Abstract
Ligand-induced dimerization of the epidermal growth factor receptor (ErbB-1/EGFR) involves conformational changes that expose an extracellular dimerization interface. Subsequent alterations within the cytoplasmic kinase domain, which culminate in tyrosine phosphorylation, are less understood. Our study addressed this question by using two strategies: a chimeric receptor approach employed ErbB-3, whose defective kinase domain was replaced by the respective part of EGFR. The implanted full-length kinase, unlike its subdomains, conferred dimerization and catalysis. The data infer that the kinase function of EGFR is restrained by the carboxyl tail; once grafted distally to the ectopic tail of ErbB-3, the kinase domain acquires quasi-dimerization and activation. In an attempt to alternatively refold the cytoplasmic tail, our other approach employed kinase inhibitors. Biophysical measurements and covalent cross-linking analyses showed that inhibitors targeting the active conformation of EGFR, in contrast to a compound recognizing the inactive conformation, induce quasi-dimers in a manner similar to the chimeric ErbB-3 molecule. Collectively, these observations unveil kinase domain-mediated quasi-dimers, which are regulated by an autoinhibitory carboxyl tail. On the basis of these observations, we propose that quasi-dimers precede formation of ligand-induced, fully active dimers, which are stabilized by both extracellular and intracellular receptor-receptor interactions.—Bublil, E. M., Pines, G., Patel, G., Fruhwirth, G., Ng, T., Yosef Yarden. Kinase-mediated quasi-dimers of EGFR.
Keywords: growth factor, kinase inhibitor, oncogene, receptor tyrosine kinase, receptor dimerization
The ErbB family of receptor tyrosine kinases comprises 4 receptors, ErbB-1 (also called EGFR) through ErbB-4, in charge of conveying signals emanating from 11 different ligands, all sharing an epidermal growth factor (EGF) domain (1). Ligand binding to the extracellular domain of a receptor induces extensive structural changes that detach a preformed molecular tether, and thus expose a dimerization arm, which promotes receptor interactions with other family members (2, 3). Dimerization of the extracellular domains is relayed across the plasma membrane in an incompletely understood manner and culminates in kinase domain activation. This is followed by phosphorylation of tyrosine residues located at the tail of the partnering receptors. The newly modified phosphotyrosine residues serve as docking sites for signaling molecules, which dock onto the receptor and underlie propagation of the signal further downstream. Despite stringent control circuits, compromised ErbB regulation is manifested in anomalous enzyme activity, which is implicated in several types of human cancer (4, 5). Accordingly, intercepting ErbB family members using antibodies or small-molecule kinase inhibitors is of clinical interest.
Unlike the well-established mode of ectodomain-mediated dimerization and receptor activation, amply supported by the resolved crystal structures of the ectodomains of all ErbB family members (3, 6–8), kinase domain activation remains less understood. Kuriyan and colleagues (9) proposed that the mode of kinase activation of ErbB-1 is equivalent to that of cyclin-activated kinases. In essence, following ligand-stimulated ectodomain dimerization, the cytoplasmic kinase domains are brought into close proximity, thus allowing the C lobe of one kinase domain (denoted the activator or donor kinase) to bind to the N lobe of the other (denoted the acceptor or receiver kinase), and hence activate the receiver kinase. This kind of kinase interactions is referred to as asymmetric. More recent studies (10, 11) reported that the juxtamembrane domain of the receiver participates in stabilizing the asymmetric dimer by binding to the C lobe of the activator. Interestingly, this model also provides an explanation of ErbB-3's mode of action. ErbB-3 is unique among the ErbB family members due to its silenced kinase domain (12–15) and an inability to form homodimers (16, 17). Zhang et al. (9) noted that amino acids comprising the N-lobe interface of the kinase domain are different in ErbB-3, as compared to the canonical ErbB interface, and thus ErbB-3 lacks the capacity to serve as a receiver. However, since its C-lobe interface is intact, ErbB-3 can serve as an activator toward other family members.
We envisioned that because of the defects within the kinase domain of ErbB-3, it might be utilized as a scaffold to study the regulation of kinase activation. Accordingly, we swapped parts of the kinase domain of ErbB-3 with the respective regions of ErbB-1/EGFR. The data obtained suggest that refolding of the cytoplasmic tail unlocks an inactive conformation and enables kinase-mediated dimer formation followed by phosphorylation. Apparently, this mode of ligand-independent dimerization and activation does not rely on the extracellular domain, but involves a kinase-kinase interface. Hence, this type of partial dimerization is denoted here as “quasi-dimerization.” To independently approach formation of quasi-dimers, we referred to previous studies that reported on the ability of a kinase inhibitor to induce dimerization of ErbB-1/EGFR (18, 19). According to one interpretation, the carboxyl tail of ErbB-1/EGFR refolds when the nucleotide-binding site is occupied by a tyrosine kinase inhibitor (TKI). TKIs are low-molecular-weight compounds, which penetrate across the plasma membrane and target the catalytic domain of tyrosine kinases (20). For example, gefitinib and erlotinib are directed against the tyrosine kinase domain of ErbB-1/EGFR, and both have been approved as therapies for lung cancer. Lapatinib, which targets both the kinase domains of ErbB-1 and ErbB-2, is used to treat ErbB-2-overexpressing mammary tumors (21). Cocrystals of the ErbB-1/EGFR kinase domain bound to each of these inhibitors (22–24) revealed that gefitinib and erlotinib stabilize an active conformation, but lapatinib stabilizes the inactive conformation. In line with these observations, we show here that TKIs that recognize the active kinase conformation can induce quasi-dimers analogous to the complexes formed by the aforementioned chimeric ErbB-3 molecule. These findings are discussed in the framework of a quasi-dimerization step, which may precede formation of fully active dimers of ErbB-1/EGFR.
MATERIALS AND METHODS
Antibodies and reagents
Antibodies against phospho-ErbB-3 and phospho-ErbB-2 were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies against phosphotyrosine, ERK, and the intracellular and extracellular domains of ErbB-3 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). An antibody for immunoprecipitation of ErbB-3 was purchased from Upstate Biotechnology (Lake Placid, NY, USA). An antibody against ErbB-1 was purchased from Alexis Biotechnology (Lausen, Switzerland). The antibody against the phosphorylated form of ERK was generously provided by the Rony Seger's laboratory (Weizmann Institute of Science, Rehovot, Israel). The antibody against HA peptide tag was purchased from Roche Applied Science (Manheim, Germany). The tyrosine kinase inhibitors erlotinib, gefitinib, and lapatinib were acquired from LC laboratories (Woburn, MA, USA). AG1478 was purchased from Calbiochem (Gibbstown, NJ, USA). Cross-linking experiments were carried out using bis-sulfosuccinimidyl-suberate (BS3; Pierce, Rockford, IL, USA).
Cell lines and tissue culture
Chinese hamster ovary (CHO) cells were cultured in DMEM/F12(HAM) (1:1), supplemented with 10% heat-inactivated FBS (Life Technologies, Carlsbad, CA, USA), l-glutamine, and a mixture of penicillin and streptomycin. Michigan Cancer Foundation-7 (MCF-7) cells were cultured in DMEM, supplemented with 10% heat-inactivated FBS (Sigma-Aldrich, Dorset, England), l-glutamine, and a mixture of penicillin and streptomycin.
