Abstract
The glioblastoma genome displays remarkable chromosomal aberrations, which harbor critical glioblastoma-specific genes contributing to several oncogenetic pathways. To identify glioblastoma-targeted genes, we completed a multifaceted genome-wide analysis to characterize the most significant aberrations of DNA content occurring in glioblastomas. We performed copy number analysis of 111 glioblastomas by Digital Karyotyping and Illumina BeadChip assays and validated our findings using data from the TCGA (The Cancer Genome Atlas) glioblastoma project. From this study, we identified recurrent focal copy number alterations in 1p36.23 and 4p16.3. Expression analyses of genes located in the two regions revealed genes which are dysregulated in glioblastomas. Specifically, we identify EGFR negative regulator, ERRFI1, within the minimal region of deletion in 1p36.23. In glioblastoma cells with a focal deletion of the ERRFI1 locus, restoration of ERRFI1 expression slowed cell migration. Furthermore, we demonstrate that TACC3, an Aurora-A kinase substrate, on 4p16.3, displays gain of copy number, is overexpressed in a glioma-grade-specific pattern, and correlates with Aurora kinase overexpression in glioblastomas. Our multifaceted genomic evaluation of glioblastoma establishes ERRFI1 as a potential candidate tumor suppressor gene and TACC3 as a potential oncogene, and provides insight on targets for oncogenic pathway-based therapy.
Keywords: glioblastoma, genomics, copy number, 1p36, 4p16, ERRFI1, TACC3
INTRODUCTION
Cancer cells undergo continuous acquisition of heritable genetic variation, manifested as mutations of single base pairs, large or small deletions or insertions, chromosomal translocations, and gain or loss of entire chromosomal regions [1,2]. The accumulation of these somatic genomic changes results in a highly heterogeneous and complex cancer genome. Glioblastoma is the most aggressive and frequently occurring type of brain tumor [3,4]. Recent comprehensive analyses have opened the door to understanding the genetic and molecular alterations that characterize the glioblastoma genome. These studies identified sequence mutations and alterations of DNA copy number, gene expression, and methylation status that may contribute to a distinct network of oncogenic signaling pathways, including receptor tyrosine kinase (RTK), growth factor and phosphatidylinositol-3-OH kinase (PI3K), p53 and retinoblastoma (RB1) pathways [5,6].
Identification of a coding sequence alteration, amplification, or homozygous deletion can help define unequivocal driver genetic alterations in cancers [6]. Several genes in the central glioblastoma pathways were identified through dramatic copy number alterations, such as amplification of the oncogenes, EGFR, c-MYC, CDK4, PDGFRA, MDM2, and MDM4, and deletion of the tumor suppressor genes, CDKN2A, CDKN2B, and PTEN [5-9]. Recent studies have validated the significance of well-known copy number alterations and have proposed additional candidates which may contribute to the development of glioblastomas [5,6,10-12]. Conversely, several core glioblastoma genes have been found to be exclusively altered by sequence mutation, such as PI(3)K, RAS, ERBB2, and IDH1/IDH2 [5,13]. However, a large proportion of recognized glioblastoma driver genes, including EGFR, TP53, CDKN2A, PTEN, NF1 and RB1, are targeted by both sequence and copy number alterations. In addition, gene expression signatures, which are an essential component of global genomic studies, have been used to exploit disease-specific signaling pathways and as clinical prognostic factors in many cancers [14-17]. However, despite major advancements, the current understanding of glioblastoma genetics is still inadequate, and additional molecular targets are urgently needed to be used in the development of clinically proven therapies.
In this study, we used Digital Karyotyping (DK) and Illumina BeadChip assays (Illumina, Inc., San Diego, CA) to identify genomic loci that are recurrently targeted by focal copy number alterations in 111 glioblastomas. Using these data and data from TCGA, we identified frequent gene copy number changes in1p36.23 and 4p16.3. We further demonstrated that ERRFI1 and TACC3 in 1p36.23 and 4p16.3, respectively, are potential glioblastoma-targeted genes.
