Abstract
The Myc family of proto-oncogenes plays a central role in tumorigenesis, yet identifying the specific transcriptional targets required for its oncogenic function remains a challenge. Given Myc’s broad role in transcriptional regulation, it seems unlikely that there exists one or even a small set of Myc effectors strictly required for transformation. Over the last decade or so, it has become clear that Myc can drive several metabolic pathways associated with cell growth. There is compelling evidence that Myc regulates these pathways directly and that their regulation is not an epiphenomenon. As such, for understanding Myc’s pleiotropic role in cell growth, cell division, and cell death, it may be fruitful to focus more broadly on Myc-regulated pathways than on specific targets. Myc was first shown to regulate glycolysis, but it is now clear that Myc regulates many biosynthetic pathways required for cell growth and division. A related family of transcriptional regulators, the Mondo family, has recently been discovered that may interact with members of the Myc family to control cell growth. The Mondo family is a key sensor of intracellular bioenergetic charge, and one function appears to be in controlling the availability and utilization of intracellular glucose. Here we focus on the metabolic pathways regulated by Myc and Mondo and speculate on the largely unexplored question of their cooperation in controlling cancer cell metabolism.
Keywords: glycolysis, glutaminolysis, MondoA, transcription, Myc
Cancer Cell Metabolism
A near-universal feature of cancer cells is their upregulation of aerobic glycolysis. First observed by Otto Warburg in the 1920s, the mechanisms that drive this molecular conversion are now being dissected in molecular detail. Furthermore, the metabolic pathways dysregulated in cancer show promise for developing new targeted agents for treatment of cancer. These two topics have been recently covered by several excellent reviews1–5 and are not discussed in detail here. We focus primarily on how Myc controls glycolysis, biosynthetic pathways, mitochondrial function, and glutaminolysis—all key pathways required to support cell growth. We then focus on the less well-characterized Mondo family and its role in sensing nutrients and controlling cell growth.
A central question in cancer biology is, what competitive advantage do cancer cells gain from their elevated level of aerobic glycolysis? Again, others have recently reviewed this topic.6,7 Briefly, these benefits appear to be cell autonomous and cell nonautonomous (Figure 1). From the cell-autonomous perspective, even though glycolysis produces less ATP per molecule of glucose than that of respiration, it is kinetically much faster than respiration and can provide sufficient ATP to support the bioenergetic needs of rapidly dividing cancer cells.5 Furthermore, elevated glycolysis provides carbon skeletons required for the biosynthetic reactions that produce the macromolecules required for high rates of cell division.8 Glucose-derived carbons feed the pentose phosphate pathway, which generates the ribose sugars required for nucleotide biosynthesis. Additionally, the pentose phosphate pathway generates NADPH, which is required for the reductive biosynthesis of nucleotides and fatty acids. Glucose-derived carbons enter the TCA cycle via pyruvate, where they support a number of biosynthetic reactions. For example, fatty acids—required for membrane biosynthesis—are derived from the TCA cycle intermediate citrate, and several amino acids (e.g., alanine and aspartate) are derived from TCA cycle intermediates.8,9 Finally, several reports have demonstrated that a high rate of aerobic glycolysis results in inactivation of PTEN, leading to the activation of the PI3K-AKT growth and survival pathway.10,11 In summary, elevated aerobic glycolysis can provide tumor cells with ATP and macromolecules. Furthermore, aerobic glycolysis provides cancer cells with a growth and survival advantage.
Figure 1.
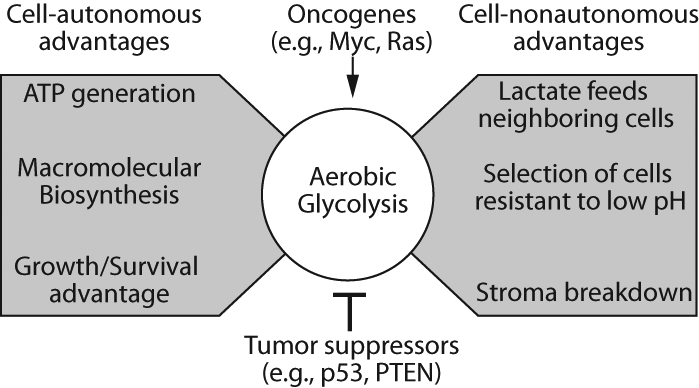
Growth advantages provided to cancer cells by aerobic glycolysis, which can be upregulated by oncogenes and inhibited by tumor suppressors.5 Activation of aerobic glycolysis provides growth advantages through cell-autonomous and cell-nonautonomous mechanisms, as indicated.
From the cell-nonautonomous perspective, elevated aerobic glycolysis provides several advantages. For example, lactate—the termination point of glycolysis—is effluxed from tumor cells via monocarboxylate transporters. Several functions have been proposed for secreted lactate and the subsequent reduction in pH of the extracellular milieu. Lactic acidosis may help degrade the tumor cell stroma, providing tumor cells with increased opportunities for migration, invasion, and metastasis. Furthermore, low pH is a potent cell stress that can trigger apoptosis. Thus, lactic acidosis likely provides a selective pressure that drives outgrowth of cell clones resistant to apoptosis, possibly through inactivation of the p53 tumor suppressor.7 In addition, lactate from highly glycolytic cells in the hypoxic regions of tumors may provide carbons to support cell growth in the aerobic regions of the tumor.12,13 Finally, hypoxic tumor cells can trigger angiogenesis via upregulation of key angiogenic target genes. Researchers generally accept that increased blood supply provides additional avenues to supply the tumor with nutrients; however, others have proposed that increased vascularization provides an avenue by which some potentially deleterious effects of lactic acidosis in the tumor microenvironment can be dissipated.6 It is interesting that a lactic acidosis–driven gene signature in the absence of elevated glycolysis portends a better prognosis in breast cancer.14 It is possible that early in tumorigenesis, tumor cells sense lactic acidosis as a stress and so adapt by restricting glycolysis. Later in tumorigenesis, these controls may be subverted by secondary events.