Transfection, covalent cross-linking, and cell lysis
Transient receptor expression in CHO cells was accomplished by transfection of 0.5–3 μg of plasmid DNA using Lipofectamine (Invitrogen, Carlsbad, CA). At 48 h after transfection and depending on the experiment, cells were treated with 25 ng/ml neuregulin β-1 (NRG) or with TKIs (2–30 μM). Thereafter, cells were washed with cold PBS (Life Technologies, Carlsbad, CA, USA) and solubilized in lysis buffer containing 50 mM HEPES (pH 7.5), 10% glycerol, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 10 mM NaF, 1 mM Na3VO4, 30 mM β-glycerol phosphate, and a phosphatase-inhibitor cocktail. In cross-linking experiments, the lysates were incubated with the cross-linker BS3 at 4°C, and the cross-linking reaction was terminated using 20 mM of glycine (final concentration). Lysates were cleared by centrifugation prior to addition of gel sample buffer. Samples were then heated, separated on SDS-PAGE, and transferred onto a nitrocellulose membrane. Membranes were blocked for 1 h in TBS-T buffer (0.02 Tris-HCI, pH 7.5; 0.15 M NaCl; and 0.05% Tween 20) with 10% v/v milk, and then incubated for 1 h or overnight with the primary antibody. Secondary anti-mouse or anti-rabbit antibodies linked to horseradish peroxidase were used to detect immunoreactive bands utilizing the enhanced chemoluminescence reaction.
Fluorescence resonance energy transfer (FRET) determination by multiphoton fluorescence lifetime imaging microscopy (FLIM) measurements
FLIM was performed with a custom-built multiphoton system constructed around an upright 90i fluorescence microscope (Nikon, Tokyo, Japan). For FRET/FLIM analysis, pixel-by-pixel lifetime determination was achieved using a modified Levenberg-Marquardt (MLM) fitting technique, as described previously (25). The FLIM images obtained with the lifetime microscope were batch analyzed overnight by running in-house exponential fitting software (TRI2) written in CVI. The program outputs files where all the fitting parameters are recorded for each image. These files are then analyzed to produce a distribution of lifetime and an average lifetime (25).
Construction of the ErbB-3/ErbB-1 chimeric molecules
Mutagenesis was carried out in the pcDNA3 plasmid by using a mutagenesis kit (Stratagene, La Jolla, CA). All restriction enzymes and buffers were purchased from New England Biolabs (NEB;a Beverly, MA, USA). Constructs generated in this study were all verified and confirmed by nucleotide sequencing. The DNA corresponding to full-length ErbB-3 was amplified by PCR to generate a fragment flanked by restriction sites for EcoRV and NotI (forward primer 5′-GCGTAGATATCATGAGGGCGAACGACGCTCTG-3′, reverse primer 5′-GATCAAGGCGGCCGCTTACGTTCTCTGGGCATTAGCC-3′). The PCR fragment was thereafter digested with EcoRV and NotI, and cloned into the multiple cloning site (MCS) of the pcDNA3 vector, which was digested with the same enzymes. To generate two XbaI restriction sites to flank the N lobe of ErbB-3, the TCTGGA codons at the sequence coding for N-terminal end of the kinase domain were mutated to TCTAGA (point mutation underscored) and the GCTGGA codons at the C-terminal end of the N lobe were mutated to TCTAGA. A third restriction site, for Kpn, was introduced into the sequence coding for the C-terminal end of ErbB-3s C lobe by mutating the GGTATC codons to GGTACC. An additional vector (V2 vector), containing only the second XbaI site and the Kpn site was generated as well. Introduction of the restriction sites into the vector was carried out without altering the protein amino acid sequence. Other XbaI and Kpn restriction sites at the MCS of pcDNA3, and a Kpn site within the ErbB-3 sequence, were eliminated by site-directed mutagenesis.
A PCR fragment encoding the N lobe of ErbB-1 (forward primer 5′-GATCGATCTAGACCCCAGTGGAGAAGCTCCCAAC-3′, reverse primer 5′-GGTACGTCTAGAAGGCAGCCGAAGGGCATGAGC-3′) and of the full-length ErbB-1 kinase (forward primer 5′-GATCGATCTAGACCCCAGTGGAGAAGCTCCCAAC-3′, reverse primer 5′-GGTACGGGTACCGCTGGGGGTCTCGGGCCATTTTG-3′), incorporating the appropriate overhang ends following enzyme digestion, were cloned into the pcDNA3-ErbB-3 vector digested with XbaI or XbaI and Kpn, to generate the N1C3 and N1C1 chimeric ErbB-3/ErbB-1 molecules, respectively. In addition, the V2 vector was digested with XbaI and Kpn, and PCR-amplified fragments of the C lobe of ErbB-1 (forward primer 5′-GATCGATCTAGATTATGTCCGGGAACACAAAGAC-3′, reverse primer 5′-GGTACGGGTACCGCTGGGGGTCTCGGGCCATTTTG-3′) containing the appropriate overhang ends following digestion, were cloned into the V2 vector to produce the N3C1 chimera.
To generate the N1C1T1 chimera, the plasmid encoding N1C1 was digested with NotI and Kpn and a PCR-amplified fragment encoding the ErbB-1 tail (forward primer 5′-CGCTAGGTACCTTGTCATTCAGGGGGATGAAAG-3′, reverse primer 5′-CGCTAGCGGCCGCTCATGCTCCAATAAATTCACTGC-3′), digested with the same enzyme was cloned into the cut vector.
Site-directed mutagenesis
Site-directed mutagenesis to construct the receiver- and activator-impaired ErbB-1 receptor, was carried out using the QuikChange site-directed mutagenesis kit (Stratagene). In short, mutations were introduced by utilizing two complementary primers containing the desired mutations to generate and amplify the mutated encoding plasmid by PCR with the PFU Turbo DNA polymerase. The restriction enzyme DPN I was used to digest the parental template. The PCR product was then transformed into a host E. coli bacteria.
In vitro kinase assay
CHO cells were transfected with plasmids encoding ErbB-3 or N1C1. After 48 h, cells were washed with cold PBS, and harvested and lysed for 20 min in lysis buffer (deprived of phosphatase inhibitors). Lysates were then cleared by centrifugation and immunoprecipitated using an anti-ErbB-3 antibody. The immunoprecipitates were incubated with kinase buffer (25 mM HEPES, pH 7.5; 5 mM MnCl2; 2 mM dithiothreitol; and 150 mM NaCl) with or without supplemental ATP (40 μM) for 1, 5, and 10 min. Thereafter, immunoprecipitates were washed with cold PBS and resolved using SDS-PAGE.
