Abstract
Accumulating evidence indicates that endocytosis plays an essential role in the nuclear transport of the ErbB family members, such as epidermal growth factor receptor (EGFR) and ErbB-2. Nevertheless, how full-length receptors embedded in the endosomal membrane pass through the nuclear pore complexes and function as non-membrane-bound receptors in the nucleus remains unclear. Here we show that upon EGF treatment, the biotinylated cell surface EGFR is trafficked to the inner nuclear membrane (INM) through the nuclear pore complexes, remaining in a membrane-bound environment. We further find that importin β regulates EGFR nuclear transport to the INM in addition to the nucleus/nucleoplasm. Unexpectedly, the well known endoplasmic reticulum associated translocon Sec61β is found to reside in the INM and associate with EGFR. Knocking down Sec61β expression reduces EGFR level in the nucleoplasm portion and accumulates it in the INM portion. Thus, the Sec61β translocon plays an unrecognized role in the release of the membrane-anchored EGFR from the lipid bilayer of the INM to the nucleus. The newly identified Sec61β function provides an alternative pathway for nuclear transport that can be utilized by membrane-embedded proteins such as full-length EGFR.
Keywords: Breast Cancer, Intracellular Trafficking, Nuclear Transport, Oncogene, Receptor Tyrosine Kinase
Introduction
Receptor tyrosine kinases (RTKs),3 including insulin-like growth factor 1 receptor, c-Met, fibroblast growth factor receptor, vascular endothelial growth factor receptor, and the entire epidermal growth factor receptor (EGFR) family, have been shown to localize in the nucleus (1–6). Among these, both EGFR and ErbB-2 are suggested to be involved in transcriptional regulation, cell proliferation, DNA repair, DNA replication, and chemo and radio resistance (7–14). Nuclear EGFR is associated with poor clinical prognosis for breast cancer, ovarian cancer, and in oropharyngeal and esophageal squamous cell carcinomas (15–19). In addition, nuclear EGFRvIII, a constitutively activated EGFR variant, is also correlated with poor patient outcome in prostate cancer (20). In the canonical model of nuclear import, nuclear localization signal (NLS)-bearing molecules form a complex with importin α/β or importin β alone. Importin β is responsible for nuclear translocation through nuclear pore complexes (NPCs) by directly associating with the nucleoporins (21). Several studies have shown that importin β and NLS are involved in the nuclear transport of many cell surface RTKs, including EGFR (14, 22–26), ErbB-2 (27, 28), and fibroblast growth factor receptor (29). In addition, nuclear transport of RTKs is mediated by the mechanisms involving endocytosis and endosomal sorting by associating with early endosomal proteins in the nucleus (24, 27). However, the exact mechanisms by which RTKs embedded in the endosomal membrane translocate into the nucleus through NPCs and exist as non-membrane-bound receptors in the nucleus are still largely unknown.
In eukaryotes, the membrane system of the endoplasmic reticulum (ER) is contiguous with the nuclear envelope (NE), a lipid bilayer that forms the boundary of the nucleus and separates the nucleoplasm (NP) from the cytoplasm. The prominent components of the NE are the outer nuclear membrane (ONM) and inner nuclear membrane (INM). The ONM is contiguous and functionally related to the ER membrane, whereas the INM has a protein composition different from that of the ONM and is associated with the underlying chromatin and lamins (30). The spatial connection between these two membranes is provided by the perinuclear space and is joined at the NPCs, which form aqueous channels embedded in the NE. The NPCs regulate the bidirectional trafficking of facilitated transport for macromolecules (>40 kDa) and passive diffusion for ions and small molecules (≤40 kDa) (31, 32). Recently, authors reported that large INM proteins, initially inserted into the ER membrane, travel into the INM through the ONM and NPCs (33–36). In the ER membrane, the heterotrimeric Sec61 complex comprises three transmembrane subunits (Sec61α, Sec61β, and Sec61γ in mammals) and forms protein-conducting channels, collectively termed a translocon (37). Localization of the Sec61 translocon is well documented to be in the ER and ER-Golgi intermediate compartment (38). In addition, the role of the Sec61 translocon in the ER is known to be bidirectional for both protein import, which inserts transmembrane and secretory preproteins into the ER during protein synthesis, and protein export, which retrotranslocates misfolded proteins from the ER to the cytoplasm for degradation as part of the ER-associated degradation (ERAD) pathway (39, 40). Sec61α is known to be stabilized by Sec61γ and mainly responsible for the translocation activity in the ER (41). In contrast to the other two subunits, Sec61β can be stable on its own, and its function is not as well defined (42, 43).
In this study, we found that cell surface EGFR translocates to the INM through the NPCs, which is mediated by importin β. However, unexpectedly, we discovered a previously unrecognized role of Sec61β in the INM, which is required for the release of EGFR from the INM to the nucleus. This pathway may provide a general mechanism for trafficking of membrane-bound proteins, including full-length cell surface receptors, from the cell surface to the nucleus through the nuclear membrane.
EXPERIMENTAL PROCEDURES
Biotinylation of Cell Surface Proteins
Cell surface proteins in MDA-MB-468 cells were biotinylated using 1 mm Sulfo-NHS-LC-Biotin (Pierce) at room temperature for 30 min and then treated with or without EGF (50 ng/ml) at 37 °C for 30 min. The biotinylation reaction was quenched with phosphate-buffered saline (PBS) containing 100 mm glycine.
Cellular Fractionation
For cellular fractionation, non-nuclear and nuclear fractions were prepared as described previously (25). Cells were lysed in Lysis buffer (20 mm HEPES, pH 7.0, 10 mm KCl, 2 mm MgCl2, 0.5% Nonidet P-40, protease inhibitor mixture). After incubation on ice for 10 min, the cells were homogenized using 30 strokes with a Dounce homogenizer. After brief centrifugation, the resulting supernatant was collected as a non-nuclear fraction, and the pelleted nuclei were further washed three times with Lysis buffer to remove any contamination from the cytoplasmic membranes. To extract nuclear proteins, the isolated nuclei were resuspended in NETN buffer (150 mm NaCl, 1 mm EDTA, 20 mm Tris, pH 8.0, 0.5% Nonidet P-40, protease inhibitor mixture) and sonicated (Sonics Vibra-Cell, amplitude 30; Sonics & Materials, Newtown, CT). The nuclear fraction was then collected after centrifugation at maximum speed.