The Myc Superfamily of Transcription Factors
Myc is a member of the basic region helix loop helix leucine zipper (bHLHZip) family of transcriptional regulators. The Myc family comprises 3 core members: c-, N-, and L-Myc. For simplicity, here we generically refer to the family as Myc. Like many members of this class of transcription factor, Myc interacts with another family member to bind DNA and activate—or, in some cases, repress—gene expression. Myc interacts with the bHLHZip protein Max to form a functional heterocomplex that binds CACGTG or closely related E-box elements in the promoters of target genes. Numerous microarray studies and genomewide localization studies have implicated Myc:Max complexes in the direct regulation of thousands of protein-coding target genes. Myc:Max complexes are capable of regulating a number of miRNAs, often with important outcomes in cellular transformation.15 If the vast number of Pol II–dependent targets were not confounding enough, determining how Myc transforms cells is further complicated by the fact that it can directly participate in transcription by Pol I and Pol III,16 that it appears to have a role in regulating global chromatin architecture,17 and that, at least in Drosophila, it can function independently of Max.18 Myc’s role in nucleotide biosynthesis, ribosome biogenesis, glycolysis, mitochondrial function, and, most recently, glutaminolysis is of interest from the standpoint of understanding cancer cell metabolism. We highlight a selection of these pathways here to illustrate Myc’s broad role in regulating metabolism and how it might interact with the Mondo family of glucose sensors.
Myc and Aerobic Glycolysis
One of the first hints that Myc might play an important role in regulating aerobic glycolysis was the seminal demonstration by the Dang lab that Myc could regulate the rate-limiting glycolytic enzyme lactate dehydrogenase A (LDH-A).19 Since that time, it has been established that Myc directly activates transcription of virtually every glycolytic enzyme gene, including several rate-limiting enzymes, such as hexokinase II. Myc can also directly control the availability of intracellular glucose by activating transcription of the glucose transporters GLUT-1, GLUT-2, and GLUT-4 (Figure 2).20,21 These findings lend support to the hypothesis that Myc is a necessary and sufficient regulator of aerobic glycolysis.
Figure 2.
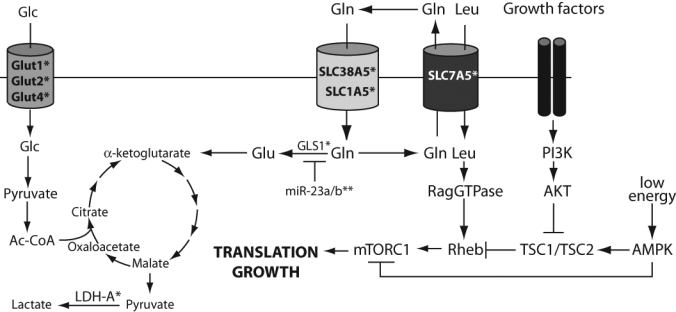
Myc regulates glycolysis and glutaminolysis. Myc upregulation leads to increased energy production through glycolysis and increased growth through glutaminolysis. The growth-promoting activity of the mTORC1 complex is controlled by flux through growth factor signaling pathways and intracellular bioenergetic charge via the AMP-dependent protein kinase AMPK. We propose that increased leucine uptake in Myc-overexpressing cells could lead to mTORC1 activation. Glc = glucose; Gln = glutamine; Glu = glutamate; Leu = leucine; Ac-CoA = acetyl Co-A; GLS1 = glutaminase 1; LDH-A = lactate dehydrogenase. *Myc-activated target. **Myc-repressed target.
Another family of transcriptional regulators has been identified that activates glycolytic target genes—hypoxia-inducible factors 1α and 2α (HIF-1α and HIF-2α), collectively referred to as HIF-α—which are transcription factors classically identified as being stabilized under low oxygen conditions.22 An additional role for HIF-1α during normoxic cell growth has recently been described.23 The genetic and biochemical interactions between Myc and the different HIF-α are complex and depend on the oxygen tension, the transformation state of the cells, and the promoter context of shared targets.21 Like Myc, HIF-α can regulate the expression of most genes encoding glycolytic enzymes. A comprehensive list of glycolytic targets regulated by Myc and/or HIF-α has recently been published.21 Myc and HIF-α recognize related DNA-binding sequences, so the possibility exists that Myc and HIF-α may coordinate the expression of glycolytic targets through overlapping sites in the promoters in these genes, although this model has not been broadly tested. Another possibility for Myc–HIF-α cooperation is that HIF-2α can stabilize transcriptionally active Myc:Max complexes on target promoters.24 The cooperation between Myc and HIF-2α drives cell growth and transformation, but whether these phenotypes require Myc–HIF-2α interactions at shared glycolytic targets or at other targets is not known. Myc–HIF-α cooperation has been demonstrated at cell cycle and DNA repair pathway targets, so possibilities clearly exist for cooperation in growth and transformation.
Myc and Biosynthetic Reactions
In addition to having a role in driving aerobic glycolysis, Myc has a broad role in driving biosynthetic reactions required to supply growth-supporting macromolecules. Several early microarray studies suggested the connection between Myc and biosynthetic pathways. Supporting this was the discovery that serine hydroxymethyltransferase (mSHMT) can rescue the slow growth phenotype of Myc null rat fibroblasts.25 mSHMT is a direct Myc target and the source of the single-carbon units required for folate metabolism and for amino acid and nucleotide biosynthesis. A cell-labeling approach using [U-13C]glucose was more recently conducted in wt and Myc null rat fibroblasts following serum stimulation.26 This study showed broad involvement of Myc in directing glucose-derived carbons toward the biosynthesis of amino acids, ribose sugars, purine nucleotides, and lipids. Myc increased the flux through the hexosamine pathway, resulting in elevation of O-linked GlcNAc protein modification; thus, Myc drives biosynthetic reactions and protein modification. This latter point is interesting given that Myc can increase the amount of acetyl-CoA, which not only has a role in the TCA cycle but is a donor for protein acetylation. The study’s authors speculated that Myc-driven increases in acetyl-CoA could result in elevated histone acetylation, which is generally an activating mark for gene activation.27 It is plausible that the alterations in global chromatin structure observed following N-Myc deletion might be related to a reduction in the acetyl-CoA pool required for histone modification.17
Myc and Mitochondrial Function
Functional electron transport, mitochondrial oxygen consumption, and mitochondrial DNA are dispensable for cell viability. However, a functional TCA cycle is essential to the biosynthetic reactions that support cell growth.8 As such, it would make sense that mitochondrial mass and/or function would increase in transformed cells. Again, Myc seems capable of driving both these functions. Myc can activate expression of several nuclearly encoded genes required for mitochondrial function.28 Furthermore, Myc can directly activate the mitochondrial transcription factor TFAM, which is required for transcription of the genes encoded by the mitochondrial genome and for mitochondrial DNA replication. This mechanism enables Myc to broadly dictate expression of the mitochondrial genome and its copy number. Thus, in addition to driving aerobic glycolysis, Myc can dictate the utilization of glucose-derived carbons for biosynthetic reactions by controlling mitochondrial mass and function. Myc also interacts with HIF-α in controlling mitochondrial function. In this context, interactions between Myc and HIF-α proteins are generally antagonistic. For example, HIF-1α activates the expression of pyruvate dehydrogenase kinase 1 (PDK1).29,30 PDK1 phosphorylates and inhibits the activity of pyruvate dehydrogenase, which converts pyruvate to acetyl-CoA; as a consequence, PDK1 prevents glucose-derived acetyl-CoA from entering mitochondria. Furthermore, HIF-1α can disrupt Myc:Max complexes and activate the expression of the Myc antagonist Mxd2 (Mxi1).31 Both mechanisms may help limit Myc’s positive role in mitochondrial function and thus restrict cell growth under hypoxic conditions.
