Abstract
The Rho-like GTPase Rac1 induces cytoskeletal rearrangements required for cell migration. Rac activity is regulated through a number of mechanisms, including control of nucleotide exchange and hydrolysis, regulation of subcellular localization, or modulation of protein expression levels1-3. Here, we identify the Small Ubiquitin-like MOdifier (SUMO) E3-ligase, PIAS3, as a new Rac1 interactor required for increased Rac activity and optimal cell migration in response to Hepatocyte Growth Factor (HGF) signalling. We show that Rac1 can be conjugated to SUMO-1 in response to HGF and that SUMOylation is enhanced by PIAS3. Moreover, we identify non-consensus sites within the polybasic region of Rac1 as the main locations for SUMO conjugation. We demonstrate that PIAS3-mediated SUMOylation of Rac1 controls its GTP-bound levels and its ability to stimulate lamellipodia, cell migration and invasion. This is the first time that a Ras superfamily member is found to be SUMOylated, providing a new insight into the regulation of these critical mediators of cell behaviour. Moreover, our data reveal a previously undescribed role for SUMO in the regulation of cell migration and invasion.
To identify proteins regulating Rac1 in the context of cell migration, we performed tandem affinity purification (TAP) of native protein complexes containing TAP-tagged Rac1 from MDCKII cells following HGF treatment (a ligand for the growth factor receptor c-Met and a migration stimulus) at a time when Rac1 is active and cells begin to migrate4 (Supplementary Information, Fig. S1a-c). Following TAP, protein complexes were analyzed by mass spectrometry (MS) (Supplementary Information, Fig. S1d and Table I). Amongst known and putative Rac1 interactors, we identified PIAS3 (Protein Inhibitor of Activated STAT3) as a new Rac1-binding protein detected only in HGF-treated samples. PIAS3 interacts with Rac1 in vitro and in vivo (Fig 1a, b; Supplementary Information, Fig. S1e, f) and this interaction was more efficient when Rac1 was in its active form (Fig. 1c; Supplementary Information, Fig. S1g). In addition, activation of HGF signalling using an oncogenic form of the Met receptor (Trp-Met)5, 6 increased this interaction (Fig. 1d).
Figure 1.
PIAS3 is a new Rac1-binding protein required for optimal Rac activation and cell migration in response to HGF. (a) GST protein alone or GST-Rac1 were incubated with cell extracts containing FLAG-PIAS3 and protein pulldowns (PD) were analyzed by Western blot (WB). (b) Epitope-tagged proteins were expressed in HEK293T cells and GFP immunoprecipitation (IP) performed. Purified proteins were detected by WB. IN, input. (c) GST-Rac1 beads divided in two samples were incubated with GDP or γGTP for 1h at 30°C and then with equal amounts of cell lysate containing FLAG-PIAS3 for 1 hour at 4°C. PIAS3 interaction was detected by WB. (d) HEK293T cells were transfected with the indicated plasmids and subjected to GFP immunoprecipitation. Samples were analyzed by WB. (e) Individual cells were tracked for at least 24 hours in the presence of HGF. Data shown is average speed of migration of cells from at least three independent experiments (at least 30 cells per experiment). *p<0.001, ** p<0.05, ns indicates no significant difference, two-tailed Student’s t-test. Values are means ± standard deviation. (f) MDCKII cells inducibly expressing pias3 or scr shRNAs were treated with doxycycline (dox), serum was then removed for 18 hours before cells were treated with HGF for 30 minutes and Rac activity measured. Bands were quantified and normalized intensities were calculated relative to control. Values are means ± standard deviation (n=3). (g) MDCKII cells inducibly expressing pias3 or scr shRNAs were treated as above but Rac activity was measured at later time points. (h) COS7 cells transfected with MYC-PIAS3 or empty vector were serum starved for 18 hours before 10 ng/ml of HGF was added for 30 minutes and Rac activity measured. (i) Bar chart shows quantification of the normalized relative amounts of Rac-GTP of (h) determined by scanning densitometry. Error bars indicate standard deviation for 3 independent experiments. ***p<0.001, two-tailed Student’s t-test. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
Next we examined if PIAS3 was necessary for cell migration in response to HGF. We generated stable MDCKII cell lines carrying doxycycline-inducible short hairpin interfering RNAs (shRNAs) to PIAS3 (two independent targets – #1 and #2) or control scrambled shRNA (Scr) (Supplementary Information, Fig. S1h). Downregulation of PIAS3 resulted in cells migrating slower than controls (Fig. 1e) and impaired lamellipodia-membrane ruffle formation (Supplementary Information, Fig. S1i). From the reduced migration and lamellipodia formation, we reasoned that PIAS3 might be influencing Rac-GTP levels. Significantly, while Rac-GTP levels increased in control cells following HGF treatment, no increase was observed in PIAS3 downregulated cells, despite normal activation of the ERK MAPK pathway, indicating a specific requirement of PIAS3 for Rac activation (Fig. 1f). Consistent with this defect in Rac activation, phosphorylation (and hence activation) of p38, a known downstream mediator of Rac signalling7 was impaired in PIAS3 downregulated cells (Supplementary Information, Fig. S1j). HGF stimulates epithelial cell motility initially inducing spreading of cell colonies starting with lamellipodia/membrane ruffle formation, followed by disruption of cell-cell junctions and subsequent cell scattering; Rac1 is involved in both processes8. PIAS3 was required not only for optimal initial Rac activation following HGF-treatment, but also for optimal Rac activation at later time points (Fig. 1g). This continuous involvement of PIAS3 in Rac activation could explain the defect in migration observed throughout the 24 hours of HGF treatment. Rac-GTP levels were elevated in cells overexpressing PIAS3 and also treated with HGF compared to control transfected cells (Fig. 1h, i), confirming a positive role for PIAS3 in augmenting Rac-GTP levels following HGF treatment. In addition to HGF, the modest activation of Rac by EGF or serum was also impaired in PIAS3 downregulated cells (Supplementary Information, Fig. S1k and data not shown). This indicates that the requirement of PIAS3 for optimal Rac activation is not restricted to a particular upstream stimulus. However, downregulation of PIAS3 did not affect Cdc42-GTP levels (Supplementary Information, Fig. S1l). Collectively these data imply that PIAS3 is required for the induction or more likely the maintainenance of Rac activity (since the Rac-PIAS3 interaction is stronger for Rac-GTP over Rac-GDP or Rac1V12 over Rac1 WT) following growth factor treatment and in this way PIAS3 is required for optimal cell migration.