INM Purification
INM purification was performed as described previously with a slight modification (44). The isolated nuclei in the nuclear fractions extracted using cellular fractionation were suspended in buffer A containing 0.25 m sucrose, 50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 1 mm dithiothreitol, and protease inhibitor mixture (Sigma). The resulting suspended nuclear pellet was incubated with 1% (w/v) sodium citrate at 4 °C with gentle rotation for 30 min and centrifuged at 500 × g for 15 min. The pellet suspended in buffer A was digested with DNase I (250 mg/ml; Sigma) at 4 °C for 14 h. After centrifugation at 10,000 × g for 2 h, the supernatant was collected as an NP portion, and the digested pellet was then submitted to recentrifugation at 100,000 × g for 20 min on a sucrose gradient to obtain purified INM fractions. The membrane fraction collected at the 0.25–1.60 m sucrose interface was the purified INM. Another set of digested pellet was resuspended in NETN buffer and sonicated. The INM portion was then collected after centrifugation at maximum speed.
ER Purification
Purification of the ER was performed using the OptiPrep density gradient medium with a slight modification (Sigma). Cultured cells were harvested and resuspended in a homogenization buffer (10 mm Tris-HCl, pH 7.5, 250 mm sucrose, protease inhibitor mixture). Cells were homogenized using 20 strokes with a Dounce homogenizer in the same buffer and then centrifuged at 12,000 × g for 20 min at 4 °C. The resulting supernatant was further centrifuged at 100,000 × g for 45 min at 4 °C. After centrifugation, the supernatant was collected as a non-nuclear/non-microsomal fraction, and the microsomal pellet was resuspended in the homogenizing buffer. The resulting mixture of 6.67 volumes of the microsomal suspension with 3.33 volumes of the OptiPrep density gradient medium was transferred to tubes (1 ml/tube) and centrifuged overnight at 200,000 × g. The ER fractions were then collected.
Confocal Microscopy and Immunoelectron Microscopy (Immuno-EM)
Confocal and Immuno-EM were performed according to standard procedures. See the supplemental Experimental Procedures for further details.
RESULTS
Cell Surface EGFR Translocates to the INM in Response to EGF
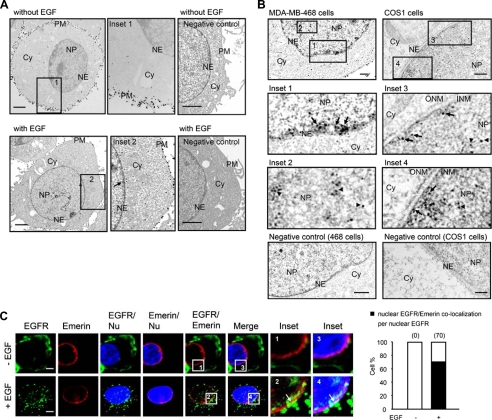
To investigate trafficking mechanisms of EGFR from the cell surface to the nucleus, we first performed three-dimensionally reconstructed z-stack images using confocal microscopy (supplemental Fig. S1) and ultrastructural studies using immuno-EM (Fig. 1, A and B) to confirm the nuclear localization of EGFR. Consistent with previous reports, the confocal images clearly demonstrated that EGF induced EGFR translocation to the nucleus (supplemental Fig. S1). The immuno-EM studies in human breast carcinoma MDA-MB-468 cells also showed that EGFR was mainly localized on the cell surface plasma membrane (PM) without EGF treatment and that after EGF stimulation, EGFR could be detected in the NE (Fig. 1A, Inset 2, arrow). Furthermore, the nuclear localization of EGFR was inside the NE in cells treated with EGF (Fig. 1B, Inset 1, arrows) and in the NP as expected (Fig. 1B, Inset 2, arrowheads). In COS1 monkey kidney cells in which the NE structure could be better visualized to distinguish the INM and ONM, EGFR was clearly detected in the INM upon EGF treatment (Fig. 1B, Insets 3 and 4, arrows). In addition, the merged image representing co-localization of EGFR and the INM marker emerin were detected upon EGF treatment (Fig. 1C, Insets 2 and 4 versus Insets 1 and 3; also confirmed in supplemental Fig. S4B), suggesting the localization of EGFR to the INM after EGF stimulation.
FIGURE 1.
Localization of EGFR to the INM. A, EGF-induced nuclear translocation of EGFR was analyzed using immuno-EM. MDA-MB-468 cells were treated with or without EGF for 30 min and subjected to immuno-EM. PM, plasma membrane; Cy, cytoplasm. Bar, 2 μm. B, localization of EGFR to the INM was analyzed using immuno-EM. MDA-MB-468 or COS1 cells were treated with EGF and subjected to immuno-EM. Secondary antibodies labeled with 10-nm gold particles were used to indicate EGFR. Bar, 2 μm. C, EGF-dependent co-localization of EGFR and the INM marker emerin. MDA-MB-468 cells were immunostained with EGFR and emerin and analyzed using confocal microscopy. Bar, 5 μm. The bar diagram indicates the percentage of cells with co-localization of EGFR and emerin calculated from a pool of 50 cells, which were positive for nuclear localization of EGFR under EGF stimulation. Nu, nucleus.
