Abstract
The Caenorhabditis elegans mitochondrial (Mit) mutants have disrupted mitochondrial electron transport chain (ETC) functionality, yet, surprisingly, they are long lived. We have previously proposed that Mit mutants supplement their energy needs by exploiting alternate energy production pathways normally used by wild-type animals only when exposed to hypoxic conditions. We have also proposed that longevity in the Mit mutants arises as a property of their new metabolic state. If longevity does arise as a function of metabolic state, we would expect to find a common metabolic signature among these animals. To test these predictions, we established a novel approach monitoring the C. elegans exometabolism as a surrogate marker for internal metabolic events. Using HPLC-ultraviolet-based metabolomics and multivariate analyses, we show that long-lived clk-1(qm30) and isp-1(qm150) Mit mutants have a common metabolic profile that is distinct from that of aerobically cultured wild-type animals and, unexpectedly, wild-type animals cultured under severe oxygen deprivation. Moreover, we show that 2 short-lived mitochondrial ETC mutants, mev-1(kn1) and ucr-2.3(pk732), also share a common metabolic signature that is unique. We show that removal of soluble fumarate reductase unexpectedly increases health span in several genetically defined Mit mutants, identifying at least 1 alternate energy production pathway, malate dismutation, that is operative in these animals. Our study suggests long-lived, genetically specified Mit mutants employ a novel metabolism and that life span may well arise as a function of metabolic state.—Butler, J. A., Ventura, N., Johnson, T. E., Rea, S. L. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism.
Keywords: aging, metabolomics, isp-1, mev-1, hif-1
Within the species Caenorhabditis elegans there exists a collection of genetic and RNA interference (RNAi)-defined mutants that have disruptions in one of several mitochondrial electron transport chain (ETC) components, yet they are long lived (1–3). These animals are collectively known as mitochondrial (Mit) mutants (3–6).
Almost all Mit mutants (∼40) have been identified in large-scale RNAi screens for extended life span (2, 7). Genes such as nuo-2, cyc-1, cco-1, and atp-3, which encode the NDUSF3 subunit of complex I, cytochrome c1 of complex III, the COX5B subunit of complex IV, and the ATP5O/OSCP subunit of complex V, respectively, illustrate that, among the set of identified target genes, there is no obvious connection between life-span extension and disruption of any particular ETC complex.
Only a small number of Mit mutants have been defined genetically, including clk-1(qm30), isp-1(qm150), tpk-1(qm162), and lrs-2(mg312) (8). clk-1 encodes a demethoxyubiquinone monoxygenase necessary for the synthesis of ubiquinone (9). The qm30 allele produces no detectable protein product (10); however, clk-1 mutants supplement their ubiquinone deficiency, in part, via nutritional sources (11). isp-1 encodes the Rieske iron-sulfur protein of complex III, and the qm150 allele encodes a missense point mutation that reduces function of the protein product (12). tpk-1 encodes thiamine pyrophosphokinase (TPK), and the qm162 allele encodes a missense mutation (13). Deficiency of TPK disrupts cellular thiamine levels and hence the ketoacid dehydrogenases that operate the mitochondrial tricarboxylic acid cycle and feed electrons into the ETC. lrs-2 encodes mitochondrial leucine tRNA synthetase. The mg312 allele is maternal-effect viable, and the allele encodes a truncated, nonfunctional protein (2). So far, only 2 genetically defined mitochondrial ETC mutations that shorten life span in C. elegans have been described: mev-1(kn1), a missense allele in the gene encoding the cytochrome b subunit of complex II (14), and gas-1(fc21), which encodes the 49-kDa subunit of complex I (15).
A recent study (16) has suggested that the mechanisms responsible for life extension in RNAi-induced Mit mutants differ from those in their genetically defined counterparts. Specifically, RNAi treatment against isp-1 or the complex I gene nuo-6 both invoked a mitochondrial unfolded protein response, along with an autophagic response. Genetically defined versions of these Mit mutants, namely isp-1(qm150) and the newly defined nuo-6(qm200), showed neither response. It was speculated that genetic mutants inhibited mitochondrial ETC flux without causing ETC supercomplex disruption and that this triggered a second, undefined mechanism of life extension.
In its natural soil environment, C. elegans is normally exposed to bouts of hypoxia (17). In contrast to RNAi-defined Mit mutants, ATP levels in genetically defined Mit mutants appear to be maintained at or above normal levels (16). We have previously proposed that genetically defined Mit mutants utilize ATP production pathways normally reserved for survival during times of oxygen deprivation (4, 18). Moreover, use of such alternate metabolic pathways may, in and of themselves, be responsible for life extension in these animals; that is, the precise configuration of metabolic processes used to operate Mit mutant cells may be the ultimate determiner of Mit mutant life span.
In the following work, we establish novel methodology to monitor metabolism in live C. elegans. We test whether genetically defined Mit mutants utilize nonoxygen requiring ATP production pathways and whether their extended life span is associated with a metabolic configuration that is unique.
MATERIALS AND METHODS
Nematode strains and maintenance
The following C. elegans strains were used: N2 Bristol, CL2166 [dvIs19pAF15(gst-4::GFP::NLS)III], CB1370 [daf-2(e1370)III], CB4876 [clk-1(e2519)III], DA465 [eat-2(ad465)II], GR1371 [lrs-2(mg312)I], NL1832 [ucr-2.3(pk732)III], MQ130 [clk-1(qm30)III], MQ770[tpk-1(qm162)III], MQ887 [isp-1(qm150)IV], TJ564 [isp-1(qm150)IV;(gst-4::gfp)III], SLR0036 [age-1(hx546)Bx6II], SLR0037[clk-1(qm30)Bx5III], TJ5032 [clk-1(qm30)III;(gst-4::gfp)III], and TK22 [mev-1(kn1)III]. All strains were maintained on standard NGM agar supplemented with Escherichia coli (OP50), as described previously (19).
RNAi
Feeding RNAi constructs targeting small fumarate reductase (F48E8.3), nuo-2, and ucr-2.3 were obtained from the Ahringer RNAi library collection (20). For feeding RNAi, 2 methods were employed, our routine method, which we have described in detail previously (3), and one based on the method of Timmons et al. (21) and designed for greater RNAi efficacy. For this method, RNAi clones were cultured for 16 h at 37°C in Luria broth supplemented with 100 μg/ml ampicillin and 12.5 μg/ml tetracycline. A 250-μl aliquot was then seeded into 25 ml 2× YT broth (containing both antibiotics) and cultured for an additional 4 h at 37°C, after which isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 0.4 mM. The culture was then grown an additional 4 h at 37°C, after which the bacteria were pelleted, 21 ml of the supernantant was discarded, and fresh ampicillin, tetracycline, and IPTG were added to a final concentration of 100 μg/ml, 5 μg/ml, and 1 mM, respectively. Then, 200-μl bacterial aliquots were seeded onto NGM agar plates containing analagous antibiotic and IPTG concentrations, dried overnight, and stored at 4°C. For both methods, the Po generation was established by allowing 1-d-old N2 adults to lay eggs onto NGM/OP50 for 4 h, after which eggs were manually transferred to fresh RNAi lawns.
Anoxia resistance
Synchronous worm populations were established via a 4 h limited lay on NGM/OP50 plates. One-day-old gravid adults were then transferred to NGM/OP50 agar plates containing 25 μM 5-fluoro-2′-deoxyuridine (FuDR) and incubated overnight to block progeny production. Animals were then collected and washed in S-basal medium (100 mM NaCl and 50 mM KH2PO4, pH 6.0), resuspended in 200 μl S-basal containing 100 μg/ml ampicillin, 25 μM FuDR, and in relevant instances 1 mM fumarate, 1 mM succinate, 1 mM malate, or 1 mM glucose. Animals were subsequently placed inside a sealed chamber (22) and continuously perfused with wet 0.05% CO2/N2. At various times, 120 animals/condition were removed, and percentage survival was scored following an overnight recovery period at 20°C on NGM/OP50 plates containing 25 μM FuDR.