RESULTS
Construction of chimeric ErbB-3/ErbB-1 receptors
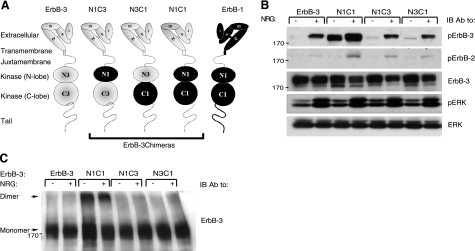
ErbB-3 conforms to the structural features defining other ErbB family members, namely a 4-domain extracellular part followed by a short segment traversing the plasma membrane, and a juxtamembrane sequence preceding a kinase domain and a carboxyl-terminal tail (Fig. 1A). It was previously suggested that the extremely low-kinase activity of ErbB-3, which differentiates ErbB-3 from its family members (12, 26) is due to multiple replacements within the canonical catalytic site (12–14). Nevertheless, restoring several motifs by mutations did not recover kinase activity (27), implying an intricate mechanism that silences ErbB-3. This may relate to a noncanonical N-lobe interface that impedes catalytic activation (9). Hence, we envisioned that ErbB-3 might be used as a scaffold: substituting elements of the silenced kinase domain with the respective functional elements of ErbB-1/EGFR may unveil mechanistic features governing ErbB activation. To this end, we generated the N1C3 chimeric molecule, in which the N lobe of ErbB-3 (Pro689–Asp797) was replaced by the respective lobe of ErbB-1 (Pro694–Asp800). We similarly generated the reciprocal N3C1 chimera (His798–Ile978 of ErbB-3 replaced by Tyr801–Ile981 of ErbB-1), as well as the N1C1 chimera, in which the whole kinase domain of ErbB-3 (Pro689–Ile978) was replaced by the corresponding domain of ErbB-1(Pro694–Ile981), as schematically depicted in Fig. 1A.
Figure 1.
The N1C1 chimera is constitutively phosphorylated and dimerized. A) Schemes present domain organizations of wild-type ErbB-3 (gray), ErbB-1/EGFR (black), and their chimeric derivatives. N denotes the N lobe and C denotes the C lobe of the respective tyrosine kinase domain. The 4 subdomains of the extracellular regions are denoted I–IV. Dashed line represents the cytoplasmic tail of ErbB-3; solid line represents tail of ErbB-1. Three chimeras are outlined. B) Plasmids encoding ErbB-3 (wild type), N1C1, N1C3, or N3C1 were cotransfected with a plasmid encoding a kinase-dead mutant of ErbB-2 (KD-B2) into CHO cells. After 48 h, cells were stimulated for 5 min without or with NRG (25 ng/ml). Thereafter, cells were detached using a scraper and lysed. Lysates were cleared by centrifugation, resolved by electrophoresis, and then transferred onto a nitrocellulose membrane. Anti-phopsho-ErbB-3 and anti-phospo-ErbB-2 antibodies were used to monitor receptor phosphorylation, whereas an anti-phospho-ERK antibody was used to assess the activation of ERK. An anti-ErbB-3 antibody was used to verify ectopic expression levels, and an antibody against ERK was utilized to verify equal loading. C) Plasmids encoding wild-type ErbB-3, N1C1, N1C3, or N3C1 were cotransfected into CHO cells together with a plasmid encoding the KD-B2 molecule. Cells were stimulated as in B and then washed with cold PBS. Thereafter, cells were detached using a scraper and extracted for 20 min at 4°C in lysis buffer containing the cross-linker BS3 (2 mM). The reaction was terminated by the addition of glycine (20 mM). Lysates were processed as in B. Dimers and monomers (arrows) were detected using an anti-ErbB-3 antibody.
The N1C1 ErbB-3 chimera is basally phosphorylated and dimerized
Because kinase activation requires prior dimerization and since ErbB-3 tends not to be incorporated into homodimers (16, 17), but rather into heterodimers with ErbB-2 (28), we cotransfected the ErbB-3 fusions with ErbB-2. In addition, to avoid transphosphorylation by ErbB-2, a kinase-dead mutant (denoted KD-B2) was used, such that tail phosphorylation may only reflect the catalytic function of the chimeric receptors. To detect tyrosine phosphorylation, the three chimeras and wild type ErbB-3 were separately cotransfected, along with KD-B2, into Chinese hamster ovary (CHO) cells, which express minute amounts of rodent ErbB-2, but no other ErbB family members (28). Cells were stimulated with an ErbB-3 ligand, neuregulin-beta1 (NRG), and extracts were immunoblotted with antibodies specific to the phosphorylated forms of either ErbB-2 or ErbB-3. As expected, NRG stimulation triggered an increase in phosphorylation of ErbB-3 and of ErbB-2 (Fig. 1B), but their relative levels cannot be inferred because of the use of different antibodies. Because the ectopically expressed receptors are devoid of kinase activity, ErbB-3 phosphorylation was attributed to heterodimerization with the endogenous ErbB-2 of CHO cells. The levels of tyrosine phosphorylation of N1C3 and N3C1 were comparable to the level observed with wild-type ErbB-3, suggesting that neither chimera gained kinase activity. Clearly, separately grafting the N lobe or C lobe of ErbB-1 into the framework of ErbB-3 is insufficient for reconstitution of kinase activity.
In sharp contrast to N1C3 and N3C1, the N1C1 chimera was highly phosphorylated even in the absence of a ligand. In addition, stimulation of N1C1 with NRG resulted in a substantial increase in receptor phosphorylation, beyond the level displayed by the similarly abundant wild-type ErbB-3 (Fig. 1B). It is notable that unlike the relatively high basal phosphorylation of N1C1, the basal (ligand-independent) phosphorylation of ErbB-2 remained undetectable. These observations suggested that basal phosphorylation of the N1C1 fusion protein occurs through formation of homodimers, rather than heterodimers with ErbB-2.
Next, we set out to test the possibility that the constitutive activation of N1C1 is associated with a propensity to form dimers in the absence of a stimulatory ligand. CHO cells were treated with NRG, or left untreated, and dimers were covalently stabilized by utilizing the bifunctional agent BS3. As shown in Fig. 1C, dimerization correlated with the tail's phosphotyrosine content: in contrast to the very low-dimerization signal obtained with untreated or with NRG-stimulated ErbB-3, N1C3, and N3C1, the N1C1 chimera displayed ligand-independent constitutive dimers, which were weakly affected by NRG, in similarity to their tyrosine phosphorylation. Thus, grafting the full kinase domain of ErbB-1 into ErbB-3 reconstituted the defective functions of this receptor, namely, an ability to autophosphorylate and undergo homodimerization. Notably, the stimulatory ligand exerted only small effects on these functions, implying close to maximal basal activation of N1C1.
The N1C1 chimeric receptor forms homodimers, rather than heterodimers with ErbB-2
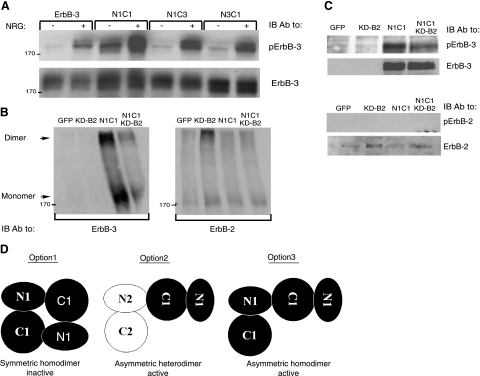
For the reason that ErbB-3 cannot form homodimers (16, 17) but may engage in heterodimers and higher oligomers (29, 30), we tested the nature of the dimers displayed by the N1C1 chimera. As a first test, we monitored basal and NRG-induced phosphorylation in cells transfected with the N1C1 plasmid alone, without the ErbB-2-encoding plasmid (KD-B2). As shown in Fig. 2A, N1C1 displayed relatively high basal activity even in the absence of KD-B2, implying that the kinase-dead mutant of ErbB-2 may not engage into N1C1-containing dimers. In line with this possibility, transfection of the N1C1 chimera alone, in the absence of KD-B2, was sufficient for formation of dimers (Fig. 2B; left panel). Because these dimers contained ErbB-3 but undetectable ErbB-2, and cotransfection with KD-B2 did not alter the monomer-to-dimer ratio (Fig. 2B; right panel), we favored the possibility that they represent homodimers. This notion was further supported by the existence of KD-B2 homodimers (Fig. 2B; right panel) and by the undetectable phosphorylation of ErbB-2 in cells coexpressing N1C1 and the kinase-dead mutant of ErbB-2 (KD-B2; Fig. 2C).