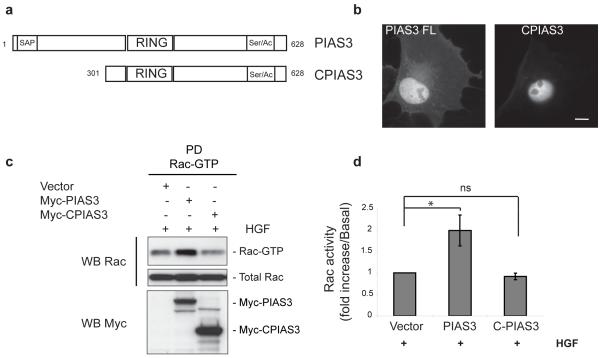
Most PIAS3 substrates reported so far are nuclear proteins, frequently transcription factors9, 10, although PIAS3 can also be detected in the cytoplasm. In contrast, Rac is mainly cytoplasmic, although nuclear Rac has been reported11, 12. To identify whether PIAS3 regulates Rac activity in the cytoplasm, we used a truncated version of PIAS3, CPIAS3, that is exclusively nuclear (Fig. 2a, b)13. In contrast to overexpression of full-length PIAS3, no increase in Rac-GTP levels occurred following CPIAS3 overexpression in response to HGF (Fig. 2c, d), even though Rac1 interacts with CPIAS3 in vitro and in vivo (Supplementary Information, Fig. S2a-c). Furthermore, PIAS3 co-localizes with Rac1 in the leading edge of cells and translocates from the cytosol to the membrane in response to HGF (Supplementary Information, Fig. S2d, e). Together, these results suggest that PIAS3 regulates Rac activity in the cytoplasm, and most likely at sites of membrane protrusion.
Figure 2.
PIAS3 regulates Rac1 activity in the cytoplasm. (a) Schematic illustration of PIAS3 full-length and CPIAS3. (b) Representative images of PIAS3 and CPIAS3 localization in COS7 cells. Scale bar, 20 μm. (c) COS7 cells transfected with empty vector, MYC-PIAS3 or CPIAS3 were serum starved for 18 hours before 10 ng/ml of HGF was added for 30 minutes and Rac activity measured. (d) Bar chart shows quantification of the normalized relative amounts of Rac-GTP determined by scanning densitometry of (c). Error bars indicate standard deviation for 3 independent experiments. * p<0.05, ns indicates no significant difference, two-tailed Student’s t-test. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
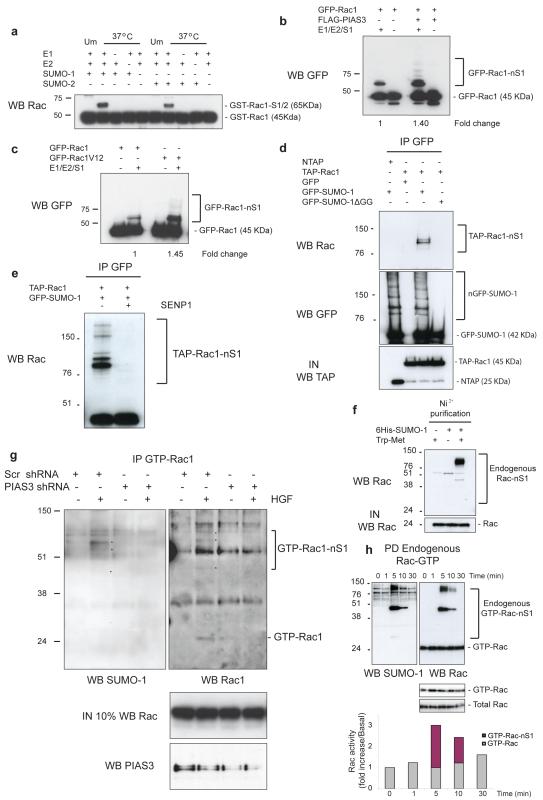
In higher eukaryotes PIAS proteins act as SUMO E3-ligases10. SUMOylation is a covalent modification leading to the attachment of SUMO to specific lysine residues of target proteins14. In vitro SUMOylation followed by Western blotting revealed a band consistent with the attachment of SUMO-1 (S1) or SUMO-2/3 (S2/3) to Rac1, which was absent when E1, E2, or SUMO protein was not present (Fig. 3a). Furthermore, SUMOylation was enhanced by increasing the E2-conjugating enzyme Ubc9 (Supplementary Information, Fig. S2f). In most cases, SUMOylation of a substrate is enhanced by a SUMO E3-ligase10, 15. Rac1 SUMOylation was enhanced by increasing amounts of PIAS3 (Supplementary Information, Fig. S2g). Other PIAS family members also enhanced in vitro Rac1 SUMOylation, although PIAS3 was the most efficient (Supplementary Information, Fig. S2g). Additionally, in vitro SUMOylation of GFP-Rac1 immunoprecipitated from HeLa cells was enhanced upon co-transfection of FLAG-PIAS3 (Fig. 3b). Conversely, downregulation of PIAS3 decreased in vitro SUMOylation of GFP-Rac1 (Supplementary Information, Fig. S2h). These results indicate that PIAS3 works as a SUMO E3-ligase for Rac1. Furthermore, consistent with the finding that PIAS3 binds more efficiently to GTP-bound Rac1 (Fig. 1c; Supplementary Information, Fig. S1g), Rac1V12 was a better substrate for SUMOylation (Fig. 3c).
Figure 3.