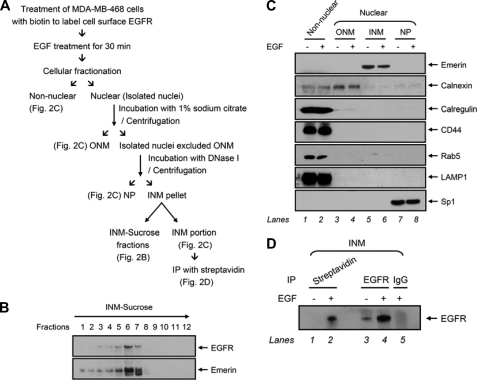
Next, we asked whether EGFR translocates into the nucleus from the cell surface to the INM. To answer this question, we analyzed proteins in the INM using cellular fractionation methods adapted from established procedures (45) (Fig. 2A). Briefly, cell surface EGFRs were labeled with biotin, and then the biotinylated EGFRs were biochemically separated into various fractions, including non-nuclear and nuclear fractions. The nuclear fraction was further separated into the ONM, NP, and INM pellet. To investigate whether EGFR can be detected in the INM by biochemical methods, we subjected the INM pellet to centrifugation on a sucrose gradient (INM-sucrose). Immunoblotting analysis of the INM-sucrose fractions with an anti-emerin antibody indicated the recovery of INM at the sucrose interface represented two major fractions (6 and 7). We found that EGFR was consistently distributed in the fractions in which we detected emerin, indicating the localization of EGFR in the INM fractions (Fig. 2B). In addition, the purity of various fractions was validated by another set of biotinylated lysates, which was subjected to subsequent subnuclear fractionation to extract the INM portions as described under “Experimental Procedures.” The INM portions (Fig. 2C, lanes 5 and 6) had undetectable cross-contamination during cellular fractionation as evident from the absence of the ER markers calnexin and calregulin, cell surface protein CD44, early endosome protein Rab5, late endosome protein LAMP1, and nuclear protein Sp1 in the INM portion. In these INM portions, the biotinylated EGFR precipitated using streptavidin-agarose beads increased significantly after EGF stimulation (Fig. 2D, lane 2 versus lane 1), and similar results were obtained using anti-EGFR antibodies to immunoprecipitate EGFR (Fig. 2D, lane 4 versus lane 3). These results strongly suggest that EGF induced the translocation of EGFR from the cell surface to the INM.
FIGURE 2.
Cell surface EGFR is targeted to the INM for EGF response. A, schematic description of cellular fractionation of biotinylated cell surface proteins in MDA-MB-468 cells. IP, immunoprecipitation. B, EGFR was distributed to the INM. INM-sucrose fractions were purified using sucrose gradient as described in A and subjected to immunoblotting with the indicated antibodies. The arrow above the panels indicates the direction of the gradient from top to bottom. C, INM portions had undetectable cross-contamination with the process of cellular fractionation. Biotinylated cell surface proteins of MDA-MB-468 cells were isolated using cellular fractionation as described in A and subjected to immunoblotting with the indicated antibodies. D, cell surface EGFR was translocated to the INM upon EGF stimulation. The purified INM portions in C were immunoprecipitated using streptavidin-agarose beads and anti-EGFR. Immunoprecipitation performed with IgG was used as a negative control.
EGFR Transport to the INM Is Regulated by Importin β through the NPCs
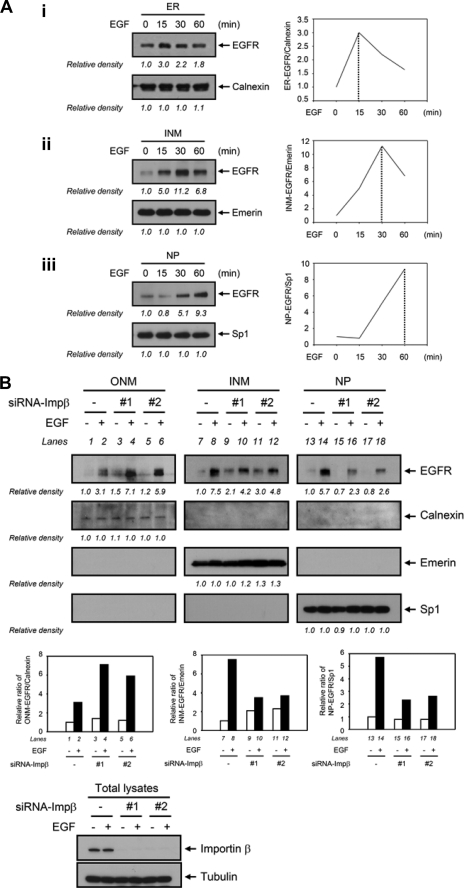
Recently, large INM proteins have been reported to be initially inserted into the ER membrane and targeted to the INM through the NPCs (33, 34). We asked whether the translocation of membrane-bound EGFR to the INM may be through the ER, similar to the INTERNET (integral trafficking from the ER to the NE transport) pathway used by recently reported large INM proteins (46). To this end, we analyzed the EGF-dependent kinetics of EGFR translocation from the ER-INM to the NP; the peaks to reach the ER and INM were 15 and 30 min, respectively, and in the final step, NP, it continued to increase at 60 min after EGF stimulation (Fig. 3A, panels i–iii, respectively). The kinetics supported the order of ER-to-INM-to-NP for the EGF-induced EGFR nuclear translocation. We then asked whether importin β, which is involved in the nuclear translocation of EGFR (24), also regulates EGFR transport to the INM through the ER/ONM. To address this issue, we knocked down importin β expression using two individual small interfering RNAs (siRNAs) targeting importin β (siRNA-Impβ-1 and siRNA-Impβ-2) and then analyzed the EGFR localization in the ONM, INM, and NP portions. Indeed, knocking down importin β expression significantly accumulated EGF-dependent EGFR translocation in the ONM (Fig. 3B, lanes 4 and 6 versus lane 2) and inhibited that to the INM (Fig. 3B, lanes 10 and 12 versus lane 8). Consistent with the previous studies, EGF-dependent EGFR nuclear translocation was inhibited upon down-regulation of importin β expression (Fig. 3B, lanes 16 and 18 versus lane 14). These results strongly suggest that importin β is responsible for the EGFR trafficking to the INM and the nucleus. In addition, it has been reported that interaction between importin β and the nuclear pore protein Nup62, a nucleoporin that lines the central regions of NPCs, plays a pivotal role in nuclear import of proteins and maintenance of the structural integrity of NPCs (47). We next asked whether Nup62 is also involved in the nuclear import of EGFR to the INM through the NPCs. The results showed that down-regulation of Nup62 expression using the siRNA approach clearly inhibited EGF-dependent EGFR translocation in the INM and NP (supplemental Fig. S2, lane 4 versus lane 2, lane 8 versus lane 6), suggesting that EGF could not enhance EGFR translocation to the INM when the structure of the NPCs was disrupted. Taken together with the previous report (25), these results support the notion that cell surface EGFR translocates to the INM and the NP, which is regulated by importin β, through the NPCs in response to EGF.