Azide resistance
One-day-old adult worms were placed into a 20-μl droplet of 500 mM NaN3 (5 animals at a time), and the time taken for each animal to become fully paralyzed was recorded. This concentration of NaN3 was chosen because it represented a balance between onset of paralysis and assay length.
Microscopy
Nomarski images were collected using a Zeiss Axioskop connected to a charge-coupled diode camera (Carl Zeiss, Oberkochen, Germany), as described previously (3).
Life-span analyses
Collection and analysis of life-span data have been described in detail by us previously (3). Animals that bagged, desiccated, or suffered gonad extrusion were scored as censored individuals. Such animals were included in life-span analyses up to the point of censorship and were weighted by half in mortality calculations.
Exometabolism analysis
We standardized our collection and analysis of the C. elegans exometabolism using the following procedure (biological replicates, or triplicates, were obtained for each strain).
Step 1: large-scale nematode cultivation
The 25,000 synchronous eggs were prepared without the use of bleach, as described previously (23), and then spotted onto NGM/OP50 plates supplemented with twice the standard peptone and OP50 concentrations (5000 eggs/plate; ref. 19). Eggs were cultivated at 20°C until reaching the first day of adulthood.
Step 2: exometabolism collection
Adult worms were collected and washed extensively in S-basal to remove worm debris and bacteria (1 g, 5×50 ml). Bacteria attached to the cuticle were removed using the sucrose flotation method of Foll et al. (24). After being washed extensively (400 g, 3×15 ml) to remove sucrose, worms were resuspended in S-basal (1.4 ml final total volume) and then transferred to a 3.5-cm glass petri dish (Kimax; Kimble Chase, Vineland, NJ, USA). Worms were placed inside a hydration chamber on an orbital shaker (100 rpm) and exposed for 18 h to either ambient conditions (22°C) or severe oxygen deprivation (continuous perfusion with 0.05% CO2/N2). At the end of the incubation period, the exometabolome-rich supernatant and worm fractions were separated by centrifugation (400 g), the supernatant was then filtered through a 0.2-μm nylon filter (6502-413X; Life Science Products, Frederick, CO, USA), and both fractions were stored at −80°C until further use.
Step 3: HPLC-ultraviolet (UV) data collection
On the day of analysis, 150 μl of each supernatant was thawed on ice and acidified with 0.5 μl of phosphoric acid (final pH≤2.5). Metabolites were then separated on a Waters 600 HPLC system (Waters Corp., Milford, MA, USA) using a Prevail organic acid column (5 μm particle size, 4.6×150 mm; Grace Discovery, Deerfield, IL, USA) with a Prevail organic acid guard column (5 μm particle size). Specific run conditions were 100-μl manual sample injection, mobile phase: 25 mM KH2PO4 acidified to pH 2.5 with phosphoric acid (Sigma, St. Louis, MO, USA), 1 ml/min flow rate (∼1000 psi), 30.0°C column temperature, 210-nm UV detection (Waters 486 detector monitored by Clarity v.2.4.4.105 software; DataApex, Prague, Czech Republic), and 20 min run length (with 40 min column reequilibration between consecutive runs).
Step 4: chromatogram normalization
UV chromatograms were normalized against total worm protein. Briefly, the pellets in step 2 were vacuum desiccated, resuspended in 1 ml boiling 1% SDS, boiled for 5 min, and then vortexed and reboiled for an additional 5 min. Total protein was quantified using the BCA colorimetric assay (23225; Pierce, Rockford, IL, USA). All chromatograms were scaled using Clarity software to the equivalent of 1 g total worm protein.
Step 5: chromatogram cropping
Normalized chromatograms were exported into the program SpecAlign (v2.3; Jason Wong, Oxford University, Oxford, UK) and then cropped to run times between 1.5 and 20 min.
Step 6: chromatogram alignment
HPLC experiments were performed over the course of 18 mo. Due to minor differences in chromatographic runs, chromatograms had to be aligned before peak decomposition and integration could be performed for multivariate analysis. Alignment was accomplished using SpecAlign. Briefly, a representative chromatogram from each strain was chosen against which all replicates were aligned. Representative templates were then sequentially aligned against each other. Alignment was intensely supervised. In general, the combined RAFFT/Peak algorithm of SpecAlign worked best for peaks eluting before 10.5 min and the RAFFT algorithm worked best for those eluting later.
Step 7: chromatogram baseline subtraction
Baselines were subtracted from chromatograms using the program Fityk (http://www.unipress.waw.pl/fityk/). Negative values were removed by writing a new function in the function editor of Fityk [Y=y−min(y)].
Step 8: peak modeling and integration
Most chromatogram peaks were modeled using the Pseudo-Voigt algorithm of Fityk, which incorporates elements of gaussian and lorentzian curves. Fityk's Split-Gaussian curve-fitting function was used for large, early-eluting peaks (<1.8 min). Closely eluting peaks were manually fitted.
Step 9: generation and alignment of a combined peak table
Peak tables were combined into a single spreadsheet, then peak retention times were manually fine aligned. Specifically, peaks assigned by SpecAlign that eluted within 3 s of each other were binned under a single retention time. A total of 900 peak entries was defined (summed across all samples; untransformed peak data are presented as Supplemental Data).
Step 10: spurious peak filtering
Peaks that were only observed in 1 sample (among all replicates) were considered spurious and removed from further analysis.
Step 11: missing data estimation
Not all peaks were present in every strain; 285 (out of 900) positions in the spreadsheet of peak areas contained “missing data.” Four types of peaks with missing data were encountered: 1) peaks that were unique to 1 strain; 2) peaks that were restricted to specific strains but which were absent in some replicates; 3) peaks that were spuriously present in multiple strains; and 4) peaks that were reliably present in every strain except one. Eighty percent of the missing peak data fell into categories 1 and 3. For the purposes of this study, we assumed that all missing data fell below the limits of detection. (We note there could have been instances where a large neighboring peak may have enveloped a peak of interest and hence masked our ability to resolve it in step 8. These are possibly the cause of category 2 missing peaks). Missing data were treated in one of two ways, depending on the analysis being performed. For cluster analysis, data was imputed by inserting a value equal to 0.1 the signal of the smallest peak area above background. For PCA, missing data were handled as described in step 13.
Step 12: cluster analysis
Cluster analysis was performed using hierarchical clustering in the GenePattern toolbox (25). Pearson correlation and pairwise complete-linkage settings were chosen for all analyses.
Step 13: data transformation
The relative abundance of metabolites differed markedly within each strain (peak amplitude variation). The degree of variation of each metabolite, across strains, also varied (peak variance was not equal). For principal component analysis (PCA), these differences were minimized by transforming peak area data. After testing several strategies, we found that a modified rank-ordering procedure, followed by mean centering of the transformed data, gave the most satisfactory results in terms of not unduly inflating variance. Briefly, for each metabolite, peak area data was rank ordered across strain. For those metabolites with missing data, empty cells were assigned an equal rank Xj according to Eq. 1:
| (1) |
where n is the number of missing data points for the jth peak. All remaining nonzero peak areas were assigned a rank starting from (nj+1). The transformed data were then mean centered, ready for PCA. We discovered that this method obviated the need for unit variance transformation, since rank transformation constrained the variance, and Eq. 1 had the effect of slightly down-weighting peaks restricted to 1 or, at most, 2 strains.
Step 14: PCA
PCA was performed using the function prcomp in the statistical program R (v2.6.0, http://www.r-project.org/). Numerical results are provided as Supplemental Data.