Rac1 is SUMOylated in vitro and in vivo. (a) GST-Rac1 protein was incubated at 37°C in vitro in the presence of complete SUMOylation assay components (E1, E2, SUMO-1 (S1) or SUMO-2 (S2)) and resolved by SDS-PAGE. Unmodified sample (Um) contains all the assay components but without incubation at 37°C. Rac1-S1/2 indicates Rac1 modified by SUMO-1 or -2. (b) GFP-Rac1 was immunoprecipitated from HeLa cells in the presence or absence of PIAS3 and in vitro SUMOylation assay was performed as in (a). Rac1 was detected by Western blot (WB). (c) Immunoprecipitated GFP-Rac1 or Rac1V12 was subjected to in vitro SUMOylation as in (a) and Rac1 was detected by WB. (d) HeLa cells transfected as indicated were lysed in the presence of NEM, and GFP was immunoprecipitated. The presence in immunoprecipitates of exogenous Rac1 or GFP was detected by WB analysis. IN, input. (e) GFP-SUMO-1 was immunoprecipitated from HeLa cells transfected as indicated, divided into two and incubated in the presence or absence of SENP1 for 2 hours at 25°C. (f) HeLa cells stably expressing 6His-SUMO-1 were transfected with Trp-Met and 6His-SUMO-1-modified proteins were purified from lysates. Endogenous Rac1-SUMO-1 was detected by WB. (g) MDCKII cells stably expressing scr shRNA or pias3 shRNA were treated as in Fig. 1f in the presence of the crosslinker DSS and HGF and Rac-GTP immunoprecipitation (IP) was performed. GTP-Rac-SUMO-1 bands were detected by WB. (h) MDCKII cells were grown to confluency and incubated for 18 hours in low calcium medium. Rac activity was measured after calcium re-addition for the indicated times. Pulldows (PD) of active Rac were analyzed by WB for Rac and SUMO-1. Blots at the bottom of (h) represent part of the blot above showing the unmodified Rac-GTP band at lower exposure, as well as levels of total Rac. Quantification of the Rac-GTP bands, both SUMO-modified and unmodified, is depicted in the histogram normalized to total Rac. Rac-nS1 in (b-h) is multi-mono-SUMOylated Rac1. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
We next evaluated whether Rac1 could be SUMOylated in vivo. SUMOylation is reversed by the SUMO-specific proteases SENP1-3 and SENP5-716; therefore we added N-ethylmaleimide (NEM) to the lysis buffer to inhibit these SUMO-deconjugating enzymes17. This allowed detection of higher molecular weight Rac1 bands potentially representing SUMO-modified Rac1 from cells co-expressing 6-His-SUMO-1 and GFP-Rac1 (Supplementary Information, Fig. S2i). To further demonstrate in vivo SUMOylation of Rac1, TAP-Rac1 was expressed with GFP-SUMO-1 or a GFP-SUMO-1 mutant lacking the diglycine C-terminal motif required for its conjugation to substrates (SUMO-1ΔGG). Probing GFP immunoprecipitates for Rac revealed up to three bands in lysates from GFP-SUMO-1, but not GFP-SUMO-1ΔGG expressing cells (Fig. 3d). Further, incubating the above GFP immunoprecipitates with the active domain of SENP118 resulted in loss of the higher molecular weight bands (Fig. 3e), confirming that these represented SUMO conjugates. Since SUMO-1 is not thought to form polymeric chains in vivo19 the presence of multiple bands suggested that Rac1 could be SUMOylated at multiple sites.
Since PIAS3 was identified as a Rac interactor in response to HGF, we analyzed if activation of the Met-HGF pathway induced the appearance of endogenous Rac-SUMO-1 conjugates. Endogenous Rac-SUMO conjugates were visualized in HeLa cells stably expressing 6His-SUMO-120 only following transfection with Trp-Met (Fig. 3f). Furthermore, we detected co-localisation of endogenous SUMO-1 and endogenous Rac1 in membrane ruffles of HGF-treated MDCKII cells (Supplementary Information, Fig. S3a). To visualize SUMOylation of endogenous Rac1 by addition of HGF alone and to determine the role of PIAS3 in this process, MDCKII cells were treated with HGF in the presence of a chemical cross-linker (DSS) to preserve SUMO-conjugated Rac during immunoprecipitation with a Rac-GTP specific antibody or CRIB-pull-down of GTP-Rac. Endogenous GTP-Rac1-SUMO-1 was detected in control cells following HGF treatment, but not in PIAS3 downregulated cells (Fig. 3g; Supplementary Information, Fig. S3b). We next determined whether SUMOylation of endogenous Rac occurs under other conditions that induce Rac activation. Calcium-switch stimulates cell-cell adhesion in confluent epithelial cells and Rac1 activation21. Following calcium re-addition and endogenous Rac-GTP pull-down we detected endogenous Rac-SUMO conjugates (Fig. 3h). Quantitation of SUMO-conjugated and unmodified Rac-GTP indicated that the SUMO-modified fraction is a significant proportion of Rac-GTP (histogram, Fig. 3h).