FIGURE 3.
Importin β-mediated INTERNET membrane trafficking regulates EGF-dependent EGFR nuclear transport. A, EGF-dependent kinetics of EGFR nuclear translocation from the ER-INM to the NP. Calnexin, emerin, and Sp1 were used as markers for the ER, INM, and NP, respectively. The diagrams indicate the relative densities of the immunoblots as quantified using the ImageJ software program (version 1.38x; National Institutes of Health). B, knockdown of importin β (Impβ) by two individual siRNAs targeting importin β (siRNA-Impβ-1 and siRNA-Impβ-2) in HeLa cells down-regulated EGF-dependent EGFR translocation to the INM and NP. The relative density by quantification is plotted diagrammatically as shown in the middle panel. Similar results were obtained in 3 independent experiments.
EGFR Associates with the Translocon Sec61β in the INM
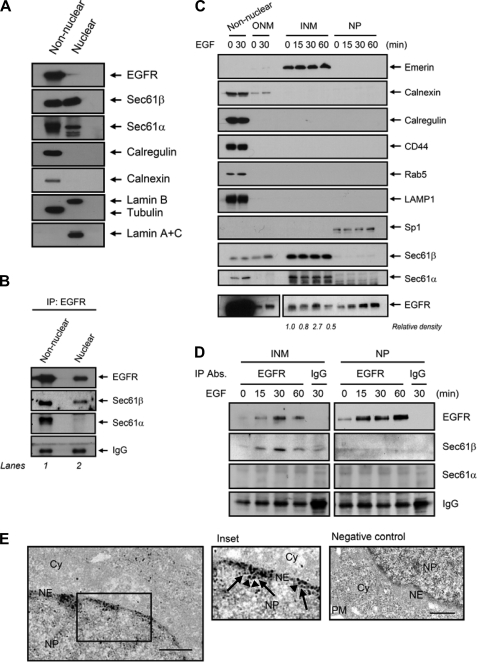
It has been reported that a sorting importin captures newly synthesized INM proteins co-translationally at the ER translocon Sec61α for the route from the ER to the INM (33). In addition, EGFR was shown to associate with Sec61β in the ER (48). Thus, we asked whether the translocon Sec61 may be involved in the translocation of membrane-associated EGFR from the ER to the INM/nucleus via the INTERNET model (46), similar to translocation of INM proteins (33). To further investigate the molecular mechanism of ER-to-INM of EGFR, we performed a co-immunoprecipitation assay to confirm that the EGFR in the ER membrane is associated with the translocon Sec61β (48). As expected, Sec61α and Sec61β were expressed in the non-nuclear fraction containing the ER as evident from the ER markers calregulin and calnexin, however, unexpectedly; they were also detected in the nuclear fraction that includes the INM (Fig. 4A). Consistent with the previous report (48), we detected interaction of EGFR and Sec61β in the non-nuclear fraction including the ER (Fig. 4B, lane 1). Additionally, we detected the EGFR/Sec61β interaction in the nuclear fraction (Fig. 4B, lane 2), whereas we only detected the EGFR/Sec61α interaction in the non-nuclear fraction. It is well known that Sec61α and Sec61β reside in the ER serving as translocon. The association of EGFR with Sec61α and Sec61β in the non-nuclear fraction that contains the ER suggests that EGFR associates with the translocon in the ER, consistent to the previous report (48). The results further demonstrated that EGFR associates with only Sec61β but not Sec61α in the nuclear fraction containing both the INM and the NP.
FIGURE 4.
EGFR associates with the translocon Sec61β in the INM. A, A431 cells maintained in a serum-starved medium for 24 h were treated with EGF followed by cellular fractionation. B, proteins in A were immunoprecipitated (IP) using anti-EGFR followed by immunoblotting. C, INM portions had undetectable cross-contamination with the process of cellular fractionation. MDA-MB-468 cells maintained in a serum-starved medium for 24 h were treated with EGF in a different period followed by cellular fractionation as described in the legend for Fig. 2 and subjected to immunoblotting with the indicated antibodies. The relative density of the INM-EGFR immunoblotting at zero time was defined as 1 after subtraction of the background by using the ImageJ software program (version 1.38x) to quantify the signals. The EGFR blotting of the left panel has five times shorter exposure than that of right panel. D, EGFR associated with Sec61β in the INM portions but not the NP portions in response to EGF in a time-dependent manner. The purified INM and NP portions in C were immunoprecipitated using the indicated antibodies (Abs.) followed by immunoblotting. E, co-localization of EGFR and Sec61β in the INM was analyzed using immuno-EM. An ultrathin section of MDA-MB-468 cells treated with EGF was immunostained with EGFR (goat anti-mouse IgG, 1-nm gold particles, arrows) and Sec61β (goat anti-rabbit IgG, 10-nm gold particles, arrowheads). Bar, 1 μm. PM, plasma membrane; Cy, cytoplasm.
