Abstract
A method for estimating the positional displacement of protein bound gold nanoparticles is presented and used to estimate the rigidity of linkage of Au144 nanoparticles bound to a tetrameric model protein. We observe a distribution of displacement values where most Au144 clusters are immobilized to within 3Å relative to the protein center of mass. The shape of the distribution suggests two physical processes of thermal motion and protein deformation. The application of this and similar rigid gold nanoparticle/protein conjugates in high resolution single particle electron cryo-microscopy is discussed.
INTRODUCTION
Metalloproteins – proteins incorporating in a precise manner one or more metal atoms -- comprise 30% of the human proteome1. Notable examples include the zinc finger proteins involved in transcription regulation, the iron-sulfur cluster ferredoxin proteins implicated in biological electron transfer processes, and ferritin proteins implicated in iron storage. These examples can incorporate a precise number of metal atoms in a precise geometry and location relative to the protein center of mass.
Functional combinations of proteins with metal clusters or nanoparticles may also be synthesized in vitro, enabling new or enhanced functionality such as enhancement of enzyme turnover2 and remote control of DNA hybridization3. Conjugates of metal clusters or nanoparticles with proteins have found most widespread application in structural biology4, where gold clusters and colloids have been used extensively for immunolocalization5, subunit identification within multiprotein complexes 6,7, and deriving phase information in single crystal x-ray studies of large biomolecule crystals8. The conjugation of gold nanoparticles with proteins can leave protein function and presumably structure intact, depending on site of label within a protein and chemical composition of the gold nanoparticle monolayer9-12. Rigid, specific and discretely immobilized protein/gold cluster conjugates have potential to improve the structural biology technique of single particle cryomicroscopy13.
In contrast to naturally occurring metalloproteins, metal cluster / protein conjugates synthesized in vitro are not well defined. The poor definition generally arises from uncertainty in two aspects: First, the uncertainty of the exact composition and structure of the inorganic cluster or nanoparticle component. Second, the uncertainty of the spatial relationship of the inorganic cluster or nanoparticle relative to the conjugate biomolecule, defined here as rigidity. We have previously created conjugates of antibody fragments and monolayer protected gold clusters in which all components were defined with respect to molecular formula14,15. In this previous work, the linkage between gold cluster and protein was direct, through a gold-sulfur bond formed between the thiolate functional group of a cysteine residue in a single chain Fv (scFv) antibody fragment to the core of an Au144(p-mercaptobenzoic acid)60 monolayer protected gold cluster (MPC). In our previous work, we engineered, expressed and tested 12 cysteine replacement mutants of the NC10 scFv to determine the optimal position of the bioconjugate cysteine residue. This work showed that a C-terminal cysteine residue separated from the last crystallographically structured residue, A112, of the scFv by a 2 lysine residue linker allowed high yield formation of functional bioconjugates with Au144(p-mercaptobenzoic acid)60. Other bioconjugate cysteine positions resulted in longer presumably less rigid linkers, lower labeling yield, or diminished scFv function. We speculated that the bioconjugation of this optimized C115 mutant to Au144, shown schematically in figure 1, would allow rigid coupling of MPCs to proteins. We included in our previous publications a characterization of the Au144 particle by TEM, HAADF-STEM, and UV-VIS spectroscopy.
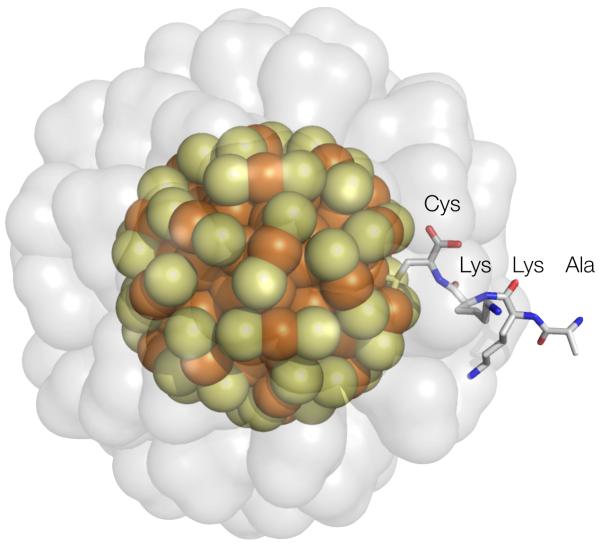
Figure 1.