Myc and Glutaminolysis
The essential role for glucose in supporting cell growth is complemented by the additional requirement for glutamine in cell growth. Glutamine is generally considered a nonessential amino acid because it is synthesized by most cell types; however, the demand for glutamine cannot typically be satisfied by de novo synthesis. The high demand for extracellular glutamine stems from its requirement for the synthesis of several biomolecules required for cell growth. For example, in addition to being required for protein synthesis, glutamine is required for DNA synthesis, to provide nitrogen for synthesis of purines and pyrimidines. Furthermore, glutamine can supply carbons to support TCA-mediated biosynthesis and generate NADPH. Glutamine enters the TCA cycle by being metabolized to glutamate and, subsequently, the TCA intermediate α-ketoglutarate (α-KG). Given the pleiotropic role of glutamine in different biosynthetic reactions, it is not surprising that glutamine uptake and utilization increase in a variety of tumor types.32 Several recent studies pointed to Myc as a key regulator of glutaminolysis, which supports Myc’s broad command over cellular biosynthetic reactions.
The first hint that Myc might contribute to glutaminolysis was the demonstration that Myc overexpression in normal diploid fibroblasts causes glutamine addiction; that is, Myc-expressing cells undergo apoptosis when glutamine is removed from the medium.33 These studies did not identify the precise cause of cell death; however, glutamine removal correlated with depletion of TCA intermediates, and addition of the TCA intermediate oxaloacetate blocked apoptosis. Two recent publications provided insight into the mechanisms by which Myc drives glutaminolysis. The first study demonstrated that Myc can directly regulate the expression of the glutamine transporters SLC38A5 and SLC1A5, thereby increasing glutamine uptake into cells (Figure 2).34 Furthermore, Myc can activate the expression of glutaminase 1 (GLS1), which deamidates glutamine to produce glutamate. Flux through glutaminolysis may also increase by the Myc-dependent activation of lactate LDH-A, which converts glutamine-derived pyruvate to lactate.34 The second study showed that Myc regulates GLS1 but by a completely different mechanism. In this case, Myc overexpression caused repression of the microRNAs miR-23a and miR-23b, which target the 3′ untranslated region of the GLS1 message to block its translation.35 Thus, Myc overexpression lifts the microRNA-dependent translational repression of GLS1 and increases mitochondrial glutaminase activity. Collectively, Myc can regulate GLS1 expression by transcriptional and translational mechanisms. Which mechanism predominates is likely dictated by cell type or context. Together, these studies provided the mechanistic framework for understanding how Myc controls glutamine utilization.
Can Myc-Dependent Glutaminolysis Activate mTORC1 Complexes?
Myc’s role in glutaminolysis is proposed to stimulate cell growth by providing glutamine-derived carbons for biosynthetic reactions. However, a recent publication hinted at an additional mechanism by which elevated Myc-dependent glutaminolysis can stimulate cell growth. It is well known that the mTOR complex 1 (mTORC1) is a key regulator of protein translation and growth and is activated by high nutrient levels and flux through growth factor signaling cascades.36 mTORC1 activity is also stimulated by the availability of amino acids—especially, leucine—via their activation of the Rag-GTPases.37 Glutamine efflux via the glutamine antiporter SLC7A5 is required for efficient uptake of essential amino acids such as leucine and for mTORC1 activation.38 Myc drives glutamine uptake via upregulation of the glutamine transporters SLC38A5 and SLC1A5, but the glutamine antiporter SLC7A5 is upregulated in Myc-overexpressing cells.35 Thus, we speculate that glutamine efflux—and, consequently, leucine uptake—may be elevated in Myc-overexpressing cells (Figure 2). In this model, increased intracellular leucine provides another avenue to activate the growth-promoting activities of mTORC1. A clear prediction of this model is that mTORC1 activity in Myc-overexpressing cells should be hypersensitive to knockdown or inhibition of the glutamine antiporter SLC7A5. If so, SLC7A5 may represent a new therapeutic target for the treatment of Myc-dependent tumor cells. This model for Myc in regulating protein translation is consonant with other findings showing that Myc can regulate expression of ribosomal RNA, tRNAs, and ribosomal protein.21,39 Together these data suggest a central role for Myc in regulating cell growth by broadly regulating protein translation.
MondoA and ChREBP: New Members of an Extended Myc Superfamily?