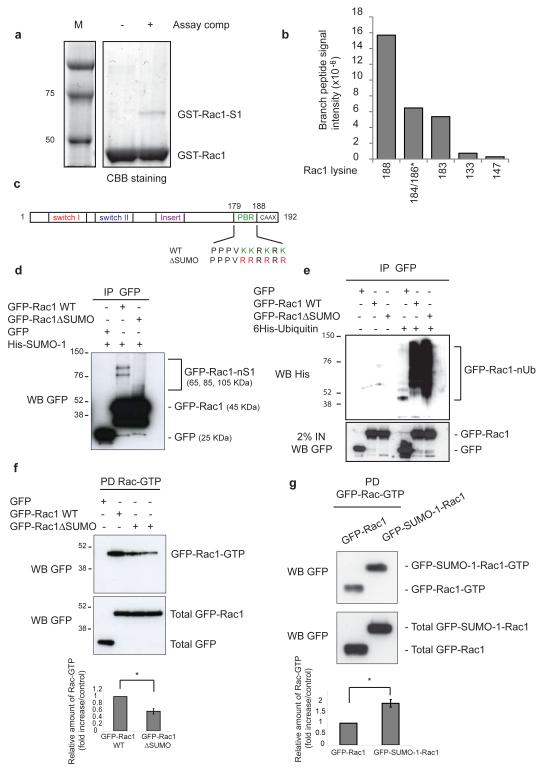
Frequently, SUMO targets the consensus sequence ΨKXD/E, where Ψ represents a hydrophobic residue and K the acceptor lysine; however, non-consensus sites have been reported22, 23. Rac1 lacks a consensus motif; therefore we adopted a MS approach to identify SUMO-modified site(s). GST-Rac1 was in vitro SUMOylated and the band corresponding to GST-Rac1-SUMO-1 (Fig. 4a) was subjected to in gel trypsin digestion and branched peptides identified by MS. Data was processed using MaxQuant24, and label free intensities for the branched peptides obtained which allowed for approximation of relative abundances. Although 10 of the potential total 17 lysine-SUMO-1 branched peptides were identified, almost 95% of the SUMO-1 modified Rac1 was conjugated at lysines 188, 183 and 184 or 186 (Fig. 4b) present within the C-terminal polybasic region (PBR) of Rac1 (Fig. 4c).
Figure 4.
Rac1 is SUMOylated in the polybasic region and SUMOylation affects its GTP-levels. (a) GST-Rac1 was incubated at 37°C in vitro in the presence (+) or absence (−) of complete SUMOylation assay components and resolved by SDS-PAGE. (b) The GST-Rac1-SUMO-1 band from (a) was excised and subjected to in gel trypsin digestion. The resultant peptides were analysed by mass spectrometry and MaxQuant data processing to detect modified and unmodified peptides. Column chart indicates the absolute intensity of SUMO-Rac1 branched peptides detected. * indicates that discrimination between lysines is not possible. (c) Schematic illustration of putative Rac1 SUMOylation sites and the mutations created. (d) HeLa cells stably expressing 6His-SUMO-1 were transfected as indicated, lysed in the presence of NEM, GFP immunoprecipitated and analysed by WB. Rac-nS1 indicates multi-mono-SUMOylated Rac1. (e) HeLa cells were transfected as indicated, lysed in the presence of NEM, GFP immunoprecipitated and analysed by WB. GFP-Rac1-nUb indicates Rac1 modified by poly-Ubiquitin. (f-g) COS7 cells transfected with the indicated plasmids were serum starved for 18 hours before 10 ng/ml of HGF was added for 30 minutes and assayed for Rac1 activity. Bar graphs depict quantification of the normalized relative amounts of Rac-GTP determined by scanning densitometry from at least three independent experiments. Error bars indicate ±SEM. * p<0.05, two-tailed Student’s t-test. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
Replacement of the putative SUMOylation lysines within the PBR with arginines (same charge) abolished most of the Rac1 SUMOylation in vitro (Supplementary Information, Fig. S3c-e). Moreover, lysines 96, 123, 130, 132, and 133 were not confirmed as SUMO-sites (Supplementary Information, Fig. S3f and data not shown). Interestingly, the in vitro SUMOylation assay showed one SUMO-Rac1 band, however, MS analysis identified more than one lysine modified by SUMO-1. This suggested that SUMOylation can occur on any of the lysines within the PBR. The monomeric SUMOylation of Rac1 in vitro is in contrast to the multiple SUMO-conjugate bands detected in vivo, presumably resulting from multi-monoSUMOylation. This difference implies the existence of additional factors in vivo that support multi-monoSUMOylation.
To confirm that the lysines present in the PBR were SUMOylated in vivo we generated a GFP-Rac1 mutant in which the four lysines of the PBR were replaced with arginines (GFP-Rac1ΔSUMO) (Fig. 4c). The Rac1ΔSUMO mutation was sufficient to decrease Rac1 SUMOylation (Fig. 4d) but not ubiquitylation (Fig. 4e; Supplementary Information, Fig. S3g). The PBR of Rac1 is implicated in binding certain effectors25-27, as well as plasma membrane and nuclear localization11, 28, 29. Rac1ΔSUMO mutation did not disrupt membrane or nuclear localization of Rac (Supplementary Information, Fig. S4a-c), nor did it affect Rac interaction with PIAS3, the effector IQGAP, or the Guanine nucleotide Exchange Factors (GEFs) β-Pix and Tiam1 (Supplementary Information, Fig. S4d-g).
We next investigated the connection between Rac1-GTP levels and SUMO modification. Interestingly, GFP-Rac1ΔSUMO was less GTP-bound than wild-type (Fig. 4f), suggesting that Rac SUMOylation was required to increase or maintain part of its activity. Consistent with this, a Rac1 chimaera mimicking constitutive SUMOylation (GFP-SUMO-1-Rac1; Supplementary Information, Fig. S4h) was more GTP-bound than wild-type Rac1 (Fig. 4g). Moreover, GFP-SUMO-1-Rac1 was localized similarly to wild-type GFP-Rac1 (Supplementary Information, Fig. S4i, j). These results suggest that Rac1 SUMOylation was not necessary for its localization but was required for its optimal GTP loading.