A schematic diagram showing the final structured A112 residue of the NC10 scFv, the linker comprised of K113 and K114 and the cysteine residue C115 whose thiolate functional group has become integral to the Au144(p-mercaptobenzoic acid)59 ligand layer. In orange and yellow are the gold and sulfur atoms of a computed model of Au144(SR)60. In transparent grey is a modeled p-mercaptobenzoic acid ligand layer, rendered as a solvent accesable surface. Sulfur and gold are rendered as spheres except for the cysteine sulfur, which is shown in its anticipated mode of bonding. The Au144 core model was taken from ref 22.
Here we develop a general mathematical method for assessing the rigidity of MPC/protein linkages, and use this method to assess the rigidity of the scFv bound Au144 MPCs. Our result removes the uncertainty of the spatial relationship of the inorganic cluster or nanoparticle relative to the antibody fragment, and allows us to define the Au144/protein complex with a structural and chemical definition approaching that of well defined naturally occurring metalloproteins. The observed positional displacement of the protein bound Au144 clusters is theoretically small enough to allow extension of cryo-EM reconstruction resolution of low symmetry biomolecules beyond what is possible without such markers.
Previous studies establish some precedent for our analysis and conclusions, although to the best of our knowledge, nothing resembling the combination of our analysis and conclusions has appeared. With regard to mathematical analysis of electron micrographs of biomolecule immobilized gold nanoparticles, Alivisatos and colleagues made a pair distribution function analysis of DNA/gold colloid conjugates. This analysis demonstrated that DNA/gold colloid conjugates can be assembled into well defined structures16. With regard to assessing protein orientation relative to an inorganic nanoparticle, fluorescence resonance energy transfer (FRET) was used to assess the spatial relationship between maltose binding protein that was site-specifically labeled with both organic fluorophores and a ~6nm cadnium selenide-zinc sulfide core-shell quantum dot17. This study concluded that the MBP has a preferred orientation relative to the QD, which is similar to the conclusion of our study of the ‘nano-bio’ interface between Au144 and the NC10 scFv.
METHODS
Nanoparticle preparation & NC10 production
Au144(p-mercaptobenzoic acid)60 MPCs were synthesized and conjugated to NC10 scFv antibody fragments as described previously12. Tern N9 neuraminidase was purified as described elsewhere18.
Complex assembly and purification
Equimolar amounts of tern N9 neuraminidase protomer and NC10 scFv/Au144 were mixed at an approximate final protein density of 1 mg/mL. This mixture was fractionated using an Ettan FPLC (Pharmacia / GE Health) equipped with a Superdex 200 gel filtration column. The first protein containing peak eluting from the column, as identified by UV spectroscopy was identified as the neuraminidase/NC10 ScFv/Au144 MPC conjugate by its apparent molecular weight and characteristic coffee color, resulting from Au144, which contains optical absorbance maxima at both 280nm(protein) and 241nm (Au144 MPC). Analysis of the hydrodynamic radius of the Au144 MPC labeled complex and unlabeled complexes confirming their biochemical integrity may be found in the supporting information.
The purified assembly was concentrated in a CentriCon centrifugal concentrator (10kd cutoff) (Millipore) in borate buffer (pH 8.0) to an approximate 0.5mg/mL protein density as assayed by the optical extinction of the gold nanoparticles at 241nm.