Step 15: electrospray ionization (ESI) mass spectrometry
Peaks that were flagged by PCA for further analysis were reisolated following steps 1–3 above using HPLC-grade water (pH 2.5, using Trace grade HCl). Fractions from 3 runs were pooled, vacuum desiccated, and then immediately before analysis resuspended in methanol. All peaks were analyzed using an Applied BioSystems API QSTAR Pulsar i Hybrid LC/MS/MS (negative ESI mode, 3500 V; Applied Biosystems, Foster City, CA, USA).
Step 16: enzymatic assay
Pyruvate was detected in exometabolism samples using lactate dehydrogenase (LDH) in conjunction with HPLC. Briefly, 1 μl of 150 U/ml LDH was mixed with 10 μl of 2.25 mM NADH and 140 μl of the supernatant was collected in step 2. Samples were incubated at room temperature for 3 min, then acidified with 0.75 μl phosphoric acid, and immediately analyzed by HPLC, as described above.
Additional statistical analyses
All other statistical analyses were undertaken using either R (v2.7), Sigma Plot (v11.0; Systat Software, Inc., San Jose, CA, USA), or Microsoft Excel (v12.2.0; Microsoft, Redmond, WA, USA). Specific details are described in figure legends.
RESULTS
Mit mutants are resistant to chronic oxygen deprivation
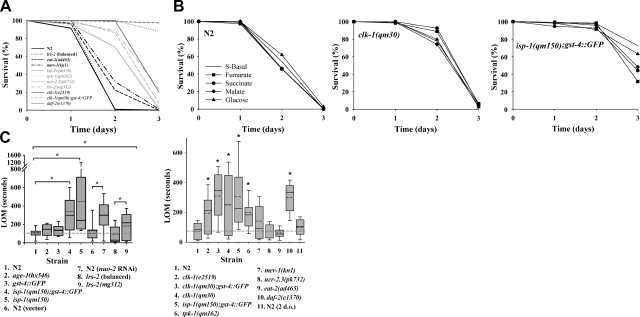
C. elegans is a facultative anaerobe, and wild-type animals can survive up to 3 d of severe hypoxia (24, 26). If Mit mutants constitutively supplement their mitochondrial ETC with alternate, non-oxygen-requiring energy-generating pathways, then we would expect them to be less sensitive to the effects of severe oxygen deprivation. Consistent with this notion, we observed that 5 genetically defined Mit mutants [clk-1(qm30), clk-1(e2519), tpk-1(qm162), lrs-2(mg312), and isp-1(qm150)] were markedly more resistant to the effects of severe oxygen deprivation than wild-type animals (Fig. 1A). After 48 h of continuous perfusion with 0.05% CO2/N2, when all wild-type animals were dead, >70% of each Mit mutant remained alive. In the case of clk-1(qm30) mutants, all animals remained alive; moreover, by 72 h (the end of the assay) only 5% of clk-1(qm30) mutants had died. Survival of clk-1(qm30) mutants was on par with long-lived daf-2(e1370) mutants, which are known to be resistant to both hypoxia (<1% oxygen) and anoxia (<0.003% oxygen; refs. 26, 27). Short-lived ucr-2.3(pk732) mutants also showed substantial oxygen-deprivation resistance (this new mutant will be described in detail below), but, surprisingly, mev-1(kn1) mutants, which are also short lived, showed only marginal resistance to severe oxygen deprivation. Reduction in physiological rates was not a prerequisite for resistance to oxygen deprivation: long-lived, dietary-restricted eat-2(ad465) mutants showed only marginal resistance to oxygen removal.
Figure 1.
Mit mutants are resistant to both severe oxygen deprivation and hypoxia. A) Wild-type (N2) animals; clk-1(e2519), clk-1(qm30), tpk-1(qm162), lrs-2(mg312), and isp-1(qm150) Mit mutants; mev-1(kn1) and ucr-2.3(pk732) Byby mutants; as well as 3 additional control strains, eat-2(ad465), daf-2(e1370), and balanced animals carrying the lrs-2(mg312) mutation, were collected in S-basal as adult worms, then continuously perfused with 0.05% CO2/N2 inside a sealed chamber for the indicated period of time (abscissa). Percentage survival was scored (120 animals/condition). Strain labels are ordered top to bottom as curves appear left to right. In A–C, Mit mutant strains containing the gst-4::GFP transgene were randomly included as controls for background effects. B) In experiments analagous to those in A, N2, clk-1(qm30), and isp-1(qm150) populations were supplemented with 1mM fumarate, succinate, malate, or glucose, and survival was scored. C) All strains described in A, as well as RNAi-induced nuo-2 Mit mutants, vector-control treated animals, gst-4::gfp transgenics, and age-1(hx546) Daf-c mutants, were tested for resistance to 500 mM sodium azide. One-day-old adults were placed in azide, and the time of last observed movement (LOM) was recorded (n=30 worms/condition). Boxes in both left and right panels delineate 25th–75th percentile. Median and mean are marked by solid and dotted lines, respectively. Whiskers on box indicate 10th and 90th percentiles. Left panel: *strain comparisons with significantly different mean LOM values (P<0.002, ANOVA on rank-transformed data, Fisher least significant difference post hoc test). Strains 6 and 8 are controls for 7 and 9, respectively. Strains 8 and 9 were 2-d-old (d.o.) adults. Right panel: *strain comparisons relative to N2 with significantly different median LOM values (P<0.05, Kruskal-Wallis 1-way ANOVA on rank-transformed data).
Fermentation substrates are not limiting for survival under chronic oxygen deprivation
We tested whether the ability of clk-1(qm30) and isp-1(qm150) Mit mutants to survive chronic oxygen deprivation was limited by the availability of fermentation substrates. We supplemented wild-type, clk-1(qm30), and isp-1(qm150) animals with various substrates during acute oxygen deprivation, including glucose, fumarate, malate, or succinate. We observed no enhancement of survival of any strain (Fig. 1B). On the contrary, isp-1(qm150) mutants appeared slightly less tolerant of severe hypoxia when cultured on any of these 4 carbon sources than when cultured in their complete absence. Nonetheless, under all 5 incubation conditions, we consistently observed enhanced survival of clk-1 and isp-1 mutants relative to wild-type animals.
Mit mutants are resistant to mitochondrial complex IV inhibition
Azide anion, N3−, is a potent inhibitor of cytochrome c oxidase, with a reported Kd for the bovine enzyme of 64 μM (28). In worms, NaN3 is widely used to induce paralysis and to simulate hypoxia (27, 29); however, millimolar concentrations are required to transit the largely impermeable worm cuticle. We selected several different Mit mutants and measured the duration it took individual 1-d-old adult animals to stop moving following exposure to 500 mM NaN3. We observed that all Mit mutants tested, including isp-1(qm150), lrs-2(mg312), clk-1(e2519), clk-1(qm30), tpk-1(qm162), and RNAi-induced nuo-2 Mit mutants, were differentially resistant to the effects of azide administration relative to control animals (Fig. 1C). These differences were significant, despite the increased variance in most Mit mutant populations. We also observed that the resistance phenotype of both isp-1 and clk-1 animals was robust to changes in genetic background (Fig. 1C, compare strains 4 and 5, left panel, and strains 3 and 4, right panel). Short-lived mev-1(kn1) mutants showed mild resitance to the effects of azide (though this did not reach significance), while ucr-2.3(pk732) mutants showed no enhancement over wild-type, and eat-2(ad465) were noticeably hypersentive to azide exposure. Consistent with previous hypoxia-resistance studies (27), daf-2(1370) mutants, but not age-1(hx546) mutants, showed marked resistance to azide treatment.
In summary, using 2 different assays, and multiple genetic lesions, we show that Mit mutants consistently show resistance to procedures that normally limit ATP production by the mitochondrial ETC. We interpret these data to suggest that Mit mutants utilize nonoxygen requiring ATP production pathways to supplement their energy needs.