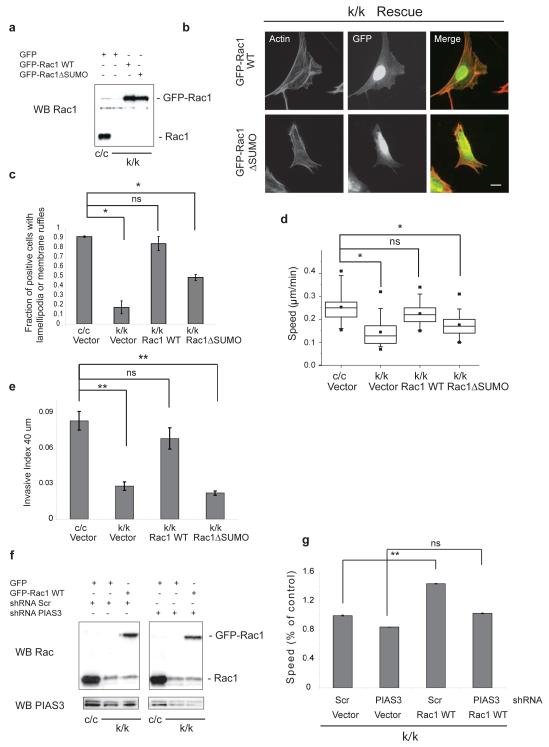
Subsequently, we addressed whether the defect in activation of non-SUMOylated mutant Rac affected its ability to promote cell migration. Rac1-null mouse embryonic fibroblasts (MEFs) derived from Rac1 conditional knock-out mice are unable to form lamellipodia-membrane ruffles and are defective in cell migration (Supplementary Information, Fig. S5a and30). We detected a higher molecular weight Rac1 species in these cells potentially corresponding to Rac1-SUMO-1 (Supplementary Information, Fig. S5b). Rac1-depleted MEFs were transfected with GFP, GFP-Rac1 WT or GFP-Rac1ΔSUMO (Fig. 5a) and GFP positive cells analyzed for the presence of lamellipodia-membrane ruffles (Fig. 5b). Whereas Rac1 WT rescued the lamellipodia-ruffle formation to around 85% of wild-type levels, non-SUMOylated Rac1 restored it to only 45% (Fig. 5c). Moreover, unlike Rac1 WT, the ΔSUMO mutant was unable to rescue significantly the migration defect of Rac1-depleted cells (Fig. 5d; Supplementary Information, Fig. S5c), or their invasion in response to HGF (Fig. 5e; see also Supplementary Information, Fig. S5d where the induction of invasion by HGF is demonstrated). However, the ΔSUMO mutant was equally competent as Rac1 WT to rescue the clonogenic defect of Rac1-depleted MEFs (Supplementary Information, Fig. S5e, f).
Figure 5.
SUMOylation of the polybasic region is required for optimal cell migration and invasion. (a) Representative Western blot (WB) showing expression levels of GFP-Rac1 WT or GFP-Rac1ΔSUMO in Rac1-depleted cells (k/k). c/c represents Rac1-WT MEFs. (b) Representative images of Rac1-depleted MEFs (k/k) reconstituted with GFP-Rac1 WT or GFP-Rac1ΔSUMO. Images were taken 24 hours later. Scale bar, 20 μm. (c) Fraction of k/k cells expressing the indicated constructs with lamellipodia-membrane ruffles 24 hours after transfection. Error bars indicate standard deviation based on n = 110-150 cells from 3 independent experiments. (d) Cells as in (a) grown on glass bottom dishes were tracked for at least 24 hours. Box-whisker plots of cell velocities of n = 90-100 cells from three independent experiments are shown. (e) Cells as in (a) were analyzed for invasion at 40 μm in the presence of HGF. Error bars indicate standard deviation based on n=5 wells. A representative from 3 independent experiments is shown. (f) Representative WB showing expression levels of GFP-Rac1 WT in Rac1-depleted cells (k/k) also stably expressing scr shRNA or pias3 shRNA. Please note lower levels of expression of exogenously expressed GFP-Rac1 WT compared to endogenous levels of Rac1 of control c/c MEFs. (g) Individual cells as in (f) were tracked for 24 hours. Data shown is average speed of migration of at least 50 cells per experiment. A representative experiment is shown here. (c-e, g) *p<0.001, ** P<0.05, ns indicates no significant difference, two-tailed Student’s t-test. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
Similarly to MDCKII, PIAS3 downregulation in MEFs resulted in reduced migration and membrane ruffle formation (Supplementary Information, Fig. S5g and data not shown). To determine whether PIAS3 was required for the ability of Rac1 WT to rescue cell migration in Rac1-depleted cells, PIAS3 was downregulated and cells then infected with GFP-Rac1 WT (Fig. 5f) before being analyzed for migration and membrane ruffle formation. Significantly, while Rac1 WT partly restored the migration and membrane ruffling of Rac1-depleted cells additionally expressing control shRNA, it was unable to do so in cells expressing shRNA targeting PIAS3 (Fig. 5g; Supplementary Information, Fig. S5h). These results suggest that PIAS3, likely through promoting Rac SUMOylation, is to a great extent responsible for the ability of Rac1 WT to rescue the phenotype of the Rac1-depleted cells. However, it can not be excluded that PIAS3 exerts some Rac-independent effects on cell motility.
Our study demonstrates that Rac1, and particularly GTP-bound Rac, can be SUMOylated. While SUMOylated-Rac represents a small fraction of total Rac, it can be a significant fraction of GTP-bound Rac (Fig. 3g, h). However, since not all Rac-GTP is SUMOylated (Fig. 3g, h), since Rac1ΔSUMO retained some GTP-binding (Fig. 4f), because PIAS3 interacts more with Rac-GTP than Rac-GDP (Fig. 1c), and also because active Rac1 is a better SUMOylation substrate than Rac1 WT (Fig. 3c), our data imply that SUMOylation is not essential for activation, but rather influences the maintenance of the active state. Consistent with this, levels of active Rac1 following HGF treatment were reduced by downregulation of PIAS3 (Fig. 1f and 3g), and augmented by PIAS3 overexpression (Fig. 1h, i and 2c, d), mirroring the effects of PIAS3 on levels of Rac SUMOylation (Figs 3b, g; Supplementary Information, Fig. S2h, S3b). Further, a SUMO-mimicking Rac1 chimaera was more GTP-bound than Rac1 WT (Fig. 4g). Potentially, Rac SUMOylation could either increase its binding to a GEF or conversely decrease binding to a GTPase activating protein (although such effects need to be demonstrated; for model see Supplementary Information, Fig. S5i). Significantly, we show that Rac SUMOylation is important for optimal lamellipodia-ruffle formation, cell migration and invasion (Fig. 5b-e), but not for clonogenicity (Supplementary Information, Fig. S5e, f). This conclusion is strengthened by the observation that PIAS3 is required for Rac1 WT to restore membrane ruffling and migration in Rac1-depleted MEFs (Fig. 5g). SUMOylation may possibly increase active Rac levels above a threshold required for lamellipodia formation, cell migration and invasion and/or target a pool of Rac towards particular biological functions through influencing binding to effector molecules, consistent with findings that SUMOylation of other proteins influences their interactions31. Overall, our study advances our understanding of how GTPases and in turn cell migration are regulated and could provide new targets for future therapeutic intervention.