Sample preparation/ Vitrification
3uL of the purified assembly was deposited on freshly glow discharged R2/4 Quantifoil EM Grid (Ted Pella Co, Redding, CA), the excess was blotted off with Watman #2 filter paper, and vitrified in liquid ethane using a custom built servo-controlled automated vitrification apparatus. The humidity in the room was maintained at 70-80% relative humidity. Samples were transported under liquid nitrogen to the microscope. Great care was taken to ensure the highest-quality amorphous ice was formed and preserved throughout sample handling. Temperatures during transportation and transfer into the microscope never exceeded –170degC.
Molecular Modeling
X-ray coordinates of the NC10/N9 complex were obtained from the Protein Data Bank, accession code 1A1419. Coordinates for the Au144 cluster were provided by Hannu Hakkinen20. The methylthiolate ligands in the provided structure were replaced with p-mercpatobenzoic acid residues were placed on the cluster using the ‘pairfit’ function in Pymol21. Ligands with steric clash were corrected manually. The Lys-Lys-Cys extension engineered into the scFv for gold cluster conjugation was added to the scFv in Pymol and the coordinates were relaxed with the MMFF molecular mechanics force field using the Molecular Operating Environment (http://www.chemcomp.com). The ligand protected cluster and relaxed mutant NC10/N9 were joined covalently in pymol. A hydrodynamic radius for the complex was calculated with Hydropro22. The position of the cluster relative to the protein complex was subsequently optimized so that calculated hydrodynamic radius values from Hydropro are consistent with measured hydrodynamic radius values from gel filtration.
Electron Cryo-Microscopy
Cryo-electron microscopy was performed using a liquid nitrogen-cooled 300kV FEI Polara G2 electron microscope equipped with a Tietz 2k-by-2k pixel high-sensitivity CCD camera, and multi-specimen holder. Great care was taken in the ensuring optimal microscope alignment for ideal imaging conditions with special emphasis on accurately setting the rotation center to minimize the variation in magnification across the image field for this linkage rigidity study. Images were acquired under low dose conditions (1.2 electrons per Å2) at 59kx magnification. Microscope control, grid targeting, image acquisition and database storage were performed using the Leginon electron microscope automation software software23. Image focal pairs were collected at close-to-focus and 2um underfocus.
Image processing
Images were examined for quality, defocus and particle distribution using the EMAN software24. From appropriate images, individual particles selected on the basis of stoichiometry (4 clusters) and planarity of projection. Particles were boxed out and centered using the the EMAN program Boxer. Boxer selected single-particle images were subjected to a low-pass filter at 1Å to remove high-frequency noise and high-pass filtered at 400Å to remove any background gradient.
A Matlab (MathWorks, Inc, Natick MA) program was developed to estimate the positional displacement of the clusters within single-particle images by calculating the deviation from perfect symmetry. The algorithm uses the selected particle images and after thresholding and converting to binary determines the centroid of each of the four nanoparticles present (Fig 2 panel D). Then the program defines two lines that span the distance between opposite centroids, forming a cross. Then the program locates the intersection of the two lines, and determines the length of all line segments from the intersection to each of the Au144 centroids. If the complex is totally symmetric, the two sets of line segments will be equal in length (Fig 4, panel B).
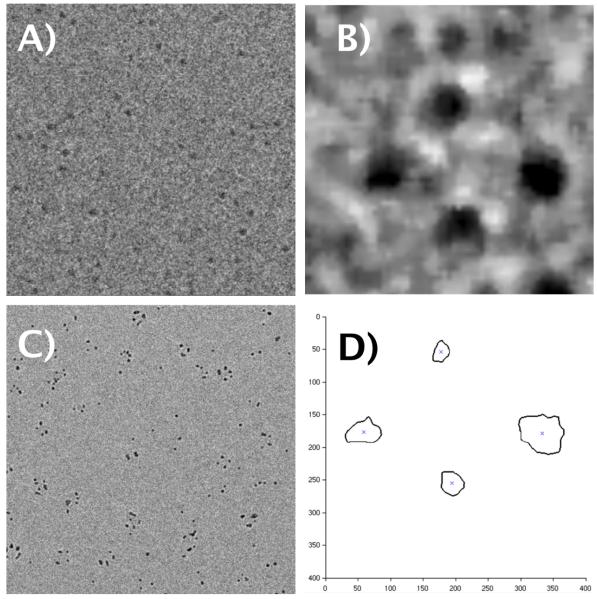
Figure 2.