Next, we then asked whether the nuclear co-localization of EGFR and Sec61β was in the INM and/or the NP. To this end, we isolated the INM portions of MDA-MB-468 cells using subnuclear fractionation and subjected them to immunoblotting analysis as described in Fig. 2A. No cross-contamination with the process of cellular fractionation was detected (Fig. 4C). Interestingly, we detected Sec61β and Sec61α in the INM portions but not the NP portions, and EGF treatment did not alter Sec61β and Sec61α protein expression in the INM (Fig. 4C). We further showed that EGF induced interaction of Sec61β but not Sec61α with EGFR in the INM (Fig. 4D) in a time-dependent manner, which was consistent with Fig. 4B. Similar results were obtained when the same experiment was performed in another cell line (supplemental Fig. S3). Furthermore, we analyzed the INM-sucrose fractions using sucrose gradient purification to show that EGFR and Sec61β, after EGF treatment, were consistently distributed in the fractions in which we detected the INM marker emerin, supporting the localization of EGFR and Sec61β in the INM fractions (supplemental Fig. S4A). In addition to the biochemical studies, we further showed the white merged image representing co-localization of EGFR, Sec61β, and emerin upon EGF treatment (supplemental Fig. S4B, Inset 2), strongly suggesting the co-localization of EGFR and Sec61β to the INM. To further support the co-localization of EGFR and Sec61β in the INM, we performed ultrastructural studies using immuno-EM with the specific primary antibodies followed by incubating with two different sized gold particle-labeled secondary antibodies, including those labeling anti-EGFR (goat anti-mouse IgG, 1-nm gold particles, arrows) and anti-Sec61β (goat anti-rabbit IgG, 10-nm gold particles, arrowheads) (Fig. 4E). The results clearly showed that EGFR and Sec61β were co-localized inside the nucleus (Fig. 4E, inset) when the specific primary antibodies against EGFR and Sec61β were treated. As a negative control, gold particles were not detected in the presence of gold particle-labeled secondary antibodies without specific primary antibodies (Fig. 4E, right panel), indicating the specificity of the detected gold particles. The gold particles labeling Sec61β were confirmed by two different specific primary anti-Sec61β antibodies obtained from Upstate Biotech Millipore (supplemental Fig. S5, upper inset panel, arrows) and from Proteintech (supplemental Fig. S5, lower panel, arrows), which demonstrated that the localization of Sec61β was primarily detectable in the INM. These results together indicate that the ER translocons Sec61β and Sec61α can localize in the INM, whereas only Sec61β associates with EGFR upon EGF treatment.
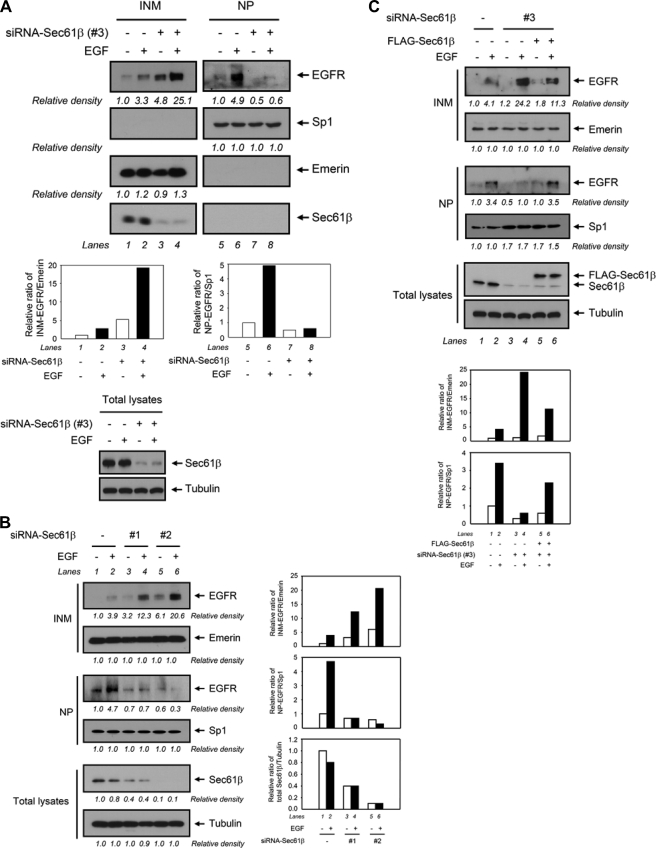
Sec61β Is Required for EGFR Nuclear Transport from the INM to the NP
The above results suggest that EGF-dependent EGFR transport to the INM involves membrane-bound trafficking and that the translocon Sec61β associates with EGFR in the INM. Together, because translocons at the ER lumen are known to be required for protein export as part of the ERAD pathway (40), we hypothesized that Sec61β in the INM plays a role resembling that of Sec61α in ERAD by releasing membrane-bound EGFR from the lipid bilayer of the INM to the NP. To this end, we knocked down Sec61β expression in HeLa cells and then analyzed the EGFR localization in the INM and NP portions. As a control (Fig. 5A), EGFR expression was indeed increased in the INM (lane 2 versus lane 1) and the NP portion (lane 6 versus lane 5) upon EGF treatment. Interestingly, once Sec61β was knocked down by siRNA, EGFR expression was significantly reduced in the NP portion (lane 7 versus lane 5, lane 8 versus lane 6) and accumulated in the INM portion (lane 3 versus lane 1, lane 4 versus lane 2), suggesting that EGFR translocation from the INM to the NP requires Sec61β. Similar results were obtained from experiments performed in A431 cells by Sec61β knockdown using different siRNAs (supplemental Fig. S6). These results support the notion that Sec61β plays a role in the INM to assist membrane-bound EGFR in releasing from the INM to the NP. In comparing two other individual siRNAs targeting Sec61β (siRNA-Sec61β-1 and siRNA-Sec61β-2), we interestingly found that the accumulation of EGFR in the INM and the decrease of EGFR in the NP mediated by the down-regulation of Sec61β expression were positively correlated with the knockdown efficiency of Sec61β (Fig. 5B, lane 2 versus lane 4 versus lane 6). Furthermore, we performed a reconstitution assay to examine the ability of an exogenous construct of FLAG-tagged Sec61β to rescue the effect of Sec61β knockdown. As shown in Fig. 5C, cells transfected with FLAG-tagged Sec61β cDNA decreased the INM-anchored EGFR induced by knockdown of Sec61β (fraction 3) upon EGF treatment (upper INM panel, lane 6 versus lane 4) and accordingly increased expression level of EGFR in the NP (upper NP panel, lane 6 versus lane 4). These results indicate that knockdown of Sec61β expression prevents EGF-dependent EGFR translocation from the INM to the NP, suggesting that transport of EGFR from the INM to the NP is regulated by the association of EGFR with Sec61β translocon in the INM.