Removal of soluble fumarate reductase enhances Mit mutant survival
Many fermentation reactions function in the absence of oxygen to generate ATP and maintain redox balance. Malate dismutation is one such process and soluble fumarate reductase (sFR) functions in this pathway to catalyze the NADH/FADH2-dependent reduction of fumarate to succinate. In the yeast Saccharomyces cerevisiae, sFR activity is essential for growth and survival during anaerobia (30).
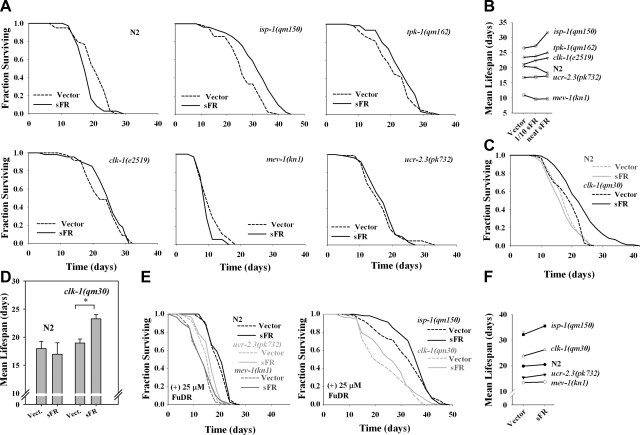
F48E8.3 encodes the sole soluble fumarate reductase in worms (18). We tested the role of this enzyme in the life-span specification of long- and short-lived mitochondrial mutants (Fig. 2). Unexpectedly, we found that removal of sFR significantly (log-rank test, P<0.005) increased the survival of isp-1(qm150) mutants relative to vector control-treated animals (Fig. 2A). tpk-1(qm162) and clk-1(e2519) mutants both showed marginally significant (log-rank test, P<0.2) life-span extension also (Fig. 2A). Wild-type animals showed a significant (log-rank test, P<0.02) life-span decrease (Fig. 2A); while mev-1(kn1) and ucr-2.3(pk732) showed either marginal life shortening or no response at all. When we inhibited sFR in clk-1(qm30) animals, using a feeding RNAi method that was more potent than in the previous assays (21), we also observed life-span lengthening (Fig. 2C). This effect was not observed in wild-type animals (Fig. 2C). To highlight the differential effects of sFR removal on Mit mutant survival, we have plotted the mean life span data from Fig. 2A, C in Fig. 2B, D, respectively. Data for an intermediate dose of sFR RNAi (1/10 dilution), which were collected at the same time as the other survival data, are also included in Fig. 2B.
Figure 2.
RNAi-mediated inhibition of soluble fumarate reductase differentially increases Mit mutant life span. Wild-type (N2) animals; isp-1(qm150), tpk-1(qm162), clk-1(e2519), and clk-1(qm30) Mit mutants; and mev-1(kn1) and ucr-2.3(pk732) Byby mutants were cultured on bacterial-feeding RNAi targeting soluble FR (undiluted or 0.1 strength) or on vector control bacteria (Vector) from the time of hatching, and life span was assessed. A) Survival curves (n=60 worms/condition, 0.1 strength sFR is not shown). isp-1 and N2 animals cultured on sFR RNAi were significantly longer and shorter lived, respectively, than their vector control (P<0.05, log-rank test). tpk-1 and clk-1(e2519) approached significance (P<0.15, log-rank test). B) Mean life-span values for each of the assays performed in A. C) clk-1(qm30) survival curves; as in A, except curves are averages of replicate experiments (n=120 worms/per condition). clk-1(qm30) cultured on sFR RNAi is significantly longer-lived than vector control (P<0.001, log rank test). D) Mean ± se life-span values for assays performed in C. *P < 0.05 vs. vector. E, F) Repeat of experiments in A and B, except life-span studies were performed in duplicate and using 25 μM FuDR (n=120 worms/condition). Mean life-span increase of isp-1 and clk-1(qm30) cultured on sFR RNAi relative to vector control trended toward significance (P<0.1). Error bars = se.
During our studies with sFR, we noted that the fertility of all tested strains appeared to be enhanced (this data was not quantified). We have previously observed, using fluorescently tagged reporter strains, that both isp-1(qm150) and clk-1(qm30) Mit mutants have what we term a “cryptic bagging” phenotype (Cry), where a single egg hatches inside the mother, and eventually the progeny emerges at a later date through the vulva. The extent to which the mother is damaged during this process remains unclear. We therefore tested whether the life-span increase we observed following sFR RNAi in isp-1(qm150) and clk-1(qm30) Mit mutants was simply a consequence of reduced cryptic bagging. Progeny production was inhibited using 25 μM 5-fluoro-2′-deoxyuridine (FuDR) and then life span was rescored for each strain. Both isp-1(qm150) and clk-1(qm30) Mit mutants differentially responded with an increase in mean, but not maximum life span (i.e., increased health span), while N2, ucr-2.3(pk732), and mev-1(kn1) mutants showed no or only marginal effects on maximum or mean life span (Fig. 2E, F). We conclude that Mit mutants utilize sFR in a manner that normally acts to limit their surival potential.
ucr-2.3 is a newly defined short-lived mitochondrial mutant
Adopting a candidate gene approach to identify ATP-producing metabolic pathways operative in Mit mutants is limited in its scope and capacity. Therefore, as detailed in the following sections, we established a novel, HPLC-based approach to test further our hypothesis that Mit mutants utilize nonoxygen requiring ATP production pathways. Simultaneously, this approach also permitted us to test aspects of our second hypothesis, that long-lived Mit mutants adopt a common metabolic state.
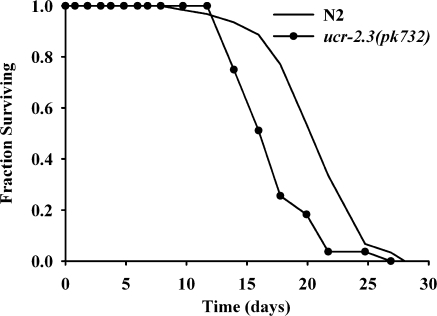
We focused our attention on long-lived clk-1(qm30) and isp-1(qm150) Mit mutants, which ultimately affect complexes II and III, respectively (31, 32), and short-lived mev-1(kn1) and ucr-2.3(pk732) mutants, which also affect complexes II and III, respectively (33). ucr-2.3 is 1 of 3 genes in C. elegans (along with ucr-2.1 and ucr-2.2) that show partial functional redundancy with each other and that encode a protein orthologous to human UQCRC2, a core subunit of mitochondrial complex III (34). The ucr-2.3(pk732) allele encodes a G51R missense mutation that was initially identified in an EMS screen for reversion of germline silencing of the Tc1 transposon (35, 36). We now report that ucr-2.3(pk732) is phenotypically indistinguishable from mev-1(kn1) in terms of its reduced life span (Figs. 2E and Fig. 3), its wild-type-like rate of growth and development, its propensity for vulval extrusion later in life, and its darker pigmentation (not shown). Similar to what we (3) have previously observed when treating N2 animals with a mev-1 feeding RNAi construct, a feeding RNAi construct targeting ucr-2.3 was also incapable of extending the N2 life span, even over 5 consecutive generations of continuous feeding (not shown). We therefore define ucr-2.3(pk732) as a novel short-lived mitochondrial ETC mutant and distinguish mev-1(kn1) and ucr-2.3(pk732) mutants from Mit mutants by describing their phenotype as “Byby” (pronounced bye-bye, in reference to their short-lived phenotype).