A): Representative low dose image; B) Representative processed single particle image C) Representative high dose image (note significant beam induced deformation). D) Representative output of Matlab algorithm identifying centroids of particles (axes units in pixels).
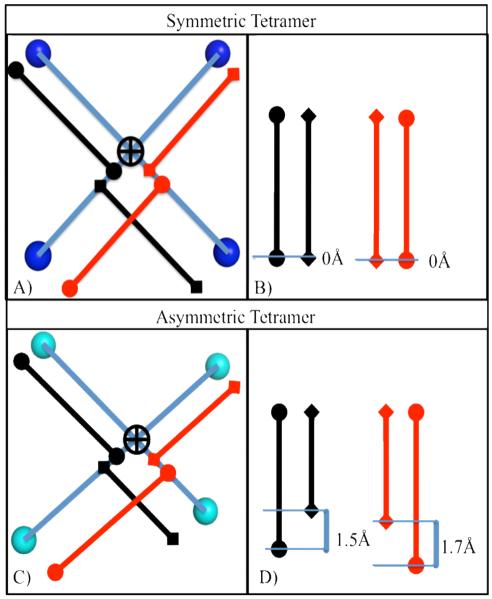
Figure 4.
Schematic of mobility assessment algorithm for any 2D projection of a D4h set of spheres. (A) A perfectly symmetric tetramer, showing lines connecting the opposing balls (blue), the calculated intersection (black cross), and the respective two sets of line segments (black and red). (B) The length of the sets of line segments is tabulated and they are subtracted to estimate the motility, which is zero for the perfectly symmetric tetramer. C) An asymmetric tetramer, showing lines connecting the opposing balls (green), the calculated intersection (black cross), and the respective two sets of lines segments (black and red). D) Substantial deviation from perfect symmetry is shown in this projection due to the magnitude of inequality in the respective vector sets.
RESULTS
Positional Displacement determination
The tern N9 influenza neuraminidase tetramer in complex with four NC10 scFv antibody fragments used in this study was chosen in part because it is a structurally determined homo-tetramer with C4 symmetry25. We have previously generated direct Au144 MPC conjugates of this complex14. In the present study, we developed a mathematical model that defines the spatial relationship of the Au144 clusters to the protein complex. To interpret the distribution of Au144 positional displacement values that resulted from this model, we combined calculated structures of Au144 and the x-ray structure of the scFv/neuraminidase complex to create a realistic all-atom model consistent with hydrodynamic radius measurement and electron microscopy of the gold labeled complex. This all atom model, depicted in its entirety in figure 3 and in portion in figure 1.
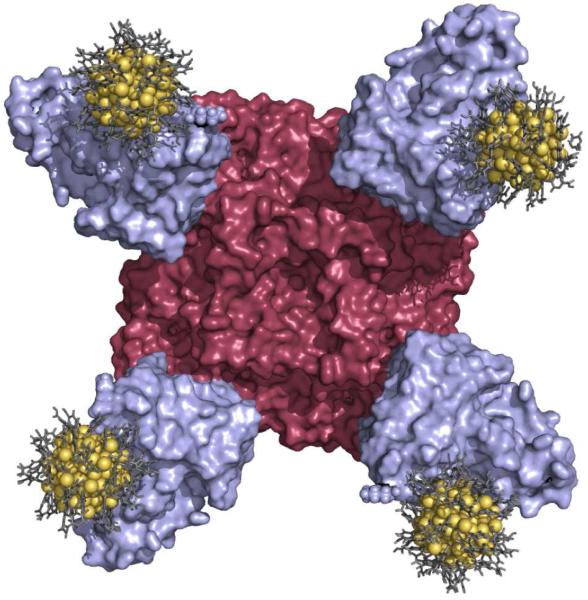
Figure 3.