About 10 years ago, 2 members of the bHLHZip family were discovered that might cooperate with Myc to control cell metabolism. Our lab discovered the first member of the Mondo family, MondoA.40 The Uyeda lab then discovered the second family member, Carbohydrate Response Element Binding Protein (ChREBP), also known as MondoB and WBSCR14.41 MondoA and ChREBP are each about 1,000 amino acids in length, more than twice the length of Myc.42 Given this difference in size between these 2 protein families, it would seem that MondoA and ChREBP are only superficially related to Myc; however, both MondoA and ChREBP dimerize with another bHLHZip protein called Max-like protein x (Mlx).40,43 The interaction with Mlx underlies the official gene names for MondoA and ChREBP: MLX interacting protein (MLXIP) and MLX interacting protein like (MLXIPL), respectively. The bHLHZip domain of human Mlx is 32% identical to that of human Max, and like Max, Mlx is ubiquitously expressed and stable.44 For MondoA and ChREBP, dimerization with Mlx is required for nuclear entry, DNA binding, and presumably, gene activation.45,46 Whether Mlx simply functions as an inert cofactor required for MondoA and ChREBP to bind their genomic targets or whether it has other regulatory functions has yet to be addressed. Though not thoroughly investigated, MondoA:Mlx and ChREBP:Mlx complexes can recognize E-box elements similar to those recognized by Myc:Max complexes, which raises the possibility that MondoA- and/or ChREBP-containing complexes may compete with or perhaps cooperate with Myc-containing complexes at some targets.
Another way that MondoA and ChREBP may interact with a more canonical Myc network is through physical cross talk via shared dimerization with Mlx. The classical Myc network has Max at its center, forming interactions with each Myc protein and with each MXD (formerly Mad) protein. Max does not appear to interact with MondoA or ChREBP. By contrast, Mlx has a restricted dimerization specificity interacting with only MXD1, MXD4, MondoA, ChREBP, and perhaps MNT; it does not interact with any of the Myc proteins.42,44 One can imagine that alterations in levels or activities of certain members of this extended protein family could easily lead to changes in the abundance of different transcriptional heterocomplexes and downstream effector programs. At present, this hypothesis remains largely unexplored.
Regulation of the Mondo Family
In contrast to Myc and MXD family members, whose abundance is primarily controlled at the transcriptional level and by high rates of protein turnover, MondoA and ChREBP appear to be relatively stable proteins. However, both seem to be latently held in the cytoplasm, and their nuclear activity is apparently dictated by cytoplasmic–nuclear partitioning. MondoA and ChREBP accumulate in the nucleus by sensing glucose-derived metabolites, but the regulatory mechanisms are quite different. For example, as we have shown, MondoA:Mlx accumulates in the nucleus and occupies target gene promoters by sensing glucose 6–phosphate (G6P).47 By contrast, ChREBP accumulates in the nucleus by sensing the pentose phosphate intermediate xylulose 5–phosphate (X5P).48 We have proposed G6P binds MondoA directly and triggers a conformational change that permits nuclear accumulation, promoter occupancy, coactivator recruitment, and target gene activation.45 X5P is proposed to work more indirectly, activating the phosphatase PP2A, which dephosphorylates 2 key residues on ChREBP that permit nuclear accumulation and DNA binding.46 This model for ChREBP is somewhat controversial49,50 and still requires rigorous testing in vivo. Regardless of the precise mechanisms, the nuclear export factor CRM1 and 14-3-3 family members appear to play important roles in controlling the nuclear–cytoplasmic abundance of MondoA and ChREBP.46,51
Another key feature that distinguishes MondoA from ChREBP is that MondoA:Mlx complexes are latently held at the outer mitochondrial membrane (OMM), whereas ChREBP:Mlx complexes seem to be generally cytoplasmic and not membrane associated.41,52 Our previous work clearly indicated a requirement for hexokinases—enzymes that catalyze the conversion of glucose to G6P—in the nuclear accumulation of MondoA.47 Interestingly, hexokinases localize to the OMM, where they have preferential access to ATP produced in the mitochondria.53 We have suggested that the colocalization of MondoA:Mlx and hexokinases to the OMM represents an effective mechanism by which cells couple sensation of glucose levels and ATP levels with their adaptive transcriptional response—that is, translocation of MondoA:Mlx complexes to the nucleus.
Whereas MondoA and ChREBP are more or less ubiquitously expressed, their mRNAs are most abundantly expressed in skeletal muscle and liver, respectively.40,46 Skeletal muscle and liver are central sites of insulin-stimulated glucose uptake following a meal, thereby suggesting key functions for MondoA and ChREBP in controlling glucose-dependent transcription and glucose homeostasis in these tissues. Accordingly, ChREBP mediates the majority of glucose-dependent transcription in hepatocytes.54 The glucose-dependent transcriptome for MondoA has not yet been determined in skeletal muscle cells, but it is required for approximately 75% of glucose-induced transcription in an epithelial cancer cell line.47 It seems likely that MondoA plays a similarly predominant role in mediating glucose-dependent transcription in muscle. In liver, ChREBP can drive the expression of genes encoding enzymes involved with lipogenesis.46 In muscle, MondoA likely regulates glycolytic target genes,52 which is in keeping with the role of muscle in ATP generation and utilization. As discussed below, MondoA and ChREBP regulate expression of thioredoxin interacting protein (TXNIP), which has well-established roles in glucose homeostasis. That MondoA and ChREBP regulate TXNIP underscores their function in dictating glucose-dependent transcription and glucose homeostasis.
TXNIP Is a Regulator of Glucose Utilization
TXNIP was initially identified as a transcript induced by vitamin D3 in HL60 cells, and it has been shown to be induced by a number of stimuli.55 As the name implies, TXNIP can interact with thioredoxin and block its redox activity. As such, cells expressing TXNIP have elevated levels of reactive oxygen species (ROS).56 TXNIP also appears to have non-ROS-dependent functions by controlling the stability of key targets that control cell growth. For example, high levels of TXNIP block cells in the G1 phase of the cell cycle, likely by stabilizing the cyclin-dependent kinase inhibitor p27.57 These functions predict a tumor suppressor function for TXNIP. This is likely the case because TXNIP knockout mice develop hepatocellular carcinoma58 and high TXNIP levels portend a good prognosis in gastric59 and breast cancer (D.E.A. and J.-T. Chi, manuscript in review).