Figure 2.
The N1C1 chimera incorporates into homodimers rather than heterodimers with ErbB-2. A) Plasmids encoding wild-type ErbB-3, N1C1, N1C3, or N3C1 were transfected into CHO cells. After 48h, cells were stimulated for 5 min without or with NRG (25 ng/ml) and then washed with cold PBS. Thereafter, cells were detached, and their lysates were cleared by centrifugation, resolved by electrophoresis and then transferred onto a nitrocellulose membrane. Anti-phopsho-ErbB-3 and anti-ErbB-3 antibodies were used to monitor phosphorylation or verify expression levels, respectively. B, C) N1C1 and KD-B2 encoding plasmids (3 μg) were transfected into CHO cells, either alone or in combination (each at 1.5 μg). After 48 h, cells were washed with cold PBS, detached using a scraper, and extracted in lysis buffer with (B) or without (C) the cross-linker BS3 (2 mM). The cross-linking reaction was carried out for 20 min at 4°C and terminated by the addition of glycine (20 mM). Thereafter, lysates were cleared by centrifugation, resolved by electrophoresis, and transferred onto a nitrocellulose membrane. presence of ErbB-3 or ErbB-2 in dimers and monomers was monitored using specific antibodies. Phosphotyrosine content of both receptors was examined by using anti-phospho-ErbB-3 or anti-phospho-ErbB-2 antibodies. D) Potential dimerization modes of the N1C1 chimera are indicated. Kinase domains of the N1C1 chimera and ErbB-2 (either KD-B2 or the endogenous receptor) are represented by solid and open shapes, respectively. Option 1 denotes a symmetric inactive homodimer of two N1C1 chimeric receptors, whereas option 2 relates to heterodimerization of the chimeric N1C1 molecule together with the endogenous or the ectopic ErbB-2 molecule. Option 3 indicates an asymmetric active homodimer of two chimeric N1C1 molecules, mediated by their kinase domains.
Several modes of dimerization of the N1C1 kinase domain may be considered in light of the resolved crystal structures of the catalytic portion of ErbB-1 (9, 10): 1) inactive, symmetric homodimers (10, 31); 2) active, asymmetric heterodimers with the endogenous or ectopically expressed ErbB-2 molecules; and 3) active, asymmetric homodimers. Option 1 may be declined, for the dimers observed undergo phosphorylation, while option 2 seems unlikely because we could not detect ErbB-2-containing heterodimers (Fig. 2B, right panel), nor could we detect considerable basal ErbB-2 phosphorylation (Figs. 1B and 2C). Hence, the results presented in Figs. 1 and 2 favor formation of active, N1C1:N1C1 homodimers, which are likely mediated by the kinase domain.
The N1C1 chimera is intrinsically active and displays sensitivity to NRG stimulation
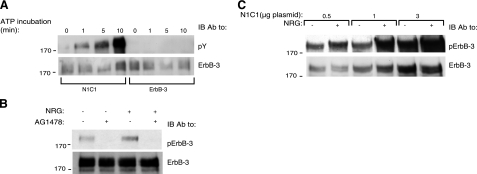
To ascertain that the kinase domain of the N1C1 chimera was intrinsically active, its function was compared to that of the wild-type ErbB-3 in an in vitro kinase assay. Immunoprecipitates of ErbB-3 and N1C1 from extracts of CHO cells transfected with the respective plasmids were incubated for increasing time intervals with a buffer supplemented with ATP. As shown in Fig. 3A, no detectable tyrosine phosphorylation was associated with ErbB-3, but phosphorylation of the N1C1 chimera was readily detectable, confirming functional intactness of the implanted kinase. This was further corroborated by the ability of an ErbB-1-specific kinase inhibitor, AG1478, to inhibit both basal and NRG-induced phosphorylation of N1C1 in intact cells (Fig. 3B). In addition, by transfecting cells with increasing amounts of the N1C1 plasmid, we noted that both the basal and the ligand-inducible phosphorylation signals increased proportionately (Fig. 3C), in line with the possibility that the basal phosphorylation of N1C1 was due to intrinsic kinase activation.
Figure 3.
Autophosphorylation of the N1C1 chimera. A) CHO cells were transfected with plasmids encoding ErbB-3 or N1C1. After 48 h, cells were washed with cold PBS and harvested. Cells were lysed for 20 min in lysis buffer (deprived of phosphatase inhibitors), cleared by centrifugation, and then immunoprecipitated using an anti-ErbB-3 antibody. Immunoprecipitates were incubated with a buffer supplemented with ATP (40 μM) for the indicated time intervals. Immunoprecipitates were then washed with cold PBS and resolved by electrophoresis and immunoblotting (IB) utilizing an anti-phosphotyrosine antibody (pY). B) A plasmid encoding the N1C1 chimera was transfected into CHO cells. After 48 h, cells were treated with AG1478 (10 mM) or left untreated for 30 min at 37°C and then stimulated for 5 min with or without NRG (25 ng/ml), and washed with cold PBS. Thereafter, cells were harvested using a scraper and lysed. Lysates were cleared, resolved by electrophoresis, and then transferred onto a nitrocellulose membrane. Phosphorylation and expression of the N1C1 chimera were detected using anti-phospho-ErbB-3 and anti-ErbB-3 antibodies, respectively. C) Cells were transfected as in A with increasing amounts of the N1C1 plasmid and stimulated with or without NRG (25 ng/ml). Thereafter, cells were lysed, and the phosphotyrosine content of the N1C1 chimera was assessed using an anti-phospho-ErbB-3 antibody. ErbB-3 levels were confirmed by stripping and reprobing the membrane with an anti-ErbB-3 antibody.
The ErbB-3-derived tail of the N1C1 chimera lacks the capacity to inhibit the kinase domain
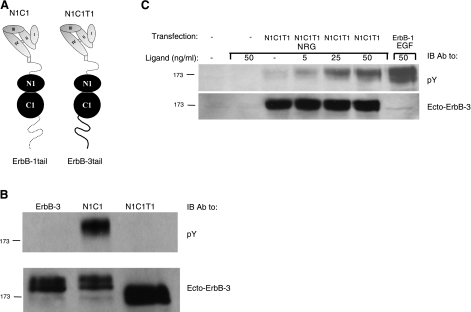
Several lines of previous evidence proposed that the tail of ErbB-1 folds over the kinase domain to impose an inhibitory effect (10, 23, 31). The N1C1 chimera incorporates the kinase domain of ErbB-1, but it harbors the tail of ErbB-3. Hence, in light of the basal activity of N1C1, we anticipated that the tail of ErbB-3 cannot interface with the kinase domain, thereby constrain the catalytic function of N1C1. To test this model, we generated a chimera similar to N1C1, except with the tail of ErbB-1 (denoted N1C1T1; Fig. 4A). Since N1C1T1 harbors a tail that potentially “fits” the kinase domain, our model predicted low if any basal phosphorylation. To test this prediction, cells were transfected with plasmids encoding ErbB-3, N1C1, or N1C1T1, and the tail's phosphorylation was monitored using an anti-phosphotyrosine antibody. As shown in Fig. 4B, no basal phosphorylation was detectable following transfection with the N1C1T1 plasmids or with a vector encoding the wild-type form of ErbB-3, in contrast with the robust basal phosphorylation displayed by N1C1. Binding of NRG to N1C1T1 was able to stimulate tyrosine phosphorylation of the chimeric receptor (Fig. 4C), in line with a restraining function of the carboxyl tail, which is released on ligand binding and receptor dimerization.