FIGURE 5.
Association of EGFR with Sec61β in the nucleus assists INM-anchored EGFR in releasing to the nucleus. A, knockdown of Sec61β prevented EGF-dependent transport of EGFR from the INM to the NP in HeLa cells. Cells were transfected with an siRNA targeting Sec61β (siRNA-Sec61β-3) (+) or a nonspecific control siRNA (−) using electroporation. Proteins from the total lysates, INM, and NP by cellular fractionation were then analyzed using immunoblotting with the antibodies as indicated. Emerin and Sp1 were used as markers for the INM and NP portions, respectively. B, knockdown of Sec61β by two individual siRNAs targeting Sec61β (siRNA-Sec61β-1 and siRNA-Sec61β-2) in HeLa cells up-regulated EGF-dependent EGFR translocation to the INM. Cells were transfected with two individual siRNAs targeting Sec61β (siRNA-Sec61β-1 and siRNA-Sec61β-2) or a nonspecific control siRNA (−) using electroporation. C, exogenous Sec61β rescued the effect of Sec61β knockdown on INM-anchored EGFR. Cells were co-transfected with a Sec61β siRNA targeting its 3′-untranslated region (UTR) (siRNA-Sec61β-3) and a 3′-UTR-deleted FLAG-tagged Sec61β using electroporation. The bar diagram indicates the relative densities of the immunoblots as quantified using the ImageJ software program (version 1.38x). The relative density by quantification is plotted diagrammatically as shown. Similar results were obtained in 2–4 four independent experiments.
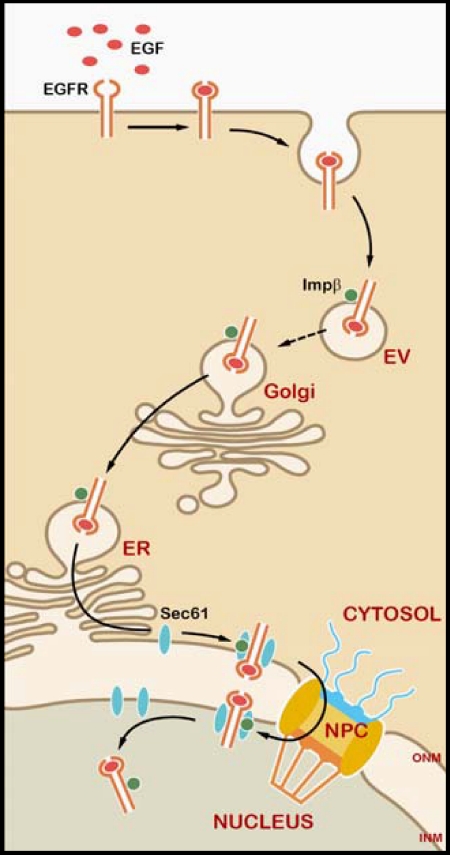
Together with the previous studies indicating that endocytosis is involved in nuclear transport of EGFR and ErbB-2 (24, 27), we proposed a model based on the current study (Fig. 6). During the trafficking of cell surface EGFR to the nucleus in response to EGF, EGFR remains in a membrane environment (Figs. 1 and 2), which after endocytosis is first embedded in the endocytic vesicles, fused to the Golgi-ER membrane via a retrograde route (49), translocated into the nucleus through ER membrane (Fig. 3) (33, 34), and released from the lipid bilayer of the INM by the association with Sec61β (Figs. 4 and 5). The INTERNET model can explain how EGFR can translocate from the ER to the nucleus; namely, membrane-associated EGFR interacts with importin β and travels from the ER/ONM to the INM via the NPCs. This way, EGFR remains embedded in the membrane from the cell surface to the NE in the entire trafficking process.
FIGURE 6.
Proposed model of EGFR trafficking from the cell surface to the nucleus. A diagram of integral trafficking of EGFR from the Golgi/ER/NE to the nucleus by EGF treatment is shown. The scale of the diagram does not reflect the relative sizes of different molecules or subcellular structures. EV, endocytic vesicle; Impβ, importin β.
DISCUSSION
In this study, we proposed a comprehensive trafficking pathway for full-length cell surface receptors to remain in a membrane-associated environment, traveling from the cell surface to the nucleus through the endosomes, Golgi, ER, NPCs, and nuclear envelope, where membrane-bound receptors escape from the lipid bilayer via the association of the translocon Sec61β (Fig. 6). It is worthwhile to mention that we frequently detected the basal level of nuclear EGFR without ligand stimulation (7, 25, 26). It is conceivable that some portion of EGFR de novo synthesized in the ER could transport to the nucleus directly instead of going to the cell surface. It supports the notion that we also detected the basal level of EGFR in the INM in the absence of EGF stimulation (such as Figs. 2D, lane 3; 3, A, panel ii; and 4C). However it should be emphasized that the cell surface EGFR can be translocated to the INM/NP in response to EGF treatment (Fig. 2D, lane 2 versus lane 1). Researchers have proposed that nuclear transport of ErbB-2 is similar to that of EGFR (3, 24, 27). Multiple full-length RTKs have been reported to be located in the nucleus, and their nuclear functions have been gradually discovered (3, 9–11, 15, 50, 51). Our proposed model (Fig. 6) provides a logical route for the nuclear translocation of EGFR from the cell surface in response to EGF and may be a general mechanism for nuclear transport of full-length RTKs or other cell surface receptors.