Supplementary Material
Acknowledgements
This study was supported by CR-UK grant C147/A6058 and an EMBO Long-Term fellowship (to SCL). MHT and EGJ were supported by CR-UK. We thank Dr Maurice (Dundee, UK) for MS analysis, Dr Matic (Martinsried, Germany) for help with SUMO-Rac1 branched peptide data interpretation, Prof. Yokosawa, Dr Trusolino and L. Vidali for reagents, N. Mack for help with the calcium switch experiments, I. Arozarena and C. Wellbrock for help with invasion assays and members of the Cell Signalling Group, A. Hurlstone, N. Divecha, J. Pérez-Martín and C. Wilkinson for critical reading of the manuscript and helpful discussions.
METHODS
Constructs
N-terminal TAP tag (N-TAP) cloned in pcDNA4/TO (Invitrogen) was a gift from Prof. Clevers (Hubrecht Laboratory, Utrecht, The Netherlands). Mouse full-length Rac1V12 was cloned in frame with N-TAP (to yield TAP-Rac1V12). This was used as a template to mutate V12 to G12 using the QuickChange site-directed mutagenesis kit (Stratagene). A mammalian expression construct encoding wild-type PIAS3 (MYC-PIAS3) was provided by Prof. Yokosawa (Department of Biochemistry, Hokkaido University, Sapporo, Japan). The Trp-Met plasmid was provided by Dr Trusolino (Institute for Cancer Research and Treatment, University of Torino Medical School, Italy). FLAG-PIAS3 was obtained by cloning Pias3 cDNA in-frame with the FLAG-epitope tag of pGloFlag vector provided by Dr Hurlstone (University of Manchester, UK). Full-length PIAS3 was fused to the N-terminus of the glutathione S-transferase (GST) gene in pGEX-KG. GST-NPIAS3 and -CPIAS3 were obtained by replacing full-length PIAS3 with the first 900 or last 981 bases respectively of PIAS3. MYC-CPIAS3 was obtained by replacing full-length PIAS3 cDNA in the MYC-PIAS3 plasmid with the last 981 bases of PIAS3. GFP-Rac1WT and GFP-SUMO-1 were obtained by cloning Rac1WT or SUMO-1 respectively into pEGFP-C1 (Clontech). The mutant GFP-SUMO-1ΔGG was obtained by deleting the last 6 amino acids from the C-terminus of GFP-SUMO-1. The GFP-SUMO-1ΔGG-Rac1 plasmid was obtained by cloning SUMO-1ΔGG into the GFP-Rac1 plasmid. GST-Rac1ΔSUMO-1, GST-Rac1 2KRa, 2KRb, 3KRa, 3KRb or GFP-Rac1ΔSUMO-1 mutants were derived from GST-Rac1WT or GFP-Rac1WT respectively using site direct mutagenesis. The generation of the single-vector tetracycline-repressor based inducible shRNA system has been described elsewhere32. DNA sense and antisense oligonucleotides were generated against the following target sequences in PIAS3: shRNA#1, CCAACACCATCGTGGTCAA; shRNA #2, CCTTCCTCCTACCAAGAAT. pRS-PIAS3 shRNA was generated by cloning shRNA#1 into pRETROSUPER.
Cell culture and cell lines
HEK293T, MDCKII, HeLa, and COS7 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) in the presence of 10% fetal bovine serum (FBS; GIBCO). MEF c/c cells, a kind gift from Dr Vidali (Biology Department, University of Massachusetts, Amherst, USA), were maintained in the same medium containing 70 μM β-mercaptoethanol. Rac1 deletion was induced by incubating cells with Ad5CMCre-eGFP (Gene Transfer Vector Core; University of Iowa). Cell lines expressing the inducible shRNA system were maintained in DMEM supplemented with 10% tetracycline-free FBS (Autogen Bioclear) and 1 mg/ml G418 (Sigma). HEK293T, HeLa and COS7 cells were transfected using Fugene 6 (Roche). Single cell clones of MDCKII cells stably expressing TAP-tagged Rac1WT or N-TAP were used for subsequent experiments. For growth factor treatment, serum was removed for 16 hours before addition of 10 ng/ml HGF or EGF (R&D Systems) for 30 min or as indicated. For COS7, cells were transfected in the presence of 10% FBS and left for 24 hours before serum removal and growth factor treatment. For the calcium switch experiment MDCKII cells were grown to confluency in complete medium, after which medium was changed to low calcium (0.02 mM) for 18 hours. Calcium containing medium (1.8 mM) was added back for the indicated times. For PIAS3 downregulation, MDCKII cells with pias3 or scr shRNAs were treated with 1 μg/μl doxycycline (dox) for four days. For the cross-linking of Rac-SUMO conjugates in vivo a range of DSS concentrations were added (Thermo Scientific) in the presence of HGF for 30 min. The reaction was quenched by addition of glycine to a final concentration of 20 mM for 15 min. Cells were washed twice in ice-cold PBS and subjected to either Rac-GTP pull-down assay or immunoprecipitation with anti-active Rac-GTP antibody (NewEast Biosciences).
TAP purification
MDCKII cells stably expressing TAP-Rac1 or N-TAP were plated at low density in five 500cm2 plates and allowed to grow in small colonies. Serum was removed for 18 hours before the addition of 10 ng/ml HGF. TAP purification was performed as previously described33.
GST pull-down assays
GST-pull-down assays were performed as previously described34.
Nickel affinity purification
6His-SUMO-1 or 6His-Ubiquitin binding proteins were purified from cell lysates using nickel-nitrilotriacetic acid agarose (QIAGEN) as previously described35.