Figure 4.
Cytoplasmic tail of ErbB-1/EGFR inhibits basal phosphorylation of the N1C1 chimera. A) Schematic representation of N1C1 in comparison with the N1C1T1 chimera. Note that the chimeras are identical except for the cytoplasmic tails. B) CHO cells were transfected with plasmids encoding the wild-type ErbB-3, N1C1, or the N1C1T1 chimera. At 48 h after transfection, cells were washed with cold PBS, detached, and lysed. Lysates were cleared, resolved by electrophoresis, and then transferred onto a nitrocellulose membrane. Anti-phosphotyrosine and anti-ErbB-3 antibodies (the latter directed against the extracellular region of the receptor) were used to monitor phosphorylation and verify expression levels, respectively. C) CHO cells were transfected with plasmid encoding the N1C1T1 chimera or they were left untreated. After 48 h, cells were stimulated for 5 min with varying concentrations of NRG, and cell lysates were analyzed as in B.
Kinase inhibitors targeting the active conformation induce homodimerization of ErbB-1/EGFR
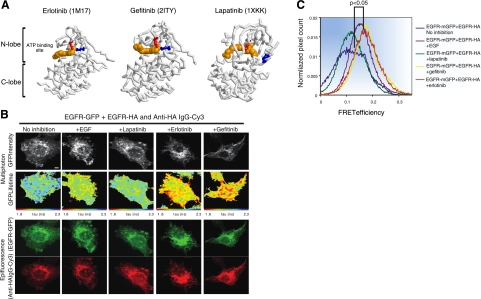
The ability of N1C1 to form catalytically active homodimers, in the absence of a stimulatory ligand, proposed that the kinase domain harbors a propensity to form functionally active dimers, once autoinhibition by the carboxyl tail is relieved. It is relevant noting that crystallization studies indicated that the nucleotide-bound catalytic domains of protein kinases, such as EGFR and the cyclin-dependent kinase, form dimeric complexes as an essential step of their activation process (9). In addition, structural analyses of the Abelson tyrosine kinase (Abl) proposed that the enzyme maintains a dynamic equilibrium of active and inactive conformations (32). Hence, the availability of clinically approved kinase inhibitors, which specifically target the active or inactive conformations of EGFR's kinase domain offers an opportunity to test direct involvement of the kinase domain in formation of ligand-independent, homodimeric complexes. Presumably, by binding to the active conformation, TKIs may displace the tail, thereby expose a molecular interface essential for asymmetric dimer formation. To test this scenario, we made use of three drugs: erlotinib and gefitinib, compounds broadly used to treat nonsmall lung cancer, and lapatinib, a molecule that targets both ErbB-1/EGFR and ErbB-2 in breast cancer (reviewed in ref. 20). It is important noting that unlike erlotinib and gefitinib, which recognize the active conformation of the kinase, lapatinib binds to the inactive conformation (23). Figure 5A depicts the 3-dimensional structure of the inactive form of ErbB-1/EGFR in a complex with lapatinib, which is distinct from the structurally similar active-like conformations containing erlotinib or gefitinib (22–24).
Figure 5.
FRET/FLIM analysis reveals that erlotinib and gefitinib, but not lapatinib, induce dimerization of ErbB-1/EGFR. A) Three-dimensional structures of the kinase domain of ErbB-1 bound to erlotinib, gefitinib, or lapatinib. Erlotinib (PDB 1M17)- and gefitinib (PDB 2ITY)-bound kinase is active (note salt bridge formed between K721, in red, and E738, in blue, which are in proximity), whereas the lapatinib-bound form (PDB 1XKK) is inactive (K721 and E738 are distant). Structural components of the kinase (N lobe, C lobe, and ATP-binding site) are labeled. B) MCF-7 cells were transfected with vectors encoding EGFR-mGFP and EGFR-HA. Cells were incubated for 24 h, serum starved for 1 h, and stimulated with EGF (50 ng/ml) for 1 h. Alternatively, cells were treated with lapatinib, gefitinib, or erlotinib as indicated (10 μM) for 1 h, prior to fixation and staining with an anti-HA antibody conjugated to Cy-3. Scale bars = 5 μm. C) Cumulative histogram of FRET efficiency between EGFR-mGFP and HA-Cy3 (bound to EGFR-HA) calculated with the following equation in each pixel and averaged per cell: FRET efficiency = 1 tau (da)/tau (control), where tau (da) is the lifetime displayed by cells coexpressing both EGFR-mGFP and EGFR-HA stained with an anti-HA IgG-Cy3, whereas tau (control) is the mean EGFR-mGFP lifetime measurement in the absence of the Cy3 acceptor. Data were obtained from 4–11 cells/treatment group and are representative of 2 independent experiments. Values of P are 0.003 and 0.025, respectively, for comparisons between untreated cells and cells treated with gefitinib or lapatinib, respectively, according to analysis of variance with post hoc testing using Tukey's honest significant test.
To test TKI-induced physical interactions, we employed FRET between two tagged forms of ErbB-1/EGFR: a receptor fused to a monomeric green fluorescent protein (mGFP; a A206K version of GFP) and another form cytoplasmically tagged with an HA peptide. Within a dimer, energy transfer from GFP to Cy3 would decrease fluorescence lifetime (tau) of mGFP. Indeed, experiments using multiphoton FLIM, as described previously (33–35), confirmed that a coexpressed ErbB-1/EGFR-HA can reduce the fluorescence lifetime of ErbB-1/EGFR-mGFP in untreated cells, but TKI-treated cells expressing ErbB-1/EGFR-mGFP alone displayed no change in lifetime (data not shown). As predicted, incubation with EGF, gefitinib, or erlotinib increased FRET efficiency compared to untreated cells (baseline control; Fig. 5B, C), in line with homodimer formation. By contrast, incubation with lapatinib did not detectably influence ErbB-1/EGFR-mGFP lifetime. These results raised the possibility that the active conformation of ErbB-1/EGFR resembles N1C1 in terms of refolding of the carboxyl tail, release of autoinhibition and dimer formation.
Biochemical assays link the active conformation of the kinase with a dimeric complex, and help define the corresponding molecular interface
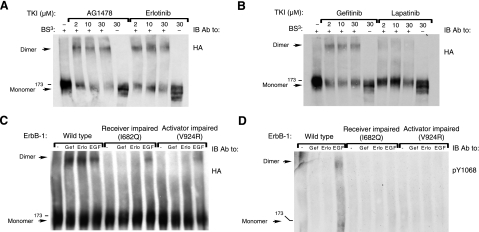
To corroborate the biophysical measurements, we biochemically analyzed the clinically approved erlotinib, gefitinib, and lapatinib, as well as a fourth kinase inhibitor, AG1478, because previous studies reported on the ability of this compound to stabilize a dimeric form of ErbB-1/EGFR (18, 19). Using covalent cross-linking we confirmed the ability of AG1478 to deplete the monomeric form of ErbB-1, while increasing the fraction of dimers (Fig. 6A). This attribute was shared by erlotinib and gefitinib, but lapatinib displayed only very weak dimerizing activity in this assay (Fig. 6A, B), in line with the observations we made using biophysical assays of receptor interactions (Fig. 5).