Figure 3.
ucr-2.3(pk732) is a new Byby mutant. Life-span analysis of NL1832 [ucr-2.3(pk732)] and wild-type (N2) worms cultured on standard NGM/OP50 plates (n=84 and 63 worms, respectively; log-rank test, P=2.2×10−4). Data represent 2 experiments, each performed by a different scorer.
Metabolic fingerprinting distinguishes Mit mutants from Byby mutants
At present, there is no clear understanding as to why genetically encoded Mit mutants are long lived. Several researchers (37, 38) have shown that there is a transriptional response activated following ETC disruption, but how this translates into long life is far from clear. A fundamental hypothesis is that longevity arises as a function of metabolic state. That is, the rerouting of metabolites through core metabolic pathways may place cells in a state that is simply conducive to a long life. If true, we would predict that the intermediary metabolism of Mit mutants should be very similar. The corollary, that “shortivity” is a function of a separate metabolic state, may also be true. To test these ideas, we established a novel approach using HPLC-UV chromatography in conjunction with multivariate analyses to monitor the exometabolism of isp-1(qm150), clk-1(qm30), mev-1(kn1), and ucr-2.3(pk732) mutants. The C. elegans exometabolism provides a proxy of internal metabolism and consists of end product metabolites. It is distinct from defecated intestinal waste products that, in our experiments, were emptied from the gut during the washing steps before analysis (refer to Materials and Methods and see also Discussion). As controls, we also collected the exometabolome of wild-type animals (N2) cultured under normoxic and, seperately, severely hypoxic (anoxic) conditions.
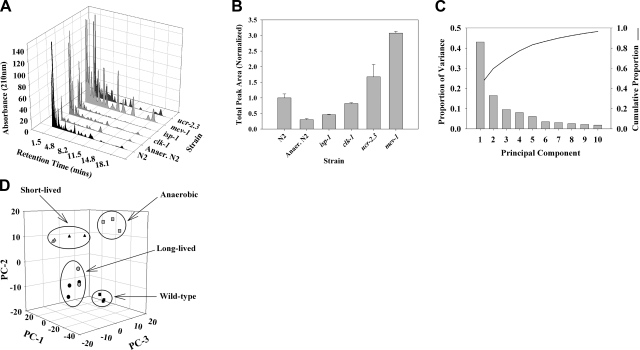
Exometabolome samples were fractionated using an organic acid HPLC column. Clear differences in chromatographic profiles across strain and condition were evident (representative exometabolome profiles are shown in Fig. 4A). Notably, differences in peak retention time or peak intensity, or both, differentiated strains. Surprisingly, when we integrated metabolite levels within each strain and used this as a proxy of metabolic activity (this approach does not take into account differences in UV absorbance between different metabolites), we observed a marked increased in the amount of metabolites excreted by Byby mutants relative to normoxic wild-type animals (Fig. 4B). In contrast, we observed a marked decrease in the amount of metabolites excreted by wild-type animals cultured under severe oxygen deprivation. The amount of metabolites excreted by clk-1(qm30) Mit mutants was relatively unchanged compared with normoxic, wild-type animals, but production was reduced by half in isp-1(qm150) animals (Fig. 4B).
Figure 4.
HPLC-UV and PCA analysis of excreted metabolites differentiates long-lived Mit mutants from short-lived Byby mutants. A) Excreted metabolites were collected and analyzed from N2 wild-type animals, long-lived isp-1(qm150) and clk-1(qm30) Mit mutants, and short-lived mev-1(kn1) and ucr-2.3(pk732) Byby mutants, using HPLC-UV, as described in Materials and Methods. Metabolites were also collected from N2 animals cultured under severe hypoxia. Representative HPLC-UV traces for each strain/condition are shown. B) A crude measure of metabolic activity in each strain was obtained by integrating the area under each trace and averaging across replicates (means±se). Anerobic N2, isp-1(qm150) and mev-1(kn1) were significantly different from N2. P < 0.05; Student's t test. C) Chromatogram peak areas in each trace were modeled using the pseudovoigt peak-fitting algorithm of the program Fityk. PCA analysis was then performed using R on the set of all peaks, over all strains. Shown is a scree plot of the resulting PCs; 70% of the variance in the excreted metabolite data is captured by the first 3 PCs. D) Plot of PC-1, PC-2, and PC-3 scores for each strain shows that long- and short-lived Mit mutants can be differentiated on the basis of their metabolic end products (solid squares, N2; shaded squares, anaerobic N2; solid circles, clk-1(qm30); shaded circles, isp-1(qm150); solid triangles, mev-1(kn1); shaded triangles, ucr-2.3(pk732).
Marked similarities in the metabolic profiles of each pair of long- and short-lived mitochondrial ETC mutants were also evident. Support for this observation became obvious using PCA. In our analysis, 70% of the variance across all samples was described by the first 3 principal components (PCs, Fig. 4C). Plotting these new variables confirmed that long-lived Mit mutants produced metabolic end products distinguishable from those produced by short-lived mitochondrial mutants and distinguishable again from those generated by wild-type animals cultured under either normoxia or severe oxygen deprivation (Fig. 4D).
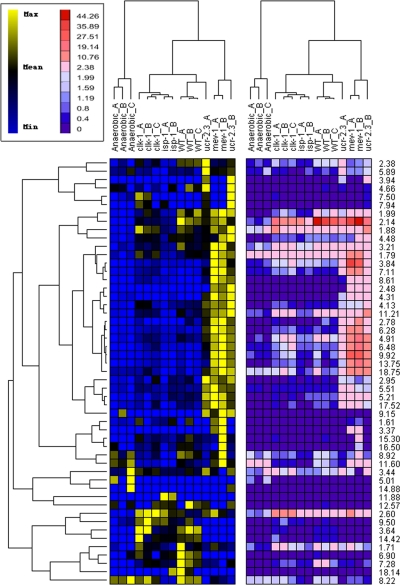
To independently confirm that the clustering in Fig. 4D did not arise simply because of differences in intensity of profiles (rather than a differential profile of peaks), we used a hierarchical-clustering approach that was insensitive to changes in peak profile intensity (Materials and Methods). We observed an identical clustering pattern of strains (Fig. 5).
Figure 5.
Hierarchical clustering confirms PCA results. A hierarchical clustering approach that was insensitive to differences in peak profile magnitude (see Materials and Methods) yielded an identical strain clustering pattern as that derived using PCA. Data are presented as 2 heat maps showing results scaled on the basis of relative metabolite abundance across samples (left panel, blue–yellow) or on the basis of globally relative metabolite abundance (right panel, blue–red). Inset illustrates scales. Branches at top and left of figure illustrate clustering hierarchies derived using data in right panel.
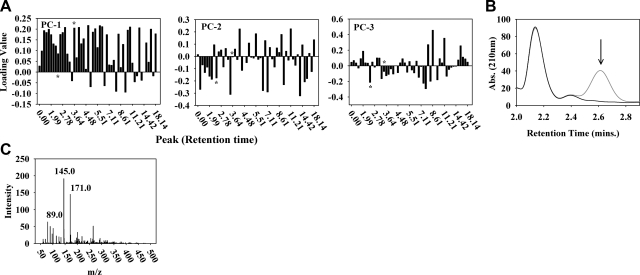
Pyruvate production is increased in Mit mutants
We sought to determine the identity of the metabolites driving the separation of Mit from Byby mutants. Figure 6A shows the relative contribution of each chromatographic peak to PC-1, PC-2, and PC-3. We selected some of the largest-contributing peaks and subjected them to analysis by ESI mass spectrometry. Most peaks contained multiple metabolites and hence require additional analysis to be further informative. Two peaks stood out in our analysis. The first was the peak of pyruvate alone, which we also confirmed enzymatically and which was significantly increased in the 2 long-lived Mit mutants relative to wild-type animals (t test, P<0.01; Fig. 6B; Supplemental Table 1). Another signficant peak was enriched in short-lived Byby mutants; it contained a complex mixture of metabolites, including lactate, α-ketoglutarate, and the dipeptide Gly-Pro (Fig. 6C).