Another way in which TXNIP may function as a tumor suppressor is by controlling glucose uptake and aerobic glycolysis. TXNIP deletion in murine embryonic fibroblasts is sufficient to stimulate glucose uptake and aerobic glycolysis, as indicated by increased lactate production.10 How TXNIP loss stimulates glucose uptake and/or aerobic glycolysis is not resolved at this point. TXNIP null cells have reduced levels of active PTEN, which occurs by a REDOX-dependent mechanism.10 The resultant stimulation of PI3K activity is sufficient to activate AKT, providing TXNIP knockout cells with a survival advantage.10 Furthermore, studies have shown that AKT activation is sufficient to drive glucose uptake.60 Finally, TXNIP deletion leads to stabilization of HIF-1α,59 and as mentioned above, HIF-α stabilization is sufficient to drive the expression of many, if not all, of the enzymes in the glycolytic pathway. Thus, multiple mechanisms likely underlie the elevation in aerobic glycolysis that accompanies TXNIP loss or downregulation. Conversely, TXNIP overexpression is a potent repressor of glucose uptake, which does not require its interaction with thioredoxin.61 Together, studies strongly implicate TXNIP as a regulator of glucose uptake and aerobic glycolysis; in addition, low TXNIP levels favor aerobic glycolysis, whereas high TXNIP levels favor energy generation via β-oxidation of fatty acids.
We have shown that MondoA:Mlx complexes are potent glucose-dependent regulators of TXNIP.45,47,62 Multiple experiments support this conclusion. For example, MondoA:Mlx complexes accumulate in the nucleus and occupy regulatory E-box elements in the TXNIP promoter as an immediate early response to elevations in intracellular glucose concentration. Furthermore, expression of a dominant active form of MondoA, ΔN237MondoA, drives high ectopic TXNIP expression. Together, data suggest a role for MondoA:Mlx complexes in the negative regulation of glucose uptake and, potentially, cell growth via their regulation of TXNIP. Consistent with this hypothesis, overexpression of ΔN237MondoA potently represses glucose uptake and restricts cell growth. Likewise, MondoA knockdown or knockout stimulates glucose uptake and stimulates cell growth. Interestingly, overexpression of ΔN237MondoA in wt and TXNIP knockout murine embryonic fibroblasts suggests that activation of TXNIP accounts for about 20% of MondoA’s potent negative regulation of glucose uptake (our unpublished data). Thus, other MondoA:Mlx effectors must function to restrict glucose uptake. One candidate is ARRDC4, which is a TXNIP paralog and is capable of blocking glucose uptake.45,61,62 As with TXNIP, we have shown that ARRDC4 is a direct MondoA target and is glucose induced. Thus, TXNIP and ARRDC4 may cooperatively function downstream of MondoA to restrict glucose uptake.
Given that MondoA knockdown stimulates glucose uptake, it is not surprising that MondoA knockdown in HA1ER cells63—primary kidney epithelial cells transformed with hTERT, SV40 early region, and oncogenic Ras—proliferate faster than control cells.62 Furthermore, consistent with TXNIP functioning downstream of MondoA, TXNIP overexpression overrides the growth advantage afforded by MondoA knockdown. It is interesting that knockdown of ChREBP in the HCT116 colon-cancer cell line restricts cell growth and reduces aerobic glycolysis.64 ChREBP knockdown is accompanied by an induction of a p53-dependent gene expression signature that likely contributes the observed growth restriction. How ChREBP knockdown signals to p53 is unclear at this time, but this finding supports current models implicating p53 as a sensor of metabolic stress.65 The HA1ER cells used in our experiments have reduced levels of functional p53 owing to expression of SV40 large T antigen. Therefore, that MondoA knockdown stimulates cell growth in p53-deficient cells whereas ChREBP knockdown reduces cell growth in p53-proficient cells suggests the intriguing possibility that the functional output of the MondoA and/or ChREBP transcriptomes may be dictated by p53 status.
MondoA and TCA Function
As alluded to above, the rapid proliferation of cancer cells relies on growth factor signaling and the abundant availability of the nutrients glucose and glutamine. Depending on cell context, removal of glucose and/or glutamine triggers apoptosis, indicating that cancer cells can become addicted to either of these growth essential nutrients.33,66 The mechanisms that coordinate glycolysis and glutaminolysis are just now becoming elucidated and have recently been reviewed.8,67
Central to the coordination of glycolysis and glutaminolysis in regulating cell growth is the synthesis of fatty acids that support membrane biosynthesis. Fatty acid biosynthesis requires citrate, which is produced from the glycolytic product pyruvate. Citrate is effluxed from mitochondria, resulting in a truncation of the TCA cycle. Recent studies indicate that glutaminolysis can refill this truncated TCA cycle by a process called anapleurosis.67 Glutamine-derived α-KG refills the TCA cycle, supplying carbons for the generation of oxaloacetate, which is a precursor of fatty acid and lipid synthesis. Glutamine-derived carbons also generate NADPH via the malic enzyme; NADPH is used to support lipid and nucleotide synthesis. Thus, to satisfy their bioenergetic and biosynthetic requirements, cancer cells must coordinate glycolytic and glutaminolytic fluxes. Our work suggests an important role for MondoA in sensing rates of glutaminolysis and coordinating glucose uptake via transcriptional repression of TXNIP.62
A clear demonstration of the requirement for glucose and glutamine for cell growth is the fact that BxPC-3 pancreatic cancer cells fail to divide in medium that contains glucose but lacks glutamine.62 Under these conditions, MondoA is a potent activator of TXNIP expression, which restricts glucose uptake and cell proliferation. Addition of glutamine to glucose-containing medium stimulates growth as expected. Surprisingly, in the presence of glucose and glutamine, MondoA represses TXNIP expression, resulting in a stimulation of glucose uptake. Thus, glutamine effectively represses glucose-dependent and MondoA-dependent transcriptional activation of TXNIP (Figure 3A). How glutamine converts MondoA to a transcriptional repressor is not yet understood. However, our data suggest that glutamine triggers displacement of coactivators from promoter-bound MondoA:Mlx complexes and/or the recruitment of corepressors to promoter-bound MondoA complexes. We have proposed a novel function for MondoA in coordinating glycolysis and glutaminolysis via its regulation of TXNIP. In the presence of glucose—but with subthreshold levels of glutamine (i.e., nonpermissive growth conditions)—TXNIP levels are high, restricting the uptake of glucose required for cell growth. At higher glutamine concentrations (i.e., permissive growth conditions), TXNIP levels decline; glucose uptake is stimulated; and cellular metabolic programs can be activated. Conceptually, this model suggests a metabolic checkpoint function for TXNIP, potentially restricting cell growth until levels of glucose and glutamine are sufficiently high to support growth and division.