Figure 6.
Cross-linking analysis reveals that AG1478, erlotinib, and gefitinib, but not lapatinib, induce dimerization of ErbB-1/EGFR. A, B) CHO cells were transfected with a plasmid encoding HA-tagged ErbB-1, and 48 h later, they were incubated for 60 min at 37°C with increasing concentrations of AG1478, erlotinib, gefitinib, or lapatinib, as indicated. Cells were then washed with cold PBS and harvested in lysis buffer containing the BS3 cross-linker. After 20 min at 4°C, the cross-linking reaction was terminated by the addition of glycine (20 mM). Thereafter, lysates were cleared by centrifugation, resolved by electrophoresis, and transferred onto a nitrocellulose membrane. Dimers were detected using an antibody against the HA tag. C, D) Cells were transfected with plasmids encoding wild-type ErbB-1/EGFR, a receiver-impaired (I682Q) mutant, or an activator-impaired (V924R) mutant of ErbB-1/EGFR. After 48 h, cells were treated with or without gefitinib (10 μM), erlotinib (10 μM), or EGF (50 ng/ml) and then lysed and cross-linked as in A. Dimers and monomers were detected using an antibody against the HA tag, and tyrosine phosphorylation was monitored using an antibody to the phosphorylated form of EGFR's tyrosine 1068.
Previous mutational and crystallographic studies concluded that kinase activation requires formation of an asymmetric dimer, in which the C lobe of the activator kinase interfaces with the N lobe of the receiver kinase (9, 10). To test the likelihood that the TKI-induced dimers we observed were structurally similar to the asymmetric active dimers induced by EGF, we introduced into the corresponding interface two mutations known to block kinase activation, namely: I682Q, a receiver-impairing mutation, and V924R, an activator-impairing mutation (9). As shown in Fig. 6C, introduction of either mutation almost abolished the ability of gefitinib and erlotinib to induce receptor dimers. By contrast, both the receiver- and the activator-defective mutants partly retained EGF-induced dimerization (Fig. 6C). As expected, this partial EGF-induced dimerization was associated with no phosphorylation signals (Fig. 6D).
In summary, the results presented in Fig. 6 confirmed the biophysical measurements of receptor interactions (Fig. 5): TKIs promote dimerization of ErbB-1/EGFR only if they recognize the active conformation of the kinase. In addition, complexes instigated by TKIs, as well as dimers induced by EGF, engage a common molecular interface. Interestingly, although mutagenesis of the dimerization interface was sufficient to completely block TKI-induced dimerization, the mutated receptors partly retained formation of (inactive) dimers in response to EGF. This observation is consistent with the possibility that quasi-TKI-induced dimers depend only on the kinase domain interface, unlike ligand-induced dimers, which are held by both intracellular and extracellular interfaces. Below, we discuss the implications of a kinase activation model that integrates ligand-induced dimerization of the extracellular part of ErbB-1/EGFR with the tail-regulated interactions between two juxtaposed kinase domains.
DISCUSSION
A clinically effective strategy to intercept ErbB signaling in cancer makes use of highly specific kinase inhibitors. Accordingly, understanding the mechanism of kinase activation and inhibition is pivotal for designing better drugs. In line with this aim, our study unraveled several mechanistic features of EGFR's kinase activation by utilizing two experimental approaches: construction of chimeric derivatives of the enzymatically defective ErbB-3 molecule, and application of TKIs able to recognize the active kinase conformation. The inactivity of ErbB-3 is partly attributed to the N-lobe interface essential for asymmetric dimerization, which deviates from the canonical structure constituting the corresponding regions in other ErbB proteins (9, 15). As we show here, replacing the N lobe of ErbB-3 with the N lobe of ErbB-1/EGFR (N1C3 chimera) failed to significantly reconstitute catalysis, probably because ErbB-3 bears additional structural defects, including a mutation of the catalytic aspartate to aspargine in the HRD motif (27). Congruently, when the whole kinase domain of ErbB-1/EGFR was implanted into the ErbB-3 framework (N1C1 chimera), we observed ligand-induced dimerization and subsequent phosphorylation (Fig. 1). Moreover, N1C1 unexpectedly displayed basal dimerization and phosphorylation, even in the absence of a stimulatory ligand. In addition, the dimers formed by N1C1 did not detectably contain ErbB-2, and thus they may represent homodimers (Fig. 2). In light of the inability of wild-type ErbB-3 to form homodimers (16, 17) (Fig. 1C) and the oligomerization-inhibitory role for a sugar moiety at asparagine 418 of the ectodomain (36), we assume that the intact kinase domain of ErbB-1/EGFR confers to N1C1 the ability to form kinase-mediated homodimers and also overcomes the intrinsic dimerization inhibitory effect of the ectodomain.
Interestingly, kinase-mediated dimers of ErbB-1/EGFR, which do not rely on the extracellular domain, have been reported (10, 31, 37). In contrast with these inactive, symmetrical dimers, the kinase domain-mediated dimers of N1C1 are catalytically active, in line with a unique mode of asymmetric dimer formation. Our results imply that the underlying mechanism involves refolding of the autoinhibitory carboxyl-terminal tail. Several previous lines of evidence indicated that the cytoplasmic tail of ErbB-1/EGFR inhibits kinase activity by folding over the catalytic domain (10, 23, 31). For example, removal of residues 965–998 of the tail increased kinase activity (9). This segment contains 2 inhibitory subdomains: residues 982–994 wrap around the backside of the kinase within the autoinhibited symmetric dimer (31, 38), whereas residues 971–979 form a short helix that blocks the active site of the kinase (23). The intramolecular interface involved corresponds to a sequence residing on the activator's C lobe: within the symmetric dimer, this interface is occupied in cis by the C-terminal tail, whereas in the context of the asymmetric dimer, it is occupied in trans by a juxtamembrane stretch (G672–I682) of the receiver (10, 39). Accordingly, once the receptor is activated by a ligand, the tail folds away from the kinase domain, thereby enabling the juxtamembrane region of the receiver to “latch” the C lobe of the activator. By contrast, we infer that in N1C1, the tail of ErbB-3 fails to function as an autoinhibitor since it is ectopic to the kinase of ErbB-1/EGFR (15). Hence, the interface is pre-exposed and allows asymmetric dimerization and subsequent activation. This mechanism is supported by the abolishment of basal phosphorylation in the context of N1C1T1, which harbors both the tail and the kinase domain of ErbB-1/EGFR (Fig. 4).
It should be noted that low-kinase activity of ErbB-3 and an ability to bind ATP were recently reported (26). Hence, we cannot exclude the possibility that the low NRG-induced ErbB-3 phosphorylation observed following transfection of wild-type ErbB-3 (Fig. 1B) is due, in part, to the kinase domain of ErbB-3.