A realistic all-atom model of the Au144/scFv/N9 complex. The N9 neuraminidase tetramer is rendered as a red surface, the NC10 scFv fragments are rendered as blue surfaces, the Au144 cores are rendered as gold spheres, and the ligand layer protecting the gold core is shown as gray sticks.
To determine the range of possible positional displacement values for the Au144 clusters depicted in figure 3 relative to the protein, we first considered the Au144 clusters attached to the tetramer as perfect spheres such that the 4 Au144 particles attached to each N9/NC10 tetramer possess D4h symmetry. This D4h symmetry, when projected into 2 dimensions as is done when recording electron cryomicroscopy data on a 2-D CCD, results in a projection pattern with D2h symmetry. Deviations from symmetry represent flexibility of clusters relative to the protein, dissociation of the scFv antibody from its antigen, or structural anomalies in the protein complex itself.
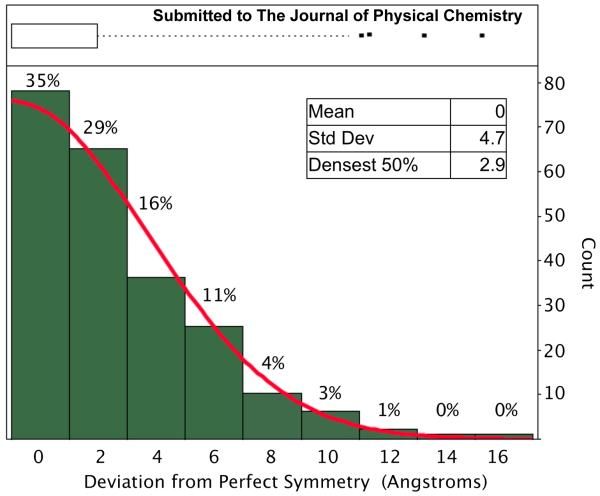
We developed a method for determining deviations from symmetry to assess positional displacement of MPCs. The method relies on first identifying sets of four particles likely to be related through attachment to a single N9/NC10 molecule. Then the method determines the center of mass of each particle, followed by construction of line segments between the centroids of opposite particles (figure 4, panel A). If the projection has perfect D2h symmetry, then the lines drawn from opposite particles will bisect. Deviations from bisection represent positional displacement of the Au144 clusters. The amount of deviation from perfect symmetry was measured for each of 224 particle projections. Figure 5, summarizes these deviations in histogram format. The densest 50% of distributions constrain the particles to within 3Å positional displacement.
Figure 5.
Histogram and box-plot showing the single-particle distribution of deviations from perfect symmetry for the MPC labeled/NC10-avian influenza neuraminidase tetramer. The top panel shows the box plot summarizing the features of the distribution. The box indicating 25th and 75th quantiles indicates the densest 50% of the data spans the range of 1Å to 3Å mobility. The dashed line extending to 9.2Å represents the 95% confidence interval and the four points at larger deviations are statistical outliers. The bottom panel shows the histogram of Au144 mobility and the fitted Gaussian distribution.
As implemented here, we underestimate the positional displacement from D2h symmetry of Au144 clusters when the plane defined by the 4 Au144 clusters is not parallel to the plane of the CCD image recorder for any individual set of 4 Au144 clusters. This underestimate arises from the shortening of interparticle spacing as the angle between the plane defined by the Au144 clusters and the plane of the CCD detector becomes larger. The amount of this error is small for the particles used in the analysis, and can be estimated by considering that a vector, when projected onto a plane, will shorten according to |L|=Cos(θ)|A| where A is the length of the vector, L is the length of the projected vector and θ is the angle between vector A and the plane of projection. Assuming that the error in positional displacement assessment depends on Cos(θ), then for particles tilted 20° from the CCD plane, the error in positional displacement assessment is 6%. If every particle included in our analysis incorporated this amount of underestimation, then the distribution would broaden slightly yielding the densest 50% of the data from zero-to-3.1Å, and a standard deviation of 5.0 Å. However, we minimized the possibility of this error by choosing particle images where the particle plane is parallel to the detection plane. By visual inspection we assert that none of the particles included in our assessment is more than 20° tilted relative to the plane of the CCD, and most particles are tilted significantly less. Therefore this underestimation error cannot affect the distribution in positional displacement in a way that could change our conclusions.