Figure 3.
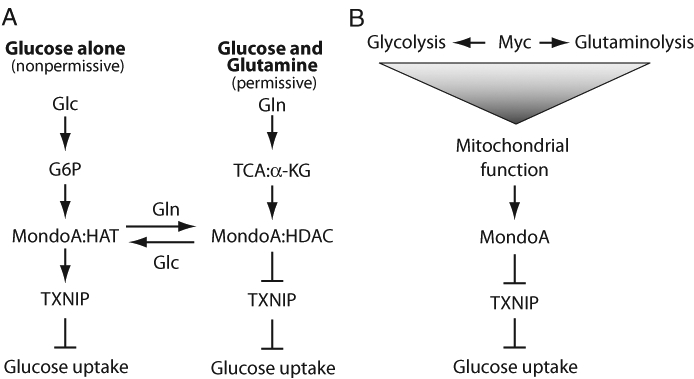
Cell growth requires coordinated glycolysis and glutaminolysis. (A) Under nonpermissive growth conditions (i.e., glucose alone), MondoA is an activator of TXNIP, which restricts glucose uptake and cell growth. Under permissive growth conditions (i.e., glucose plus glutamine), MondoA is a histone deacetylase–dependent repressor of TXNIP, allowing glucose uptake and cell growth. (B) Our model for an interaction between Myc and MondoA in coordinating glycolysis and glutaminolysis. Myc controls glycolysis, mitochondrial mass and function, and glutaminolysis. We propose that MondoA integrates information about the function of these 3 pathways by sensing TCA cycle function. When TCA cycle function is high, MondoA is a transcriptional repressor of TXNIP. Glc = glucose; Gln = glutamine; G6P = glucose-6-phosphate; TCA = tricarboxylic acid cycle; α-KG = α-ketoglutarate; HAT = histone acetyltransferase; HDAC = histone deacetylase.
Glutamine has several cellular fates, suggesting several possible avenues by which it might drive the MondoA-dependent repression of TXNIP. It is difficult to rule out the involvement of other pathways; however, cell-permeable analogs of α-KG completely phenocopy the glutamine-dependent repression of TXNIP.62 Because α-KG enters the TCA cycle, we have proposed that MondoA senses a functional TCA cycle and represses TXNIP expression as a consequence. Glutamine depletion results in a reduction of most, if not all, TCA intermediates;33 thus, we suggest that in the absence of a functional TCA cycle, MondoA is a transcriptional activator of TXNIP. By contrast, in the presence of glutamine, the TCA cycle is refilled, and we suggest that TCA-derived signals convert MondoA from a transcriptional activator to a transcriptional repressor. At this time we lack mechanistic detail on how MondoA senses a functional TCA cycle. However, the OMM localization of MondoA:Mlx complexes perfectly places them to sense TCA cycle status.52 A final interesting implication of our findings is that glutaminolysis, via MondoA-dependent repression of TXNIP, functions upstream of glucose uptake and glycolysis.
Is There Cooperation between Myc and Mondo Family Members?
Given that Myc and Mondo family members have similar bHLHZip domains and similar overall domain structure, it is possible that they cooperate to control specific biological processes or coregulate target genes. At the highest level, we observed synthetic lethal interactions following loss of the Drosophila orthologs of Myc and Mondo.42 This synthetic lethality suggests that the Myc and Mondo proteins coregulate at least one function required for development of the fruit fly. This type of analysis has yet to be conducted in higher eukaryotes and will likely require the development of conditional alleles for inactivating MondoA and ChREBP in the mouse. There are several ways by which cooperation between the Myc and Mondo families might manifest. We outline some of these below.
Myc proteins have several highly conserved sequences in their N-termini that are critical for function. In particular, Myc Box II plays a central role in cell transformation via the recruitment of different transcriptional coregulatory complexes.15 MondoA and ChREBP have a domain in their N-termini that has some sequence homology to Myc box II.42 As such, it is possible that under some circumstances MondoA or ChREBP might be able to drive transformation in isolation or cooperate with Myc in transformation. This hypothesis remains largely unexplored; however, as discussed above, ChREBP knockdown reduces cell proliferation in culture and reduces tumorigenesis in xenograft models.64 Thus, ChREBP may be necessary for transformation, but it remains to be formally tested whether MondoA and/or ChREBP is sufficient for transformation. Furthermore, it has not been tested whether MondoA and/or ChREBP preferentially interacts with Myc or whether they cross talk with other oncogenes. Given that disruption of lipid synthesis reduces growth of glycolytic tumor cells,68 it is of interest to determine whether ChREBP’s positive contribution to cell growth can be traced to its activation of lipogenic target genes. Neither MondoA nor ChREBP was identified in screens for cDNAs that could rescue the slow-growth phenotype of Myc-null fibroblasts.69,70 It is unclear whether functional full-length clones of MondoA or ChREBP were present in the retroviral libraries used in these screens. If so, this result seemingly rules out a model that MondoA and/or ChREBP can substitute for Myc.
Two recent articles suggest that there may be functional cooperation between Myc and MondoA or Myc and CHREBP at specific target genes.71,72 For example, Myc and ChREBP are required for the direct and glucose-dependent activation of liver-type pyruvate kinase (Pklr) transcription. Furthermore, Myc and ChREBP bind the Pklr promoter under hyperglycemic growth conditions, suggesting a glucose-dependent promoter co-occupancy by Myc and ChREBP. Interestingly, Myc knockdown or small molecule inhibition of Myc:Max complex formation reduces the glucose-induced binding of ChREBP to the Pklr promoter, suggesting a key role for Myc:Max complexes in the recruitment of ChREBP:Mlx complexes. Myc inactivation also blocked ChREBP recruitment to the promoters of several other glucose-dependent target genes and blocked their induction. Thus, there may be a general requirement for Myc:Max complexes at ChREBP-dependent and, presumably, MondoA-dependent glucose-induced target genes. Given that Myc expression requires the presence of growth factors, one can envision a model where glucose-dependent target genes required for growth (i.e., those regulated by MondoA and/or ChREBP) can be activated only in the presence of growth factors that drive Myc expression. Such a model represents a novel way to coordinate growth factor and nutrient availability. A key question not addressed in previous studies is whether a reciprocal regulatory relationship exists at Myc-dependent target genes—specifically, is ChREBP and/or MondoA required for induction of Myc targets? No genome-wide occupancy studies have been published for MondoA or ChREBP; however, it is of great interest to determine how widespread the codependence between Myc and MondoA or ChREBP may be.