Our other experimental arm was based on the dynamic equilibrium of tyrosine kinases between different conformations (32). We assumed that ATP binding to ErbB-1/EGFR controls the equilibrium not only through reorganization of the kinase domain's activation loop (40), but also by refolding of the carboxyl tail. This would allow the juxtamembrane region of a neighboring receptor to bind with the exposed interface within the kinase C lobe and latch an asymmetric dimer. TKIs strongly bind with the ATP-binding site of ErbB-1/EGFR; hence, they may induce (or stabilize) tail refolding and formation of catalytically disabled asymmetric dimers. In line with this sequel and with previous reports (18, 19), we observed dimers of EGFR after treatment of intact cells with TKIs (Figs. 5 and 6). Two lines of evidence imply that the TKI-induced dimers that we observed are analogous to N1C1 dimers, as well as to ligand-induced dimers of ErbB-1/EGFR: First, a TKI that prevents formation of the active conformation was unable to induce receptor dimerization (Fig. 6), and second, mutagenesis of the interface involved in asymmetric dimer formation abolished TKI-induced dimers (Fig. 6C). In summary, although they are distinct, the two arms of our study converge on a single mechanism of ligand-independent formation of conformationally active dimers, which are held by the kinase domain.
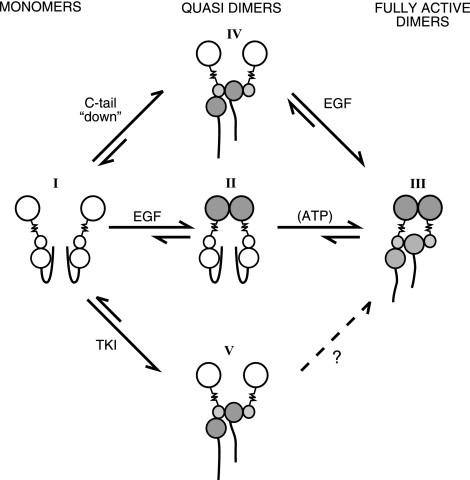
On the basis of our study and previously described mechanisms (10, 11, 41), we propose a model that integrates the contributions of both the kinase domain and the extracellular region to a 2-step dimerization process (see Fig. 7). As already discussed, the carboxyl tail of monomeric ErbB-1/EGFR molecules folds over the kinase domain, to prevent asymmetric dimer formation and kinase activation (Fig. 7; molecular species I). Ligand binding to the extracellular domain promotes extracellularly held quasi-dimers (Fig. 7; species II), which rapidly transform into catalytically active full dimers (Fig. 7; species III). We propose that ligand-independent, kinase-mediated quasi-dimers may assume a similar asymmetric configuration of the kinase by extending the tail, as in N1C1 (Fig, 7; species IV). Alternatively, binding a TKI may also induce a quasi-dimer, which is ligand-independent and catalytically disabled (Fig. 7; species V). We speculate that binding of ATP promotes short-lived quasi-dimers analogous to the TKI-induced complexes, but they dissociate once their bound nucleotide undergoes hydrolysis.
Figure 7.
Proposed mechanism of catalytic activation of ErbB-1/EGFR. EGFR is schematically presented as a large extracellular lobe connected through a transmembrane domain (zigzag line) to a bilobular kinase domain, flanked by a flexible carboxyl tail. Open and shaded lobes symbolize inactive and active conformations. Model assumes that the transition from catalytically inactive monomers (molecular species I) to active dimers (III) involves an intermediate quasi-dimer state (II). Accordingly, in the absence of a ligand EGFR assumes a monomeric inactive conformation (I), in which the cytoplasmic tail inhibits untimely kinase activation. Ligand stimulation enforces conformational changes that stabilize the extracellularly held quasi-dimer (II). Once juxtaposed, the kinase domains displace the carboxyl tails, which results in an active asymmetric dimer held by both extracellular and intracellular interfaces (III). According to the model, tail displacement and subsequent quasi-dimer formation may alternatively initiate with no involvement of the extracellular interface. For example, TKIs that recognize the active conformation of the kinase can extend the tail and form disabled quasi-dimers (V). Alternatively, a mutated terminus, or an ectopic tail (e.g., the tail of N1C1), would enable kinase activation by exposing an interface needed for asymmetric dimer formation (IV). Whether quasi-dimer formation is reversible or influenced by ATP remains unknown.
It is probable that oncogenic insults manipulate the proposed 2-step activation of ErbB-1/EGFR. Class IV EGFR mutants of brain tumors harbor internal deletions at the junction of the kinase domain and the carboxyl tail (42, 43), which likely unleash the catalytic function due to tail refolding. Similarly, the high basal kinase activity of the viral form of ErbB-1/EGFR, v-ErbB, may be attributed to sequence alterations in the C-terminal tail leading to quasi-dimer formation analogous to the mechanism underlying N1C1 activation (39). Artificially introduced mutations may also lend support to the proposed mechanism of kinase activation: mutagenesis of the segment Asp979–Asp982 of the cytoplasmic tail, which is essential for docking onto the C lobe of the kinase, resulted in substantial ligand-independent activation of ErbB-1/EGFR (10), which might be due to quasi-dimerization.
In summary, our 2-arm study uncovered the existence of quasi-dimers of ErbB-1/EGFR, and proposed their involvement as intermediate complexes in receptor activation. The emerging model raises the possibility that ligand-independent kinase activation, through formation of asymmetric quasi-dimers, may bear physiological relevance. Potentially, heterodimer formation with ErbB-2/HER2, interactions with cytokine receptors, integrins, and G-protein-coupled receptors, as well as tail phosphorylation by non-ErbB kinases, may transregulate ErbB-1/EGFR by mechanisms involving quasi-dimer formation. These possibilities, along with the reversibility and contributions of quasi-dimers to basally high phosphorylation of overexpressed or mutated ErbB-1/EGFR in human tumors remain open for future investigation.
Acknowledgments
The authors thank members of their groups for insightful comments. G.P. is supported by a Cancer Research UK Clinical Training Fellowship.