Attempts to fit the histogram of positional displacement were made with combinations of Gaussian, Laplacian and exponential distributions. The best fit was achieved with a standard Gaussian probability distribution function. The distribution was also analyzed using JMP8 software (SAS, Cary NC). Figure 5, top panel shows the results of this analysis in a box-plot summarizing the statistics of the positional displacement distribution. Since we are considering only positive displacement values, the normal distribution is centered at zero with a standard deviation of 4.9Å. The lowest half-bracket (figure 5, top panel, black box) represents the densest 50% of positional displacements, which lie in the 0-3Å range. Data points above 9.2Å positional displacement are statistical outliers. Based upon the model of figure 3, we expect that displacements greater than ~10Å can be attributed to protein dissocation or denaturation.
DISCUSSION
Three distinct chemistries have emerged over the past 40 years for making conjugates of gold nanoparticles to proteins. These chemistries result in gold nanoparticle/protein conjugates formed through linker-mediated, electrostatic/nonspecific, or direct nanoparticle/protein bonds. Monolayer protected gold clusters with magic number nuclearities of Au6826, Au10227, and Au14420 can form direct protein-gold bonds 9-11,14,15 and are especially appropriate for biological single particle electron cryomicroscopy experiments because they possess the a definition, diameter, and electron density which is discernable in the low-dose (20e-/Å2) Schertezer focus condition specified for certain electron cryomicroscopy applications28. We have previously generated the first conjugates of a structurally defined protein19 and molecularly defined Au144 gold cluster in which the clusters are directly attached to the protein through a Cys-S-Au bond14. In the present work, we define the rigidity of the Au144 cluster / protein linkage.
To define the positional displacement of the Au144 clusters and thereby the Au144/protein linkage rigidity, we developed a general method for assessing the positional displacement of clusters attached to molecular or supramolecular structures of defined symmetry. We expect this method may find widespread use among MPC chemists assessing rigidity of attachment of clusters in supramolecular assemblies. In our particular preparation of Au144/NC10/N9 we observe a distribution of positional displacement where the densest 50% of the displacement distribution is within 3.0Å of rigidity. The positional displacement data agrees well with a normal probability distribution as shown in figure 5. The Gaussian distribution and statistical outliers (Fig 5 top panel, points) can be attributed to two distinct physical phenomena. The Gaussian distribution is explained by three factors: thermal motion of the protein (protein breathing), electron beam induced motion, and stochastic mobility of the clusters relative to the protein due to flexibility of the Au144/protein linkage, resulting in Brownian oscillation the amplitude of which is determined by the rigidity of the protein/Au144 linkage. While the four identified statistical outliers cannot be explained by linker flexibility, we speculate that statistical outliers may instead be attributed to known and separate phenomenon, including dissociating scFv and deformed, degraded, or misfolded protein. Taken together these multiple phenomena contribute to a Gaussian process with a small fraction of statistical outliers.
Our analysis is intended to detect non-systematic deformations of the Au144/scFv conjugation, because they will change the symmetry of the system. Since the Au144/scFv binds the neuraminidase tetramer as determined by gel filtration and electron cryomicroscopy, any systematic deformation does not seriously affect protein function.
We may be able to isolate and quantify the contributions the observed Gaussian distribution by using larger proteins where thermal motion components would be amplified, using longer and shorter linkers between the protein and gold cluster to isolate the protein/Au144 linker contribution, and varying the electron dose per image to isolate the contribution of e-beam induced motion.
