- 18.Nie Z., Hirsch D. S., Randazzo P. A. (2003) Curr. Opin. Cell Biol. 15, 396–404 [DOI] [PubMed] [Google Scholar]
- 19.D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 20.Donaldson J. G. (2002) Methods Mol. Biol. 189, 191–198 [DOI] [PubMed] [Google Scholar]
- 21.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V. B., Brenner S. E., Batalov S., Forrest A. R., Zavolan M., Davis M. J., Wilming L. G., Aidinis V., Allen J. E., Ambesi-Impiombato A., Apweiler R., Aturaliya R. N., Bailey T. L., Bansal M., Baxter L., Beisel K. W., Bersano T., Bono H., Chalk A. M., Chiu K. P., Choudhary V., Christoffels A., Clutterbuck D. R., Crowe M. L., Dalla E., Dalrymple B. P., de Bono B., Della Gatta G., di Bernardo D., Down T., Engstrom P., Fagiolini M., Faulkner G., Fletcher C. F., Fukushima T., Furuno M., Futaki S., Gariboldi M., Georgii-Hemming P., Gingeras T. R., Gojobori T., Green R. E., Gustincich S., Harbers M., Hayashi Y., Hensch T. K., Hirokawa N., Hill D., Huminiecki L., Iacono M., Ikeo K., Iwama A., Ishikawa T., Jakt M., Kanapin A., Katoh M., Kawasawa Y., Kelso J., Kitamura H., Kitano H., Kollias G., Krishnan S. P., Kruger A., Kummerfeld S. K., Kurochkin I. V., Lareau L. F., Lazarevic D., Lipovich L., Liu J., Liuni S., McWilliam S., Madan Babu M., Madera M., Marchionni L., Matsuda H., Matsuzawa S., Miki H., Mignone F., Miyake S., Morris K., Mottagui-Tabar S., Mulder N., Nakano N., Nakauchi H., Ng P., Nilsson R., Nishiguchi S., Nishikawa S., Nori F., Ohara O., Okazaki Y., Orlando V., Pang K. C., Pavan W. J., Pavesi G., Pesole G., Petrovsky N., Piazza S., Reed J., Reid J. F., Ring B. Z., Ringwald M., Rost B., Ruan Y., Salzberg S. L., Sandelin A., Schneider C., Schönbach C., Sekiguchi K., Semple C. A., Seno S., Sessa L., Sheng Y., Shibata Y., Shimada H., Shimada K., Silva D., Sinclair B., Sperling S., Stupka E., Sugiura K., Sultana R., Takenaka Y., Taki K., Tammoja K., Tan S. L., Tang S., Taylor M. S., Tegner J., Teichmann S. A., Ueda H. R., van Nimwegen E., Verardo R., Wei C. L., Yagi K., Yamanishi H., Zabarovsky E., Zhu S., Zimmer A., Hide W., Bult C., Grimmond S. M., Teasdale R. D., Liu E. T., Brusic V., Quackenbush J., Wahlestedt C., Mattick J. S., Hume D. A., Kai C., Sasaki D., Tomaru Y., Fukuda S., Kanamori-Katayama M., Suzuki M., Aoki J., Arakawa T., Iida J., Imamura K., Itoh M., Kato T., Kawaji H., Kawagashira N., Kawashima T., Kojima M., Kondo S., Konno H., Nakano K., Ninomiya N., Nishio T., Okada M., Plessy C., Shibata K., Shiraki T., Suzuki S., Tagami M., Waki K., Watahiki A., Okamura-Oho Y., Suzuki H., Kawai J., Hayashizaki Y. (2005) Science 309, 1559–156316141072 [Google Scholar]
- 22.Mizushima S., Nagata S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen A. W., Daugherty P. S. (2005) Nat. Biotechnol. 23, 355–360 [DOI] [PubMed] [Google Scholar]
- 24.Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 25.Roth J., Berger E. G. (1982) J. Cell Biol. 93, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoffel W., Jenke B., Blöck B., Zumbansen M., Koebke J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine T. P., Munro S. (1998) Curr. Biol. 8, 729–739 [DOI] [PubMed] [Google Scholar]
- 28.Levine T. P., Munro S. (2002) Curr. Biol. 12, 695–704 [DOI] [PubMed] [Google Scholar]
- 29.Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. (2004) Nat. Cell Biol. 6, 393–404 [DOI] [PubMed] [Google Scholar]
- 30.John J., Rensland H., Schlichting I., Vetter I., Borasio G. D., Goody R. S., Wittinghofer A. (1993) J. Biol. Chem. 268, 923–929 [PubMed] [Google Scholar]
- 31.Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 32.Donaldson J. G., Honda A. (2005) Biochem. Soc. Trans. 33, 639–642 [DOI] [PubMed] [Google Scholar]
- 33.Venkateswarlu K., Cullen P. J. (2000) Biochem. J. 345, 719–724 [PMC free article] [PubMed] [Google Scholar]
- 34.Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., Wieland F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11731–11736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krauss M., Jia J. Y., Roux A., Beck R., Wieland F. T., De Camilli P., Haucke V. (2008) J. Biol. Chem. 283, 27717–27723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randazzo P. A., Hirsch D. S. (2004) Cell. Signal. 16, 401–413 [DOI] [PubMed] [Google Scholar]
- 37.Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 38.Kim S. W., Hayashi M., Lo J. F., Yang Y., Yoo J. S., Lee J. D. (2003) J. Biol. Chem. 278, 2661–2668 [DOI] [PubMed] [Google Scholar]
- 39.Hill K., Li Y., Bennett M., McKay M., Zhu X., Shern J., Torre E., Lah J. J., Levey A. I., Kahn R. A. (2003) J. Biol. Chem. 278, 36032–36040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.