G.F. is supported by the King's College London–University College London Comprehensive Cancer Imaging Centre, funded by Cancer Research UK and the Engineering and Physical Sciences Research Council, in association with the Medical Research Council and the UK Department of Health. Y.Y. is the incumbent of the Harold and Zelda Goldenberg Professorial Chair. The EGFR-mGFP plasmid was a kind gift of Melanie Keppler (Richard Dimbleby Laboratory, King's College London). The multiphoton FLIM systems with in-house acquisition/analysis software were built by Simon Ameer-Beg (King's College London), Paul Barber (Gray Institute for Radiation Oncology and Biology, University of Oxford, Oxford, UK), and Borivoj Vojnovic (Gray Institute). This work is supported by grants from the U.S. National Cancer Institute (NCI; CA072981), the German Science Foundation, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
REFERENCES
- 1.Yarden Y., Sliwkowski M. X. (2001) Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 2.Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2002) Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 3.Ogiso H., Ishitani R., Nureki O., Fukai S., Yamanaka M., Kim J. H., Saito K., Sakamoto A., Inoue M., Shirouzu M., Yokoyama S. (2002) Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110, 775–787 [DOI] [PubMed] [Google Scholar]
- 4.Hynes N. E., MacDonald G. (2009) ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 5.Bublil E. M., Yarden Y. (2007) The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr. Opin. Cell Biol. 19, 124–134 [DOI] [PubMed] [Google Scholar]
- 6.Cho H. S., Leahy D. J. (2002) Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 7.Bouyain S., Longo P. A., Li S., Ferguson K. M., Leahy D. J. (2005) The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc. Natl. Acad. Sci. U. S. A. 102, 15024–15029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Kofler M., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2003) The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol. Cell. 11, 495–505 [DOI] [PubMed] [Google Scholar]
- 9.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 10.Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. (2009) Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Red Brewer M., Choi S. H., Alvarado D., Moravcevic K., Pozzi A., Lemmon M. A., Carpenter G. (2009) The juxtamembrane region of the EGF receptor functions as an activation domain. Mol. Cell. 34, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guy P. M., Platko J. V., Cantley L. C., Cerione R. A., Carraway K. L., 3rd (1994) Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc. Natl. Acad. Sci. U. S. A. 91, 8132–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sierke S. L., Cheng K., Kim H. H., Koland J. G. (1997) Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem. J. 322, 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plowman G. D., Whitney G. S., Neubauer M. G., Green J. M., McDonald V. L., Todaro G. J., Shoyab M. (1990) Molecular cloning and expression of an additional epidermal growth factor receptor-related gene. Proc. Natl. Acad. Sci. U. S. A. 87, 4905–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jura N., Shan Y., Cao X., Shaw D. E., Kuriyan J. (2009) Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U. S. A. 106, 21608–21613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger M. B., Mendrola J. M., Lemmon M. A. (2004) ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 569, 332–336 [DOI] [PubMed] [Google Scholar]
- 17.Pinkas-Kramarski R., Soussan L., Waterman H., Levkowitz G., Alroy I., Klapper L., Lavi S., Seger R., Ratzkin B. J., Sela M., Yarden Y. (1996) Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 15, 2452–2467 [PMC free article] [PubMed] [Google Scholar]
- 18.Arteaga C. L., Ramsey T. T., Shawver L. K., Guyer C. A. (1997) Unliganded epidermal growth factor receptor dimerization induced by direct interaction of quinazolines with the ATP binding site. J. Biol. Chem. 272, 23247–23254 [DOI] [PubMed] [Google Scholar]
- 19.Gan H. K., Walker F., Burgess A. W., Rigopoulos A., Scott A. M., Johns T. G. (2007) The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J. Biol. Chem. 282, 2840–2850 [DOI] [PubMed] [Google Scholar]
- 20.Baselga J. (2006) Targeting tyrosine kinases in cancer: the second wave. Science 312, 1175–1178 [DOI] [PubMed] [Google Scholar]
- 21.Spector N. (2008) Treatment of metastatic ErbB2-positive breast cancer: options after progression on trastuzumab. Clin. Breast. Cancer 8(Suppl. 3), S94–S99 [DOI] [PubMed] [Google Scholar]
- 22.Stamos J., Sliwkowski M. X., Eigenbrot C. (2002) Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 277, 46265–46272 [DOI] [PubMed] [Google Scholar]
- 23.Wood E. R., Truesdale A. T., McDonald O. B., Yuan D., Hassell A., Dickerson S. H., Ellis B., Pennisi C., Horne E., Lackey K., Alligood K. J., Rusnak D. W., Gilmer T. M., Shewchuk L. (2004) A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 64, 6652–6659 [DOI] [PubMed] [Google Scholar]
- 24.Yun C. H., Boggon T. J., Li Y., Woo M. S., Greulich H., Meyerson M., Eck M. J. (2007) Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber P. R., Ameer-Beg S. M., Gilbey J., Carlin L. M., Keppler M., Ng T. C., Vojnovic B. (2009) Multiphoton time-domain fluorescence lifetime imaging microscopy: practical application to protein-protein interactions using global analysis. J. R. Soc. Interface 6, S93–S105 [Google Scholar]
- 26.Shi F., Telesco S. E., Liu Y., Radhakrishnan R., Lemmon M. A. (2010) ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U. S. A. 107, 7692–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prigent S. A., Gullick W. J. (1994) Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J. 13, 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzahar E., Waterman H., Chen X., Levkowitz G., Karunagaran D., Lavi S., Ratzkin B. J., Yarden Y. (1996) A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 16, 5276–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landgraf R., Eisenberg D. (2000) Heregulin reverses the oligomerization of HER3. Biochemistry 39, 8503–8511 [DOI] [PubMed] [Google Scholar]
- 30.Chen C. H., Chernis G. A., Hoang V. Q., Landgraf R. (2003) Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. U. S. A. 100, 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau M., Fleishman S. J., Ben-Tal N. (2004) A putative mechanism for downregulation of the catalytic activity of the EGF receptor via direct contact between its kinase and C-terminal domains. Structure 12, 2265–2275 [DOI] [PubMed] [Google Scholar]
- 32.Schindler T., Bornmann W., Pellicena P., Miller W. T., Clarkson B., Kuriyan J. (2000) Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289, 1938–1942 [DOI] [PubMed] [Google Scholar]
- 33.Prag S., Parsons M., Keppler M. D., Ameer-Beg S. M., Barber P., Hunt J., Beavil A. J., Calvert R., Arpin M., Vojnovic B., Ng T. (2007) Activated ezrin promotes cell migration through recruitment of the GEF Dbl to lipid rafts and preferential downstream activation of Cdc42. Mol. Biol. Cell 18, 2935–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peter M., Ameer-Beg S. M., Hughes M. K., Keppler M. D., Prag S., Marsh M., Vojnovic B., Ng T. (2005) Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys. J. 88, 1224–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makrogianneli K., Carlin L. M., Keppler M. D., Matthews D. R., Ofo E., Coolen A., Ameer-Beg S. M., Barber P. R., Vojnovic B., Ng T. (2009) Integrating receptor signal inputs that influence small Rho GTPase activation dynamics at the immunological synapse. Mol. Cell. Biol. 29, 2997–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokoe S., Takahashi M., Asahi M., Lee S. H., Li W., Osumi D., Miyoshi E., Taniguchi N. (2007) The Asn418-linked N-glycan of ErbB3 plays a crucial role in preventing spontaneous heterodimerization and tumor promotion. Cancer Res. 67, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 37.Yu X., Sharma K. D., Takahashi T., Iwamoto R., Mekada E. (2002) Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol. Biol. Cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landau M., Ben-Tal N. (2008) Dynamic equilibrium between multiple active and inactive conformations explains regulation and oncogenic mutations in ErbB receptors. Biochim. Biophys. Acta 1785, 12–31 [DOI] [PubMed] [Google Scholar]
- 39.Boerner J. L., Danielsen A., Maihle N. J. (2003) Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Exp. Cell. Res. 284, 111–121 [DOI] [PubMed] [Google Scholar]
- 40.Bose R., Zhang X. (2009) The ErbB kinase domain: structural perspectives into kinase activation and inhibition. Exp. Cell. Res. 315, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess A. W., Cho H. S., Eigenbrot C., Ferguson K. M., Garrett T. P., Leahy D. J., Lemmon M. A., Sliwkowski M. X., Ward C. W., Yokoyama S. (2003) An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell. 12, 541–552 [DOI] [PubMed] [Google Scholar]
- 42.Kuan C. T., Wikstrand C. J., Bigner D. D. (2001) EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr. Relat. Cancer 8, 83–96 [DOI] [PubMed] [Google Scholar]
- 43.Frederick L., Wang X. Y., Eley G., James C. D. (2000) Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 60, 1383– 1387 [PubMed] [Google Scholar]