The 2 previous models for Myc–MondoA cooperation are target gene–centric, but that Myc can drive glutaminolysis and that MondoA senses glutamine levels in the TCA cycle via α-KG34,35,62 suggests that Myc–MondoA cooperation may be mediated by a functional TCA cycle. Although not tested in the original publications, Myc-dependent glutaminolysis likely drives TCA anapleurosis. We have shown that a functional TCA cycle converts MondoA to a transcriptional repressor of TXNIP.62 The resulting reduction in TXNIP levels is sufficient to stimulate glucose uptake, and others have shown that TXNIP deletion is sufficient to drive aerobic glycolysis. Together, these observations suggest that in Myc-dependent tumor cells, high glutamine-derived TCA intermediates may increase the relative transcriptional repression activity of MondoA at the TXNIP promoter, resulting in elevated glucose uptake (Figure 3B). Thus, in addition to its well-established role in the direct regulation of target genes encoding glycolytic enzymes and glycolysis, Myc may stimulate glucose uptake by this second glutaminolysis-dependent mechanism. Previous reports have clearly demonstrated that Myc-overexpressing cells can be addicted to glutamine.33,34 In these reports, cell death upon glutamine removal was attributed to the direct inhibition of glutaminolysis. Given our published data and this proposed model, we suggest that glutamine removal from the medium leads to the induction of TXNIP and a restriction of glucose uptake. Thus, in addition to abrogating glutaminolysis, glutamine removal may trigger cell death by restricting glucose uptake. This model is consistent with many reports showing highly glycolytic tumor cells to be sensitive to inhibition of glycolysis.3,4 It will be of interest to examine whether TXNIP is induced upon glutamine removal in Myc-addicted tumor cells and whether the ensuing apoptosis requires TXNIP.
Summary and Perspective
It is well established that the Myc family of proto-oncogenes has pleiotropic functions in controlling cell growth and division. Myc can clearly orchestrate cell growth by controlling the expression of key enzymes in the glycolytic pathway and multiple other biosynthetic pathways. Furthermore, Myc’s direct regulation of glucose and glutamine transporters suggests that Myc, in addition to controlling nutrient utilization pathways, can control levels of intracellular nutrients required to fuel these biosynthetic pathways. The negative-feedback regulatory circuit that we have described among MondoA, TXNIP, and their potent negative regulation of glucose uptake suggests they may also control access to nutrients required for biosynthetic reactions. MondoA and ChREBP loss have, at this point, opposing roles in cell growth that seem to correlate with p53 status. Therefore, a clear goal is to understand how MondoA and ChREBP cross talk with the p53 pathway. Furthermore, it is clearly important to determine whether MondoA and ChREBP’s role in cell growth stems primarily from modulating glucose uptake or whether other MondoA and/or ChREBP transcriptional targets also participate. To address this question, we are currently determining the genome-wide distribution of MondoA:Mlx complexes under different nutrient growth conditions. This analysis will provide insight into the extent of Myc-MondoA and/or Myc-ChREBP cooperation at the programmatic level. Finally, the proposed cooperation between Myc-driving glutaminolysis and MondoA-sensing glutamine flux to repress TXNIP suggests one mechanism for coordinating glycolysis and glutaminolysis during growth and division.
We focused this review on the Myc and Mondo families in growth and proliferation. However, glucose homeostasis plays an essential role in postmitotic tissues. Myc is typically downregulated in postmitotic cells,15 so interactions between MondoA and/or ChREBP and Myc are not likely relevant in terminally differentiated or quiescent cells. Members of the MXD family are well documented to be upregulated following cell cycle exit73; thus, it is possible that MondoA and ChREBP interact genetically or biochemically with the repressive side of the Myc network in postmitotic cells. More broadly, MondoA’s and ChREBP’s high expression in skeletal muscle and liver, respectively, implies an important role in these 2 glucose responsive tissues. Whether MondoA and ChREBP contribute only to glucose homeostasis in skeletal muscle and liver or whether they have more general functions in postmitotic cells awaits further study.
Acknowledgments
We thank the members of the Ayer laboratory for comments on the manuscript.
Footnotes
The work in D.E.A.’s laboratory is supported by National Institutes of Health grants GM055668 and GM060387.