Protein analysis
For immunoprecipitation (IP) and immunoblot analysis, cell lysates were prepared in IP lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% (v/v) Triton-X-100, 10% (v/v) glycerol, 2 mM EDTA, 25 mM NaF, and 2 mM NaH2PO4) containing protease and phosphatase inhibitor cocktails (Sigma). Equivalent amounts of protein were either denatured with SDS-PAGE sample buffer (Nupage, Invitrogen) for immunoblotting, or incubated with 2-5 μg antibody pre-bound to 20 μl of GammaBind G-Sepharose (Amersham) for IP analysis. Immunoprecipitated proteins were eluted with SDS-PAGE sample buffer (Nupage, Invitrogen), resolved by SDS-PAGE, and transferred to polyvinylidene difluoride membrane (Immobilon-P; Millipore). Immunoblotting was performed using the following primary antibodies: anti-Myc (Clone 9E10, CRUK), anti-Flag (Clone M2, Sigma), anti-Rac (clone 102, BD Biosciences), anti-Rac1 (ARC03, Cytoskeleton), anti-Cdc42 (Santa Cruz), anti-Rho GDI (Clone A-20, Santa Cruz), anti-IQGAP1/2 (Clone 24, BD Biosciences), anti-GST (ab58626, Abcam), anti-PIAS3 (N-Term or C-Term, ABGENT), anti-PAP (p 1291, Sigma), anti-c-Met (Clone C-12, Santa Cruz), anti-Tubulin (Clone TAT-1, CRUK), anti-GFP (CRUK), anti-6His (631212, Clontech), anti-Tiam1 (C16, Santa Cruz), anti β-Pix (BD Biosciences), anti-ERK, anti-p-ERK (Cell Signaling), anti-p-p38, anti-p38 (Cell Signaling) and subsequently with horseradish peroxidise-conjugated anti-mouse, anti-rabbit or anti-sheep secondary antibodies (GE Healthcare) and visualised by enhanced chemiluminescence (Perkin Elmer).
Subcellular fractionation
Cells were washed and treated with ice-cold hypotonic lysis buffer (10 mM Tris-HCl pH 7.4, 1.5 mM magnesium chloride, 5 mM potassium chloride, 1 mM DTT, 0.2 mM sodium vanadate, 1 mM PMSF) containing protease and phoshatase inhibitor cocktails (Sigma) for 5 min. Lysates were homogenized by sonication at low speed. Homogenates were centrifuged at 700g for 3 min to pellet nuclei and intact cells. Supernatants were then spun at 40,000g for 30 min at 4°C. The cytosol-containing supernatant was removed and the crude membrane pellet gently washed with hypotonic lysis buffer. Equal amounts of protein from membrane and cytosol fractions were then analysed by Western blotting.
In vitro SUMOylation
In vitro SUMOylation reactions contained E1 enzyme (250 ng SAE1/SAE2), E2 enzyme (1.2 μg Ubc9), SUMO-1 or SUMO-2 protein (5.7 μg) and GST-Rac1 protein (5 μg) in SUMOylation buffer (55 mM Tris, pH 7.5, 5.5 mM MgCl2, 2.2 mM ATP, 5.5 mM DTT). SUMOylation reactions were incubated at 37°C for 4 hours. After termination with SDS-PAGE sample buffer, reaction products were separated on SDS-PAGE.
Rac and Cdc42 activity
Rac and Cdc42 activity assays were performed as previously described33.
Cell migration assay
MDCK cells stably expressing pias3 shRNAs were plated in the presence or absence of doxycycline for 4 days. Cells were seeded sparsely and images were acquired every 30 min for 24 hours at 37°C using a time-lapse system consisting of a Zeiss Axiovert 200M microscope under the control of Metamorph software (Molecular Devices, PA, USA). Single cells were manually tracked using ImageJ software (NIH).
Invasion assay
Cells were suspended in serum-free collagen I at 2.3 mg/ml to a final concentration of 105 cells/ml and HGF (10 ng/ml) added where indicated. 100μl aliquots were dispensed into 96-well ViewPlates (Perkin-Elmer; Buckinhamshire, UK) coated with 0,2% bovine serum albumin (Sigma). Plates were centrifuged at 300g (to sediment cells) and incubated at 37°C/5% CO2 for 60 min before adding 30 μl of DMEM (Invitrogen) containing 10% FBS (GIBCO) and β-Mercaptoethanol. After 24 hours, cells were fixed in 4% formaldehyde and stained with 5 mg/ml Hoechst 33258 (Sigma). Confocal z sections were collected from each well at 0 μm (bottom of well) and 40 μm with a BD ATTovision (version 1.61855). Nuclear staining was quantified with ATTovision software. Invasion indexes were calculated as the number of cells at 40 μm divided by those at 0 μm (mean values of quintuplicate samples).
Identification of proteins from TAP by LC-MS/MS analysis and database searches
Gel lanes from 1D gels (Nupage 4–12%) were manually cut into 40 × 1.5 mm bands using a razor blade. Processing of bands and mass spectrometry analysis was performed as previously described33. Product ion data were searched against the combined forward and reverse Canis familiaris protein database build 2.1 downloaded from NCBI RefSeq (ftp://ftp.ncbi.nih.gov/genomes/Canis_familiaris/) using the same criteria as previously described33. These criteria resulted in a false discovery rate (FDR) of 0.5% at the protein level for this data set. Of these identified proteins, only those that were detected in TAP-Rac1 samples in three different experiments and not in control N-TAP samples were classified as being Rac1-specific interactors.