Figure 6.
Identity of metabolites driving Mit and Byby mutant separation in the PCA. A) HPLC-UV peak loadings for PC-1, PC-2, and PC-3. Peaks are identified on the basis of their retention time (minutes, abscissa). Loading value magnitude reflects the degree a peak contributes to the listed PC (absolute value of 1=completely; 0=no contribution). Positive and negative loading values indicate opposing contributions to PC score. HPLC-UV peaks showing large absolute loading values were collected from clk-1(qm30) or mev-1(kn1) mutants for metabolite identification. Asterisks indicate 2.60- and 3.84-min peaks.B) Long-lived mutants differentially excrete pyruvate (2.60 min, arrow). Shown is a HPLC-UV trace before (gray) and after (black) incubation of clk-1 exometabolome with NADH and lactate dehydrogenase. C) Peak at 3.84 min is enriched in Byby mutants. ESI-MS fragmentation patterns match lactate, a-ketoglutarate, and the dipeptide Gly-Pro (m/z 89.0, 145.0 n, and 171.0, respectively).
DISCUSSION
In the present study, we sought to gain insight into the metabolic processes mediating life extension in the Mit mutants. We hypothesized that Mit mutants supplement their mitochondrial ETC deficiencies by exploiting alternate ATP production pathways that do not require oxygen as a terminal electron sink, and that longevity is a property that arises out of the way metabolic networks are configured. Our findings revealed that several Mit mutants, including clk-1(qm30), clk-1(e2519), tpk-1(qm162), lrs-2(mg312), and isp-1(qm150), display marked resistance to severe oxygen deprivation as well as to hypoxia. Our findings also revealed that soluble fumarate reductase played an unexpected role in significantly limiting the life-span potential of clk-1(qm30) and isp-1(qm150) Mit mutants. These findings support the idea that alternate ATP production pathways are operative in Mit mutants and that alterations in fumarate metabolism can have significant effects on life span, depending on strain background. Using the C. elegans exometabolome as a proxy for internal metabolic events, we also showed that clk-1(qm30) and isp-1(qm150) Mit mutants are metabolically more similar to each other than to either aerobically or anaerobically cultured wild-type animals. The metabolism in these mutants also appears markedly different from that operative in short-lived mev-1(kn1) and ucr-2.3(pk732) Byby mutants. Our findings support the notion that life span, per se, may indeed be a fundamental property of metabolic configuration.
In our studies on low oxygen tolerance, we tested 2 conditions that were relatively easy for us to achieve: severe oxygen deprivation and psuedohypoxia. Oxygen concentrations in our oxygen-deprivation chamber were below the limit of detection of a Clarke-type oxygen electrode. It is likely, therefore, that worms incubated under these conditions were exposed to near anoxia. Mendenhall et al. (26) showed that daf-2(e1370) mutants can survive exposure to anoxia for ≥5 d. Resistance was dependent on daf-16 as well as the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase-2 and -3 (gdp-2 and gpd-3). Surprisingly, RNAi-mediated removal of other glycolytic enzymes did not affect the anoxia resistance of daf-2(e1370) mutants. Recent microarray studies have shown that gpd-2 and gpd-3 are both elevated in clk-1(qm30), isp-1(qm150), and cco-1 (RNAi-induced) Mit mutants (37). It is likely that gpd-2/-3 played an integral role in the resistance of Mit mutants to anoxia in the current study.
Sodium azide was previously employed to simulate hypoxic conditions in worms (27). In our assay for sensitivity to hypoxia, we treated a variety of Mit mutants with sodium azide and observed that every one showed significant resistance to paralysis. Given the uniform resistance of Mit mutants to hypoxia, it is possible that HIF-1 is active in these animals. Hypoxia-inducible factor-1 (HIF-1) is a conserved transcription factor that is one of the primary effectors of hypoxia sensation in C. elegans (39). Of the 110 genes known to be upregulated following hypoxia exposure, 63 require hif-1 activity (40). At least 3 mechanisms are known to modulate hif-1 and resistance to hypoxia in worms: inhibition of the daf-2/akt-1/pdk-1/daf-16 pathway (27); germline sterility, which acts independently of hif-1 and daf-16 (41); and translational retardation, which appears to regulate the level of activation of the unfolded protein response (42). Whether inhibition of the mitochondrial ETC represents a fourth, novel pathway remains unclear. Nonetheless, our findings raise the exciting possibility that reducing flux through the mitochondrial ETC, at least in the manner employed by Mit mutants, may also prove beneficial in humans during, or while recovering from, hypoxic stress such as stroke.
To what extent hif-1 plays a role in Mit mutant life extension remains to be determined, but it is interesting that overexpression of hif-1 extends the life span of wild-type animals (43–45). Augmentation of HIF-1 transcriptional activity may in fact be the reason inhibition of soluble fumarate reductase increased the life span of clk-1(qm30) and isp-1(qm150) Mit mutants in the present study. Specifically, EGL-9 is a prolyl 4-hydroxylase that hydroxylates 2 proline residues in HIF-1 in an oxygen- and α-ketoglutarate-dependent manner. Under normoxic conditions, formation of these modified residues directs VHL-1, an E3 ubiquitin ligase, to degrade HIF-1. Fumarate is a competitive inhibitor of α-ketoglutarate in the EGL-9 reaction (46). Although a role for hif-1 in Mit mutant life extension is an intriguing notion, at this point we cannot rule out the alternate possibilty that sFR acts as a redox sink in Mit mutants and that when it is inhibited, NADH/FADH2 reducing equivalents are funneled down other metabolic pathways resulting in unknown, beneficial effects.
Presently, we remain uncertain why Byby and Mit mutants respond so differently to anoxia, hypoxia, and loss of sFR. Unlike Mit mutants, we observed that mev-1 mutants were mostly intolerant of anoxia, showed marginal resistance to hypoxia, and were unaffected by the loss of sFR. ucr-2.3 mutants, on the other hand, were quite resistant to anoxia but showed zero tolerance of hypoxia and little or response to loss of sFR. These functional differences between Mit and Byby mutants underscore their obvious differences in life span and are in agreement with our HPLC results that showed there were overt differences in the metabolites excreted by Mit and Byby mutants, both in terms of the type of metabolites they made and their overall level of metabolic activty (absolute signal abundance).
Very little is known about the life-extending mechanisms operative in Mit mutants. Recent evidence suggests p53/cep-1 is partially responsible for the process (47). Oxidative stress, bioenergetic stress, or ER or mitochondrial unfolded protein stress may all play a role in Mit mutant longevity (4, 6), although several studies now challenge the role of oxidative stress (2, 3, 48). Genetic epistasis studies have led to the tentative conclusion that Mit mutant longevity occurs independently of the daf-2/daf-16 pathway (1, 2, 47, 49) and that it is synonymous with dietary restriction or at least it uses some of the same mechanistic processes (50, 51). In the current study, we bypassed questions regarding what processes might be activated in Mit mutants to signal longevity specification and instead we focused our efforts on identifying the end result of such changes. We asked the simple question, are discrete metabolic configurations associated with different life span outcomes; do the Mit mutants adopt a metabolic configuration that is unique? We made no assumptions about whether such changes are programmed or whether they occur simply as a consequence of metabolite rerouting, although we hypothesized that the latter may well be the case. Our data support the conclusion that long-lived Mit mutants do indeed adopt a novel metabolic configuration that is different from that of short-lived Byby mutants and different again from that of wild-type animals cultured under normoxia. Surprisingly, there was very little overlap between the metabolic profile of Mit mutants and wild-type animals exposed to severe oxygen deprivation. It will be interesting to determine if the exometabolome profiles of Mit mutants and wild-type animals converge when the latter is exposed to less severe oxygen deprivation.