The constraint of the majority of positional displacements to within 3Å represents a surprisingly rigid interface, given that the Au144 cluster is bound to the protein through a two lysine linker which separates Au144 bound C-terminal cysteine residue from the final structured residue (A112) observed in protein x-ray structure of NC10. We explain remarkable rigidity by noting that the Au144 cluster may be held in place by three interactions: the direct cys-SH gold bond, a set of steric contacts due to the requirement of the labeled cys residue to integrate into the highly structured monolayer27 of the cluster, and by secondary electrostatic interactions between the negatively charged p-mercaptobenzoic acid monolayer and positively charged lysine residues within the protein proximal to the site of attachment of the Au144 MPC. The preferred orientation between the nanoparticle and the protein that is presumably enforced by one specific interaction and a set of secondary interactions between a protein and a nanoparticle has also been suggested in a study of maltose binding protein labeled quantum dots17.
Application to electron cryomicroscopy
It was previously proposed that rigid conjugates of molecularly defined metal clusters to known sites on a protein may improve attainable resolution in single particle electron cryomicroscopy of low symmetry biomolecules, perhaps enabling the revolutionary advance of atomic resolution constructions of noncrysatlline biomolecules13. The improvement will occur by exploiting rigidly attached gold clusters as selection markers that allow rejection of stoicheometrically or structurally flawed particles, and as an internal calibration metric for determining defocus, magnification, drift, contrast transfer function, and alignment parameters for each particle included in a reconstruction. Computational simulation indicates that cluster positional displacement of 3.0 Å or less is sufficient to achieve atomic resolution (3.5Å or better) in a reconstruction which includes 100,000 cluster labeled biomolecule particles29. While our study suggests that over half of the data collected may be rejected in single particle reconstruction to enhance the signal to noise ratio, the positional displacement distribution containing the densest 50% of positional displacements within 3.0Å of symmetry for this system indicates that the goal of atomic resolution reconstructions is attainable with direct conjugates of MPCs to proteinaceous cryoEM targets.
Future work may best proceed with smaller molecular versions of the MPC magic number series, such as Au6826 or Au10227, which are large enough to be readily identified in low dose, close to focus electron cryo micrographs, but will obscure less of the protein density in projection when compared to the Au144 clusters used in this study.
Supplementary Material
ACKNKOWLEDGEMENTS
The authors acknowledge and thank Roger Kornberg, in whose lab this work was initiated under the support of NIH grant 5R01GM063025-04. We thank Dave Bushnell and Bruce Ackerson for helpful conversations and critical reading of the manuscript. We thank Jerry Ebalunode for assistance with MOE. We thank Bridgette Carragher and the Leginon team for assistance with setting up the Leginon software we used for data acquisition. We especially thank Grant Jensen for inspiring this line of research. Funding for the completion of this work was provided by Colorado State University.