Nuclear transport of INM proteins is a related example of integral membrane proteins other than the EGFR family proteins (46). INM proteins located in the ER membrane can be regulated by the importin proteins and transported to the nucleus through the NLS-mediated INTERNET mechanism (33, 34). On the other hand, investigators have identified the tripartite NLS of EGFR in the juxtamembrane region within the intracellular COOH terminus of EGFR (23), and importin β is known to interact through NLSs of proteins including EGFR and ErbB-2 (24, 27). Of note is that the NLS of EGFR resembles the viral INM-sorting motif sequence, a hydrophobic transmembrane sequence of 18–20 amino acids following positively charged residues positioned within 4–8 residues of the end of the transmembrane sequence. The viral INM-sorting motif sequence can be recognized by an ER membrane-associated importin α-16 in sorting the viral INM-directed proteins to the NE (33). Given previous findings and our present results indicating that EGF-dependent EGFR translocation to the INM is reduced upon knockdown of importin β expression (Fig. 3B), this suggests that importin β recognizes the EGFR NLS and may play a critical role in translocating the EGFR from the ER to the INM.
Researchers have proposed extraction of EGFR localized in the ER from lipid layers to the cytoplasm via the ERAD pathway (48). Regarding the distribution of the core components of the Sec61 translocon, they do not permanently reside in the ER as none of the Sec61 subunits contain any known ER retention or retrieval signals normally associated with ER resident proteins. The Sec61 translocon is thus far thought to be localized in the ER and ER-Golgi intermediate compartment (38). In the present study, we unexpectedly observed a novel functional role of Sec61 localized in the INM, which functioned as an intranuclear translocon in the INM and played an ERAD-resembling translocation role in releasing the INM-bound EGFR from lipid layers of the INM to the NP (Fig. 5). In addition to the ER translocon, a yeast ERAD ubiquitin E3 ligase, Doa10, which is thought to be in the ER, has also been shown in the INM (52), further supporting the notion that the Sec61β-dependent ERAD-resembling translocation mechanism exists in the INM. A more systemic study is required to further address this interesting observation. Collectively, the current study identifies a novel pathway that allows trafficking of full-length membrane receptors in a membrane-embedded form from the cell surface to the nucleus (Fig. 6). This pathway involves a new role of ER-associated translocon Sec61β in the INM to translocate membrane-embedded proteins into the NP, which may serve as a general role for nuclear translocation in addition to the well known NPCs.
Supplementary Material
Acknowledgment
We thank Dr. Stephanie A. Miller for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 109311 and PO1 099031 (to M.-C. H.). This work was also supported by the National Breast Cancer Foundation, Inc. and the Sister Institutional fund from China Medical University Hospital and MD Anderson Cancer Center (to M.-C. H.); the Taiwan Cancer Research Center of Excellence Grant DOH99-TD-C-111-005; the Institutional Core Grant CA 16672 High Resolution Electron Microscopy Facility; and the National Science Council Taiwan Merit Postdoctoral Scholarship TMS-94-2B-001 (to Y.-N. W.).
In memoriam, Mrs. Serena Lin-Guo for her courageous battle in breast cancer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S6.
- RTK
- receptor tyrosine kinase
- EGFR
- epidermal growth factor receptor
- NPC
- nuclear pore complex
- NP
- nucleoplasm
- NE
- nuclear envelope
- INM
- inner nuclear membrane
- ONM
- outer nuclear membrane
- NLS
- nuclear localization sequence
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation.
REFERENCES
- 1.Bryant D. M., Stow J. L. (2005) Traffic 6, 947–954 [DOI] [PubMed] [Google Scholar]
- 2.Carpenter G., Liao H. J. (2009) Exp. Cell Res. 315, 1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo H. W., Hung M. C. (2006) Br. J. Cancer 94, 184–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sehat B., Tofigh A., Lin Y., Trocmé E., Liljedahl U., Lagergren J., Larsson O. (2010) Sci. Signal 3, ra10. [DOI] [PubMed] [Google Scholar]
- 5.Gomes D. A., Rodrigues M. A., Leite M. F., Gomez M. V., Varnai P., Balla T., Bennett A. M., Nathanson M. H. (2008) J. Biol. Chem. 283, 4344–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y., Venema V. J., Venema R. C., Tsai N., Caldwell R. B. (1999) Biochem. Biophys. Res. Commun. 256, 192–197 [DOI] [PubMed] [Google Scholar]
- 7.Huo L., Wang Y. N., Xia W., Hsu S. C., Lai C. C., Li L. Y., Chang W. C., Wang Y., Hsu M. C., Yu Y. L., Huang T. H., Ding Q., Chen C. H., Tsai C. H., Hung M. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massie C., Mills I. G. (2006) Nat. Rev. Cancer 6, 403–409 [DOI] [PubMed] [Google Scholar]
- 9.Wells A., Marti U. (2002) Nat. Rev. Mol. Cell Biol. 3, 697–702 [DOI] [PubMed] [Google Scholar]