We chose to analyze the C. elegans exometabolome for the current study because it is less complex than the entire worm metabolome. A recent systems biology investigation using S. cerevisiae showed that the exometabolome could be used to reliably predict internal metabolism (52). We collected exometabolites following an 18 h incubation in minimal media. Under excess food conditions, C. elegans can passage material through its gut in <2 min (53). It is almost certain that our worm populations entered a state of starvation sometime during their 18 h incubation period. It is possible that the exometabolome profiles we identified that separate Mit from Byby mutants simply reflect differences in their rate of progression into starvation. Against this idea, however, our pilot studies illustrated that between 30 min (the earliest time point we could access) and 18 h there was little change in the profiles of metabolite peaks; only the magnitude of metabolite peaks increased. Based on these findings, we favor the notion that the metabolic profiles we have observed reflect bonafide alterations in pathways that fuel life in long- and short-lived mitochondrial mutants. The differential accrual of pyruvate by Mit mutants, but of lactate, α-ketoglutarate, and Gly-Pro by Byby mutants, also supports this idea.
Three recent studies support our current observations showing that long-lived Mit mutants adopt a common metabolic configuration. Cristina et al. (37) recently explored the transcriptional alerations that occur in clk-1(qm30), isp-1(qm150), and RNAi-induced cyc-1 Mit mutants. Relative to wild-type animals, clk-1(qm30) and isp-1(qm150) mutants both showed expression changes in ∼5% of the genome. In cyc-1 mutants, some 30% of the genome displayed significant expression alterations. Remarkably, at an individual gene level, there was little overlap between the expression profiles of these 3 mutants; only 73 genes showed coalteration. Using GO term analysis, however, the researchers found that all 3 mutants likely exist in a closely related metabolic state akin to that invoked by activation of the retrograde response in petite Saccharomyces cerevisiae (54). This metabolic state was characterized at the mRNA level by elevated expression of genes involved in glycolysis, anapleuerosis, mitochondrial ETC compensation, amino acid metabolism, xenobiotic detoxification, and peroxisomal fat metabolism. Our HPLC data now provide strong support for the idea that clk-1(qm30) and isp-1(qm150) do in fact reach the same metabolic end state.
In a second study, Falk et al. (38) analyzed the transcriptional alterations occurring in several mitochondrial mutants, including the short-lived gas-1(fc21) mutant. Surprisingly, these researchers found that many of the biochemical processes transcriptionally altered in long-lived Mit mutants were also altered in gas-1(fc21) mutants. This is unexpected in light of our present findings, where we have shown that Mit and Byby mutants differ markedly at the level of their exometabolome. We can think of 3 possible interpretations to explain the gas-1(fc21) observation: the transcriptome does not accurately reflect the intracellular metabolome; the exometabolome does not accurately reflect the intracellular metabolome; and gas-1(fc21) is a Mit mutant, not a Byby mutant. With respect to the latter notion, Hartman et al. (55) reported that the maximum life span of gas-1(fc21) mutants could in fact be extended beyond that of wild-type animals simply by growing the animals at 15°C. It is possible that gas-1(fc21) represents a class of genetically defined Mit mutants that are unable to fully stablize their ETC supercomplexes, even though they have the potential to be long-lived Mit mutants. Low temperatures could enhance their ETC supercomplex formation to reveal their latent Mit phenotype. A corollary of this prediction is that other genetically defined Mit mutants should be identifiable when screened at lower temperatures. Moreover, mild hypothermia may be a novel form of treatment for some human patients with mitochondrial ETC disorders.
The third study that supports our contention that long-lived Mit mutants adopt a common metabolic configuration relates to the work of Fuche et al. (56), who used 1H nuclear magnetic resonance spectroscopy, in conjunction with microarray analyses, to map the intracellular metabolome of long-lived dauers, several daf-2 mutants, and a translation-defective ife-2 mutant. Surprisingly, all these animals presented with a metabolic profile that was almost identical. Marked changes in lipid, carbohydrate, and amino acid catabolic pathways were observed. It remains to be seen whether this longevity-associated metabolic state is the same as the one we have now detected in Mit mutants.
The concept of long-lived organisms adopting an altered metabolism has been hinted at previously by microarray studies of several organisms (53–55), as well as by proteomic (57), amino acid (49), and now metabolomic-based studies (56). The novelty of our present study is that we have used an unbiased metabolomic approach to identify the first metabolic signatures able to distinguish long-lived mitochondrial ETC mutants from short-lived mitochondrial ETC mutants. We have also been able to distinguish both classes of mutants from wild-type animals. Future studies will be aimed at fully characterizing this profile at the single metabolite level.
Supplementary Material
Acknowledgments
The authors thank Elizabeth Glasser, Justin Springett, Yvonne Penrod, and William Swan for technical assistance. The authors also thank Milena Girotti, Shylesh Bhaskaran, and Chris Link for critically reading the manuscript, as well as the anonymous reviewer who provided insightful feedback.
Financial support was provided by the National Institute on Aging (to T.E.J. and S.L.R.), the Polis Foundation (to T.E.J. and S.L.R.), the Ellison Medical Foundation (to T.E.J. and S.L.R.), and the National Ataxia Foundation (to N.V.). The nematode strain GR1371 was provided by the Caenorhabditis Genetics Centre (University of Minnesota, Minneapolis, MN, USA). NL1832 was a gift from Ron Plasterk (Utrecht University, Utrecht, Holland).