Footnotes
Supporting Information Available. Full details of the stoichiometric product purification and determination of hydrodynamic radius are available. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Waldron KJ, Robinson NJ. Nat Rev Microbiol. 2009;7:25. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- (2).Xiao Y, Patolsky F, Katz E, Hainfeld JF, Willner I. Science. 2003;299:1877. doi: 10.1126/science.1080664. [DOI] [PubMed] [Google Scholar]
- (3).Hamad-Schifferli K, Schwartz John J, Santos Aaron T, Zhang S, Jacobson Joseph M. Nature. 2002;415:152. doi: 10.1038/415152a. [DOI] [PubMed] [Google Scholar]
- (4).Jahn W. Journal of Structural Biology. 1999;127:106. doi: 10.1006/jsbi.1999.4123. [DOI] [PubMed] [Google Scholar]
- (5).Hayat MA. Colloidal gold: principles, methods and applications. Vol. 1. Academic Press; San Diego, CA: 1989. [Google Scholar]
- (6).Poglitsch CL, Meredith GD, Gnatt AL, Jensen GJ, Chang WH, Fu J, Kornberg RD. Cell. 1999;98:791. doi: 10.1016/s0092-8674(00)81513-5. [DOI] [PubMed] [Google Scholar]
- (7).Boisset N, Pochon F, Chwetzoff S, Barray M, Delain E, Lamy J. J Struct Biol. 1992;108:221. doi: 10.1016/1047-8477(92)90022-3. [DOI] [PubMed] [Google Scholar]
- (8).Weinstein S, Jahn W, Hansen H, Wittmann HG, Yonath A. J Biol Chem. 1989;264:19138. [PubMed] [Google Scholar]
- (9).Aubin-Tam ME, Hwang W, Hamad-Schifferli K. Proc Natl Acad Sci U S A. 2009;106:4095. doi: 10.1073/pnas.0807299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Aubin-Tam ME, Hamad-Schifferli K. Biomed Mater. 2008;3:034001. doi: 10.1088/1748-6041/3/3/034001. [DOI] [PubMed] [Google Scholar]
- (11).Aubin-Tam ME, Hamad-Schifferli K. Langmuir. 2005;21:12080. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- (12).Ackerson CJ, Jadzinsky PD, Sexton JZ, Bushnell DA, Kornberg RD. Bioconjug Chem. 2010;21:214. doi: 10.1021/bc900135d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jensen GJ, Kornberg R. P Natl Acad Sci USA. 1998;95:9262. doi: 10.1073/pnas.95.16.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ackerson CJ, Jadzinsky PD, Sexton JZ, Bushnell DA, Kornberg RD. Bioconjugate Chemistry. 2010 doi: 10.1021/bc900135d. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ackerson CJ, Jadzinsky P, Jensen GJ, Kornberg R. J Am Chem Soc. 2006;128:2635. doi: 10.1021/ja0555668. [DOI] [PubMed] [Google Scholar]
- (16).Zanchet D, Micheel C, Parak W, Gerion D, Williams S, Alivisatos A. Journal of Physical Chemistry B. 2002;106:11758. [Google Scholar]
- (17).Medintz IL, Konnert JH, Clapp AR, Stanish I, Twigg ME, Mattoussi H, Mauro JM, Deschamps JR. Proc Natl Acad Sci U S A. 2004;101:9612. doi: 10.1073/pnas.0403343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).McKimm-Breschkin JL, Caldwell JB, Guthrie RE, Kortt AA. J Virol Methods. 1991;32:121. doi: 10.1016/0166-0934(91)90192-3. [DOI] [PubMed] [Google Scholar]
- (19).Malby RL, McCoy AJ, Kortt AA, Hudson PJ, Colman PM. J Mol Biol. 1998;279:901. doi: 10.1006/jmbi.1998.1794. [DOI] [PubMed] [Google Scholar]
- (20).Lopez-Acevedo O, Akola J, Whetten RL, Gronbeck H, Hakkinen H. J Phys Chem C. 2009;113:5035. [Google Scholar]
- (21).deLano, W.
- (22).Garcia De La Torre J, Huertas ML, Carrasco B. Biophys J. 2000;78:719. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, Reilein A. J Struct Biol. 2000;132:33. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- (24).Ludtke SJ, Baldwin PR, Chiu W. J Struct Biol. 1999;128:82. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- (25).Malby RL, McCoy AJ, Kortt AA, Hudson PJ, Colman PM. J Mol Biol. 1998;279:901. doi: 10.1006/jmbi.1998.1794. [DOI] [PubMed] [Google Scholar]
- (26).Dass A. Journal of the American Chemical Society. 2009;131:11666–7. doi: 10.1021/ja904713f. [DOI] [PubMed] [Google Scholar]
- (27).Jadzinsky PD, Calero G, Ackerson CJ, Bushnell DA, Kornberg RD. Science. 2007;318:430. doi: 10.1126/science.1148624. [DOI] [PubMed] [Google Scholar]
- (28).Jensen GJ, Kornberg R. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9262. doi: 10.1073/pnas.95.16.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jensen GJ. Doctoral. Stanford University; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.