- 10.Wang S. C., Nakajima Y., Yu Y. L., Xia W., Chen C. T., Yang C. C., McIntush E. W., Li L. Y., Hawke D. H., Kobayashi R., Hung M. C. (2006) Nat. Cell Biol. 8, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 11.de la Iglesia N., Konopka G., Puram S. V., Chan J. A., Bachoo R. M., You M. J., Levy D. E., Depinho R. A., Bonni A. (2008) Genes Dev. 22, 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S. C., Hung M. C. (2009) Clin. Cancer Res. 15, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosesson Y., Mills G. B., Yarden Y. (2008) Nat. Rev. Cancer 8, 835–850 [DOI] [PubMed] [Google Scholar]
- 14.Lo H. W., Cao X., Zhu H., Ali-Osman F. (2010) Mol. Cancer Res. 8, 232–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo H. W., Xia W., Wei Y., Ali-Seyed M., Huang S. F., Hung M. C. (2005) Cancer Res. 65, 338–348 [PubMed] [Google Scholar]
- 16.Hoshino M., Fukui H., Ono Y., Sekikawa A., Ichikawa K., Tomita S., Imai Y., Imura J., Hiraishi H., Fujimori T. (2007) Pathobiology 74, 15–21 [DOI] [PubMed] [Google Scholar]
- 17.Psyrri A., Yu Z., Weinberger P. M., Sasaki C., Haffty B., Camp R., Rimm D., Burtness B. A. (2005) Clin. Cancer Res. 11, 5856–5862 [DOI] [PubMed] [Google Scholar]
- 18.Hadzisejdić I., Mustać E., Jonjić N., Petković M., Grahovac B. (2010) Mod. Pathol. 23, 392–403 [DOI] [PubMed] [Google Scholar]
- 19.Xia W., Wei Y., Du Y., Liu J., Chang B., Yu Y. L., Huo L. F., Miller S., Hung M. C. (2009) Mol. Carcinog. 48, 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards J., Traynor P., Munro A. F., Pirret C. F., Dunne B., Bartlett J. M. (2006) Clin. Cancer Res. 12, 123–130 [DOI] [PubMed] [Google Scholar]
- 21.Cook A., Bono F., Jinek M., Conti E. (2007) Annu. Rev. Biochem. 76, 647–671 [DOI] [PubMed] [Google Scholar]
- 22.Hsu S. C., Miller S. A., Wang Y., Hung M. C. (2009) Am. J. Transl. Res. 1, 249–258 [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu S. C., Hung M. C. (2007) J. Biol. Chem. 282, 10432–10440 [DOI] [PubMed] [Google Scholar]
- 24.Lo H. W., Ali-Seyed M., Wu Y., Bartholomeusz G., Hsu S. C., Hung M. C. (2006) J. Cell. Biochem. 98, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 25.Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. (2001) Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
- 26.Lo H. W., Hsu S. C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J. Y., Hung M. C. (2005) Cancer Cell 7, 575–589 [DOI] [PubMed] [Google Scholar]
- 27.Giri D. K., Ali-Seyed M., Li L. Y., Lee D. F., Ling P., Bartholomeusz G., Wang S. C., Hung M. C. (2005) Mol. Cell. Biol. 25, 11005–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S. C., Lien H. C., Xia W., Chen I. F., Lo H. W., Wang Z., Ali-Seyed M., Lee D. F., Bartholomeusz G., Ou-Yang F., Giri D. K., Hung M. C. (2004) Cancer Cell 6, 251–261 [DOI] [PubMed] [Google Scholar]
- 29.Reilly J. F., Maher P. A. (2001) J. Cell Biol. 152, 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart C. L., Roux K. J., Burke B. (2007) Science 318, 1408–1412 [DOI] [PubMed] [Google Scholar]
- 31.Stewart M. (2007) Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 32.Hoelz A., Blobel G. (2004) Nature 432, 815–816 [DOI] [PubMed] [Google Scholar]
- 33.Saksena S., Summers M. D., Burks J. K., Johnson A. E., Braunagel S. C. (2006) Nat. Struct. Mol. Biol. 13, 500–508 [DOI] [PubMed] [Google Scholar]
- 34.King M. C., Lusk C. P., Blobel G. (2006) Nature 442, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 35.Kutay U., Mühlhäusser P. (2006) Nature 442, 991–992 [DOI] [PubMed] [Google Scholar]
- 36.Rexach M. F. (2006) Nat. Struct. Mol. Biol. 13, 476–478 [DOI] [PubMed] [Google Scholar]
- 37.Osborne A. R., Rapoport T. A., van den Berg B. (2005) Annu. Rev. Cell Dev. Biol. 21, 529–550 [DOI] [PubMed] [Google Scholar]
- 38.Greenfield J. J., High S. (1999) J. Cell Sci. 112, 1477–1486 [DOI] [PubMed] [Google Scholar]
- 39.Schnell D. J., Hebert D. N. (2003) Cell 112, 491–505 [DOI] [PubMed] [Google Scholar]
- 40.Römisch K. (2005) Annu. Rev. Cell Dev. Biol. 21, 435–456 [DOI] [PubMed] [Google Scholar]
- 41.Rapoport T. A. (2007) Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 42.Panzner S., Dreier L., Hartmann E., Kostka S., Rapoport T. A. (1995) Cell 81, 561–570 [DOI] [PubMed] [Google Scholar]
- 43.Römisch K. (1999) J. Cell Sci. 112,4185–4191 [DOI] [PubMed] [Google Scholar]
- 44.Klein C., Gensburger C., Freyermuth S., Nair B. C., Labourdette G., Malviya A. N. (2004) Biochemistry 43, 15873–15883 [DOI] [PubMed] [Google Scholar]
- 45.Humbert J. P., Matter N., Artault J. C., Köppler P., Malviya A. N. (1996) J. Biol. Chem. 271, 478–485 [DOI] [PubMed] [Google Scholar]
- 46.Wang Y. N., Yamaguchi H., Hsu J. M., Hung M. C. (2010) Oncogene 29, 3997–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoffler D., Fahrenkrog B., Aebi U. (1999) Curr. Opin. Cell Biol. 11, 391–401 [DOI] [PubMed] [Google Scholar]
- 48.Liao H. J., Carpenter G. (2007) Mol. Biol. Cell 18, 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y. N., Wang H., Yamaguchi H., Lee H. J., Lee H. H., Hung M. C. (2010) Biochem. Biophys. Res. Commun. 399, 498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das A. K., Chen B. P., Story M. D., Sato M., Minna J. D., Chen D. J., Nirodi C. S. (2007) Cancer Res. 67, 5267–5274 [DOI] [PubMed] [Google Scholar]
- 51.Offterdinger M., Schöfer C., Weipoltshammer K., Grunt T. W. (2002) J. Cell Biol. 157, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng M., Hochstrasser M. (2006) Nature 443, 827–831 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.