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Dillin A., Hsu A.-L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A. G., Kamath R. S., Ahringer J., Kenyon C. (2002) Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 [DOI] [PubMed] [Google Scholar]
- 2.Lee S. S., Lee R. Y., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. (2003) A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33, 40–48 [DOI] [PubMed] [Google Scholar]
- 3.Rea S. L., Ventura N., Johnson T. E. (2007) Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 5, e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rea S. L. (2005) Metabolism in the Caenorhabditis elegans Mit mutants. Exp. Gerontology 40, 841–849 [DOI] [PubMed] [Google Scholar]
- 5.Ventura N., Rea S. L. (2007) Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnology J. 2, 584–595 [DOI] [PubMed] [Google Scholar]
- 6.Ventura N., Rea S. L., Testi R. (2006) Long-lived C. elegans Mitochondrial mutants as a model for human mitochondrial-associated diseases. Exp. Gerontology 41, 974–991 [DOI] [PubMed] [Google Scholar]
- 7.Hamilton B., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S. S. (2005) A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19, 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rea S. L., Graham B. H., Nakamaru E., Kar A., Falk M. J. (2010) Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev. Disabil. Res. Rev. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rea S. (2001) CLK-1/Coq7p is a DMQ mono-oxygenase and a new member of the di-iron carboxylate protein family. FEBS Lett. 509, 389–394 [DOI] [PubMed] [Google Scholar]
- 10.Burgess J., Hihi A. K., Benard C. Y., Branicky R., Hekimi S. (2003) Molecular mechanism of maternal rescue in the clk-1 mutants of Caenorhabditis elegans. J. Biol. Chem. 278, 49555–49562 [DOI] [PubMed] [Google Scholar]
- 11.Jonassen T., Larsen P. L., Clarke C. F. (2001) A dietary source of coenzyme Q is essential for growth of long-lived Caenorhabditis elegans clk-1 mutants. Proc. Natl. Acad. Sci. U. S. A. 98, 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J., Bussiere F., Hekimi S. (2001) Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644 [DOI] [PubMed] [Google Scholar]
- 13.De Jong L., Meng Y., Dent J., Hekimi S. (2004) Thiamine pyrophosphate biosynthesis and transport in the nematode Caenorhabditis elegans. Genetics 168, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senoo-Matsuda N., Hartman P. S., Akatsuka A., Yoshimura S., Ishii N. (2003) A complex II defect affects mitochondrial structure, leading to ced-3- and ced-4-dependent apoptosis and aging. J. Biol. Chem. 278, 22031–22036 [DOI] [PubMed] [Google Scholar]
- 15.Hartman P. S., Ishii N., Kayser E. B., Morgan P. G., Sedensky M. M. (2001) Mitochondrial mutations differentially affect aging, mutability and anesthetic sensitivity in Caenorhabditis elegans. Mech. Ageing Dev. 122, 1187–1201 [DOI] [PubMed] [Google Scholar]
- 16.Yang W., Hekimi S. (2010) Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell 9, 433–447 [DOI] [PubMed] [Google Scholar]
- 17.Van Voorhies W. A., Ward S. (2000) Broad oxygen tolerance in the nematode Caenorhabditis elegans. J. Exp. Biol. 203, 2467–2478 [DOI] [PubMed] [Google Scholar]
- 18.Rea S., Johnson T. E. (2003) A metabolic model for life span determination in Caenorhabditis elegans. Dev. Cell 5, 197–203 [DOI] [PubMed] [Google Scholar]
- 19.Wood W. B. ed (1988) The Nematode Caenorhabditis Elegans, Cold Spring Harbor Laboratory, New York, NY, USA [Google Scholar]
- 20.Kamath R. S., Ahringer J. (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 [DOI] [PubMed] [Google Scholar]
- 21.Timmons L., Court D. L., Fire A. (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 [DOI] [PubMed] [Google Scholar]
- 22.Link C. D., Johnson C. J. (2002) Reporter transgenes for study of oxidant stress in Caenorhabditis elegans. Methods Enzymol. 353, 497–505 [DOI] [PubMed] [Google Scholar]
- 23.Rea S. L., Wu D., Cypser J. R., Vaupel J. W., Johnson T. E. (2005) A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 37, 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foll R. L., Pleyers A., Lewandovski G. J., Wermter C., Hegemann V., Paul R. J. (1999) Anaerobiosis in the nematode Caenorhabditis elegans. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 124, 269–280 [DOI] [PubMed] [Google Scholar]
- 25.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J. P. (2006) GenePattern 2.0. Nat. Genet. 38, 500–501 [DOI] [PubMed] [Google Scholar]
- 26.Mendenhall A. R., LaRue B., Padilla P. A. (2006) Glyceraldehyde-3-phosphate dehydrogenase mediates anoxia response and survival in Caenorhabditis elegans. Genetics 174, 1173–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott B. A., Avidan M. S., Crowder C. M. (2002) Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296, 2388–2391 [DOI] [PubMed] [Google Scholar]
- 28.Li W., Palmer G. (1993) Spectroscopic characterization of the interaction of azide and thiocyanate with the binuclear center of cytochrome oxidase: evidence for multiple ligand sites. Biochemistry 32, 1833–1843 [DOI] [PubMed] [Google Scholar]
- 29.Samara C., Syntichaki P., Tavernarakis N. (2008) Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell. Death Differ. 15, 105–112 [DOI] [PubMed] [Google Scholar]
- 30.Enomoto K., Arikawa Y., Muratsubaki H. (2002) Physiological role of soluble fumarate reductase in redox balancing during anaerobiosis in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 215, 103–108 [DOI] [PubMed] [Google Scholar]
- 31.Feng J., Bussière F., Hekimi S. (2001) Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644 [DOI] [PubMed] [Google Scholar]
- 32.Wong A., Boutis P., Hekimi S. (1995) Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii N., Takahahi K., Tomita S., Keino T., Honda S., Yoshino K., Suzuki K. (1990) A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat. Res. 237, 165–171 [DOI] [PubMed] [Google Scholar]
- 34.Nomura H., Athauda S. B., Wada H., Maruyama Y., Takahashi K., Inoue H. (2006) Identification and reverse genetic analysis of mitochondrial processing peptidase and the core protein of the cytochrome bc1 complex of Caenorhabditis elegans, a model parasitic nematode. J. Biochem. 139, 967–979 [DOI] [PubMed] [Google Scholar]
- 35.Vastenhouw N. L., Fischer S. E. J., Robert V. J. P., Thijssen K. L., Fraser A. G., Kamath R. S., Ahringer J., Plasterk R. H. A. (2003) A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 13, 1311–1316 [DOI] [PubMed] [Google Scholar]
- 36.Ketting R. F., Haverkamp T. H. A., van Luenen H. G. A. M., Plasterk R. H. A. (1999) mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of werner syndrome helicase and RNased. Cell 99, 133–141 [DOI] [PubMed] [Google Scholar]
- 37.Cristina D., Cary M., Lunceford A., Clarke C., Kenyon C. (2009) A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 5, e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falk M. J., Zhang Z., Rosenjack J. R., Nissim I., Daikhin E., Sedensky M. M., Yudkoff M., Morgan P. G. (2008) Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol. Genet. Metab. 93, 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell-Coffman J. A. (2010) Hypoxia signaling and resistance in C. elegans. Trends Endocrinol. Metab. 21, 435–440 [DOI] [PubMed] [Google Scholar]
- 40.Shen C., Nettleton D., Jiang M., Kim S. K., Powell-Coffman J. A. (2005) Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J. Biol. Chem. 280, 20580–20588 [DOI] [PubMed] [Google Scholar]
- 41.Mendenhall A. R., LeBlanc M. G., Mohan D. P., Padilla P. A. (2009) Reduction in ovulation or male sex phenotype increases long-term anoxia survival in a daf-16-independent manner in Caenorhabditis elegans. Physiol. Genomics 36, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson L. L., Mao X., Scott B. A., Crowder C. M. (2009) Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science 323, 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Shao Z., Zhai Z., Shen C., Powell-Coffman J. A. (2009) The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One 4, e6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D., Thomas E. L., Kapahi P. (2009) HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 5, e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehta R., Steinkraus K. A., Sutphin G. L., Ramos F. J., Shamieh L. S., Huh A., Davis C., Chandler-Brown D., Kaeberlein M. (2009) Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science 324, 1196–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isaacs J. S., Jung Y. J., Mole D. R., Lee S., Torres-Cabala C., Chung Y. L., Merino M., Trepel J., Zbar B., Toro J., Ratcliffe P. J., Linehan W. M., Neckers L. (2005) HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8, 143–153 [DOI] [PubMed] [Google Scholar]
- 47.Ventura N., Rea S. L., Schiavi A., Torgovnick A., Testi R., Johnson T. E. (2009) p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell 8, 380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dingley S., Polyak E., Lightfoot R., Ostrovsky J., Rao M., Greco T., Ischiropoulos H., Falk M. J. (2010) Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion 10, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakowski B., Hekimi S. (1996) Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 50.Greer E. L., Brunet A. (2009) Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakowski B., Hekimi S. (1998) The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 95, 13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mo M. L., Palsson B. O., Herrgard M. J. (2009) Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghafouri S., McGhee J. D. (2007) Bacterial residence time in the intestine of Caenorhabditis elegans. Nematology 9, 87–91 [Google Scholar]
- 54.Jazwinski S. M. (2005) The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene 354, 22–27 [DOI] [PubMed] [Google Scholar]
- 55.Hartman P. S., Ishii N., Kayser E. B., Morgan P. G., Sedensky M. M. (2001) Mitochondrial mutations differentially affect aging, mutability and anesthetic sensitivity in Caenorhabditis elegans. Mech. Ageing Dev. 122, 1187–1201 [DOI] [PubMed] [Google Scholar]
- 56.Fuchs S., Bundy J., Davies S., Viney J., Swire J., Leroi A. (2010) A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong M. Q., Venable J. D., Au N., Xu T., Park S. K., Cociorva D., Johnson J. R., Dillin A., Yates J. R., 3rd. (2007) Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.