Identification of sites of Rac1 sumoylation in vitro
GST-Rac1 was modified with SUMO-1 by in vitro conjugation (see In vitro SUMOylation above). Assay components were fractionated by SDS-PAGE and Coomassie-stained protein bands subjected to in-gel digestion with trypsin (Promega) as described previously36. Mass spectrometric analysis was performed by LC-MS/MS using a linear ion trap-orbitrap hybrid mass spectrometer (LTQ-Orbitrap, Thermo Fisher Scientific) equipped with a nanoelectrospray ion source and coupled to a Proxeon nano-HPLC system (Proxeon Biosystems). Peptides were injected into a 150 mm reverse phase C18 column and eluted with a linear 60-min gradient. Data were acquired in the data-dependent mode to automatically switch between MS and MS/MS acquisition. The 5 most intense ions with 3+ or greater charge were fragmented by collision induced dissociation and recorded in the linear ion trap (full details of HPLC gradient and MS acquisition settings are available upon request).
MS raw data files were processed using the quantitative MS processing software MaxQuant (version 1.0.12.31) with SILAC type set to ‘singlets’37,38, and using the Mascot search engine (Matrix Science, version 2.2.2). Please see 36 for further details of the algorithms and principles applied in the MaxQuant package. Enzyme specificity was set to trypsin-P. Cysteine carbamidomethylation was selected as a fixed modification and methionine oxidation and protein N-acetylation were searched as variable modifications. Data were searched against a target/decoy human IPI database (version 3.24)39 that was appended with linear mimetics of branched peptides representing all possible lysine modifications of Rac1 with the C-terminal tryptic fragment of SUMO-1. The use of linear representations of branched adducts has already been described for automated database search applications40. By this method, identified branched peptides are shortlisted as proteins in the MaxQuant output file. Initial maximum allowed mass deviation was set to 7 parts per million (ppm) for peptide masses and 0.5 Da for MS/MS peaks. The minimum peptide length was set to 6 amino acids and a maximum of four missed cleavages. 1% FDR was required at both the protein and peptide level.
Statistical analysis
Statistical differences between two groups of data were analysed with a two-tailed unpaired Student’s t-test.
Footnotes
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 2.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–70. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellenbroek SI, Collard JG. Rho GTPases: functions and association with cancer. Clin Exp Metastasis. 2007;24:657–72. doi: 10.1007/s10585-007-9119-1. [DOI] [PubMed] [Google Scholar]
- 4.Royal I, Lamarche-Vane N, Lamorte L, Kaibuchi K, Park M. Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol Biol Cell. 2000;11:1709–25. doi: 10.1091/mbc.11.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 6.Hays JL, Watowich SJ. Oligomerization-induced modulation of TPR-MET tyrosine kinase activity. J Biol Chem. 2003;278:27456–63. doi: 10.1074/jbc.M210648200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, et al. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–6. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 8.Ridley AJ, Comoglio PM, Hall A. Regulation of scatter factor/hepatocyte growth factor responses by Ras, Rac, and Rho in MDCK cells. Mol Cell Biol. 1995;15:1110–22. doi: 10.1128/mcb.15.2.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–34. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans. 2007;35:1405–8. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- 11.Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL. The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J Biol Chem. 2004;279:44197–210. doi: 10.1074/jbc.M404977200. [DOI] [PubMed] [Google Scholar]
- 12.Michaelson D, et al. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol. 2008;181:485–96. doi: 10.1083/jcb.200801047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashina K, Yamamoto H, Chayama K, Nakajima K, Kikuchi A. Suppression of STAT3 activity by Duplin, which is a negative regulator of the Wnt signal. J Biochem. 2006;139:305–14. doi: 10.1093/jb/mvj033. [DOI] [PubMed] [Google Scholar]
- 14.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci U S A. 2002;99:2872–7. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–95. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, et al. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J Biol Chem. 1999;274:31131–4. doi: 10.1074/jbc.274.44.31131. [DOI] [PubMed] [Google Scholar]
- 18.Martin SF, Hattersley N, Samuel ID, Hay RT, Tatham MH. A fluorescence-resonance-energy-transfer-based protease activity assay and its use to monitor paralog-specific small ubiquitin-like modifier processing. Anal Biochem. 2007;363:83–90. doi: 10.1016/j.ab.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Tatham MH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–74. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 20.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–71. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 21.Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. J Biol Chem. 2001;276:33305–8. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- 22.Kamitani T, et al. Identification of three major sentrinization sites in PML. J Biol Chem. 1998;273:26675–82. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 23.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 24.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J Cell Biol. 2006;172:759–69. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolias KF, Couvillon AD, Cantley LC, Carpenter CL. Characterization of a Rac1- and RhoGDI-associated lipid kinase signaling complex. Mol Cell Biol. 1998;18:762–70. doi: 10.1128/mcb.18.2.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolias KF, et al. Type Ialpha phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr Biol. 2000;10:153–6. doi: 10.1016/s0960-9822(00)00315-8. [DOI] [PubMed] [Google Scholar]
- 28.van Hennik PB, et al. The C-terminal domain of Rac1 contains two motifs that control targeting and signaling specificity. J Biol Chem. 2003;278:39166–75. doi: 10.1074/jbc.M307001200. [DOI] [PubMed] [Google Scholar]
- 29.Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003;15:1071–80. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 30.Vidali L, Chen F, Cicchetti G, Ohta Y, Kwiatkowski DJ. Rac1-null mouse embryonic fibroblasts are motile and respond to platelet-derived growth factor. Mol Biol Cell. 2006;17:2377–90. doi: 10.1091/mbc.E05-10-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 32.Rooney C, et al. The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 2010;11:292–8. doi: 10.1038/embor.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodcock SA, Jones RC, Edmondson RD, Malliri A. A modified tandem affinity purification technique identifies that 14-3-3 proteins interact with Tiam1, an interaction which controls Tiam1 stability. J Proteome Res. 2009;8:5629–41. doi: 10.1021/pr900716e. [DOI] [PubMed] [Google Scholar]
- 34.Woodcock SA, et al. SRC-induced disassembly of adherens junctions requires localized phosphorylation and degradation of the rac activator tiam1. Mol Cell. 2009;33:639–53. doi: 10.1016/j.molcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–71. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 36.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 37.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 38.Cox J, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 39.Kersey PJ, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–8. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 40.Maiolica A, et al. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol Cell Proteomics. 2007;6:2200–11. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.