The authors declare no conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11-20 [DOI] [PubMed] [Google Scholar]
- 2. Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res 2006;66:8927-30 [DOI] [PubMed] [Google Scholar]
- 3. Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008;13:472-82 [DOI] [PubMed] [Google Scholar]
- 4. Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene 2006;25:4633-46 [DOI] [PubMed] [Google Scholar]
- 5. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 2004;4:891-9 [DOI] [PubMed] [Google Scholar]
- 7. Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell 2008;134:703-7 [DOI] [PubMed] [Google Scholar]
- 8. Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 2008;18:54-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev 2009;19:32-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hui ST, Andres AM, Miller AK, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci U S A 2008;105:3921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pelicano H, Xu RH, Du M, et al. Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism. J Cell Biol 2006;175:913-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Semenza GL. Tumor metabolism: cancer cells give and take lactate. J Clin Invest 2008;118:3835-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sonveaux P, Vegran F, Schroeder T, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 2008;118:3930-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen JL, Lucas JE, Schroeder T, et al. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet 2008;4:e1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976-90 [DOI] [PubMed] [Google Scholar]
- 16. White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet 2008;24:622-9 [DOI] [PubMed] [Google Scholar]
- 17. Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J 2006;25:2723-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallant P, Steiger D. Myc’s secret life without Max. Cell Cycle 2009;8:3848-53 [DOI] [PubMed] [Google Scholar]
- 19. Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A 1997;94:6658-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim JW, Zeller KI, Wang Y, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 2004;24:5923-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 2007;12:108-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev 2007;17:71-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lum JJ, Bui T, Gruber M, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev 2007;21:1037-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 2007;11:335-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikiforov MA, Chandriani S, O’Connell B, et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase: a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 2002;22:5793-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 2009;28:2485-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacDonald VE, Howe LJ. Histone acetylation: where to go and how to get there. Epigenetics 2009;4:139-43 [DOI] [PubMed] [Google Scholar]
- 28. Li F, Wang Y, Zeller KI, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 2005;25:6225-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 2006;3:177-85 [DOI] [PubMed] [Google Scholar]
- 30. Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 2006;3:187-97 [DOI] [PubMed] [Google Scholar]
- 31. Zhang H, Gao P, Fukuda R, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 2007;11:407-20 [DOI] [PubMed] [Google Scholar]
- 32. DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010;29:313-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 2007;178:93-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A 2008;105:18782-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao P, Tchernyshyov I, Chang TC. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009;458:762-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol; 22:169-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shaw RJ. mTOR signaling: RAG GTPases transmit the amino acid signal. Trends Biochem Sci 2008;33:565-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009;136:521-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown SJ, Cole MD, Erives AJ. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics 2008;9:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol 2000;20:8845-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamashita H, Takenoshita M, Sakurai M, et al. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A 2001;98:9116-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Billin AN, Ayer DE. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. In: Eisenman RN, editor. The Myc/Max/Mad transcription factor network. Heidelberg: Springer; 2006. p. 255-78 [DOI] [PubMed] [Google Scholar]
- 43. Ma L, Tsatsos NG, Towle HC. Direct role of ChREBP-Mlx in regulating hepatic glucose-responsive genes. J Biol Chem 2005;280:12019-27 [DOI] [PubMed] [Google Scholar]
- 44. Billin AN, Eilers AL, Queva C, Ayer DE. Mlx, a novel max-like BHLHZip protein that interacts with the max network of transcription factors. J Biol Chem 1999;274:36344-50 [DOI] [PubMed] [Google Scholar]
- 45. Peterson CW, Stoltzman CA, Sighinolfi MP, Han KS, Ayer DE. Glucose controls nuclear accumulation, promoter binding, and transcriptional activity of the MondoA:Mlx heterodimer. Mol Cell Biol 2010; in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 2006;4:107-10 [DOI] [PubMed] [Google Scholar]
- 47. Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci U S A 2008;105:6912-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci U S A 2003;100:5107-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr 2007;27:179-92 [DOI] [PubMed] [Google Scholar]
- 50. Tsatsos NG, Towle HC. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem Biophys Res Commun 2006;10:449-56 [DOI] [PubMed] [Google Scholar]
- 51. Eilers AL, Sundwall E, Lin M, Sullivan AA, Ayer DE. A novel heterodimerization domain, CRM1, and 14-3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol Cell Biol 2002;22:8514-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol 2006;26:4863-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pastorino JG, Hoek JB. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr Med Chem 2003;10:1535-51 [DOI] [PubMed] [Google Scholar]
- 54. Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem 2006;281:28721-30 [DOI] [PubMed] [Google Scholar]
- 55. Kim SY, Suh HW, Chung JW, Yoon SR, Choi I. Diverse functions of VDUP1 in cell proliferation, differentiation, and diseases. Cell Mol Immunol 2007;4:345-51 [PubMed] [Google Scholar]
- 56. Muoio DM. TXNIP links redox circuitry to glucose control. Cell Metab 2007;5:412-4 [DOI] [PubMed] [Google Scholar]
- 57. Jeon JH, Lee KN, Hwang CY, Kwon KS, You KH, Choi I. Tumor suppressor VDUP1 increases p27(kip1) stability by inhibiting JAB1. Cancer Res 2005;65:4485-9 [DOI] [PubMed] [Google Scholar]
- 58. Sheth SS, Bodnar JS, Ghazalpour A, et al. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene 2006;25:3528-36 [DOI] [PubMed] [Google Scholar]
- 59. Shin D, Jeon JH, Jeong M, et al. VDUP1 mediates nuclear export of HIF1alpha via CRM1-dependent pathway. Biochim Biophys Acta 2008;1783:838-48 [DOI] [PubMed] [Google Scholar]
- 60. Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 2004;64:3892-9 [DOI] [PubMed] [Google Scholar]
- 61. Patwari P, Chutkow WA, Cummings K, et al. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J Biol Chem 2009;284;24996-5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kaadige MR, Looper RE, Kamalannaadhan S, Ayer DE. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci U S A 2009;106:14878-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hahn WC, Dessain SK, Brooks MW, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol 2002;22:2111-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tong X, Zhao F, Mancuso A, Gruber JJ, Thompson CB. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc Natl Acad Sci U S A 2009;106:21660-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cheung EC, Vousden KH. The role of p53 in glucose metabolism. Curr Opin Cell Biol 2010;22:186-91 [DOI] [PubMed] [Google Scholar]
- 66. Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci U S A 1998;95:1511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A 2007;104:19345-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hatzivassiliou G, Zhao F, Bauer DE, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell 2005;8:311-21 [DOI] [PubMed] [Google Scholar]
- 69. Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene 2000;19:3330-4 [DOI] [PubMed] [Google Scholar]
- 70. Nikiforov MA, Kotenko I, Petrenko O, et al. Complementation of Myc-dependent cell proliferation by cDNA expression library screening. Oncogene 2000;19:4828-31 [DOI] [PubMed] [Google Scholar]
- 71. Collier JJ, Doan T-TT, Daniels MC, Schurr JR, Kolls JK, Scott DK. c-Myc is required for the glucose-mediated induction of metabolic enzyme genes. J Biol Chem 2003;278:6588-95 [DOI] [PubMed] [Google Scholar]
- 72. Zhang P, Metukuri MR, Bindom SM, Prochownik EV, O’Doherty RM, Scott DK. c-Myc is required for the ChREBP-dependent activation of glucose-responsive genes. Mol Endocrinol 2010; in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol 2006;16:265-74 [DOI] [PubMed] [Google Scholar]