RESULTS
Detection of focal copy number alterations by DK and Illumina beadchips
DK is a highly quantitative copy number analysis platform that has previously been used to identify copy number events in human cancers, including glioblastomas [18-20]. Analysis of 27 glioblastoma samples revealed 52 high-copy amplification events ranging from 98kb to 6.8Mb with 12 to 205 copies per nucleus. The targeted genes within those regions include gain of EGFR, CDK4, PDGFRA, MDM2, and MDM4 (Supplemental Table S1). In addition, we identified 120 regions of homozygous deletion, ranging from 100kb to 5.1Mb. The most common loss is on chromosome 9p21, where tumor suppressor genes CDKN2A and CDKN2B are located (Supplemental Table S2).
Illumina BeadChips in conjunction with the Infinium assay have also been effectively used to examine copy number variations in human cancers at high-throughput levels [5,6,20]. First, using the highly quantitative DK data, we optimized the criteria for defining focal high-copy amplifications and homozygous deletions for Illumina high-density SNP arrays. Two glioblastoma samples, xenograft H456 and primary tumor TB2607, were analyzed by both DK and Illumina high-density SNP arrays. Using DK as a standard, we identified the values and filtering criteria (stated in Materials and Methods) that faithfully reveal amplifications and deletions from the data produced by the Illumina BeadChips. We generated Illumina high-density SNP array profiles from 84 glioblastomas samples. As controls, we also analyzed genomic DNA from normal adult and fetal brain and 3 matching blood specimens from glioblastoma patients. In total, we identified 474 focal gain events (Supplemental Table S3) and 1540 focal loss events (Supplemental Table S4).
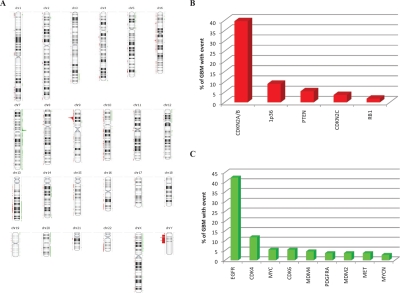
Of the genomic copy number profiles from a total of 111 glioblastoma samples assessed by DK or Illumina BeadChips, we identified a high degree of heterogeneity and copy number instability across the glioblastoma genomes (Figure 1A). Despite heterogeneity, we detected several regions which are recurrently gained or lost in glioblastomas (Figure 1B, C). The two most prevalent focal amplifications were the EGFR and CDK4 loci, occurring in 42% and 12% of all cases, respectively. The two most prevalent focal deletions were the CDKN2A/B and 1p36 loci, occurring in 40% and 9% of all samples, respectively. Additionally, we detected multiple intragenic homozygous deletions within large genes, including LRP1B, WWOX, and A2BP1.
Figure 1: Focal high copy number gains and homozygous deletions in glioblastomas.
Copy number analysis using DK and Illumina BeadChips revealed gains (indicated by green) and losses (indicated by red) spanning the genome of glioblastomas. (A) Glioblastoma copy number karyotype generated utilizing Nexus Copy Number Professional Software (BioDiscovery Inc.). The most common regions for (B) focal deletion and (C) focal gain of copy number in 111 glioblastomas. Loci are identified with reported tumor suppressor genes and oncogenes if available.
ERRFI1 on 1p36 is a candidate tumor suppressor gene, whose products regulate glioblastoma cell migration
In addition to the well-characterized glioblastoma genes, the most striking genomic changes identified in our analysis were recurrent focal copy number changes on 1p36 and 4p16. Focal deletions of 1p36 have been previously reported to occur in numerous cancer types, including glioblastoma, neuroblastoma, oligodendroglioma, and colorectal, lung, and breast cancer [21-23]. We detected focal homozygous deletions on 1p36 in 9/111 of our glioblastoma samples. With the addition of samples from the TCGA glioblastoma data set, we mapped 1p36 deletions in a comprehensive data set of a total 430 glioblastomas and found they occurred in two distinct minimal deleted regions (MDRs) on 1p36.32 (Figure 2) and on 1p36.23 (Figure 3).
Figure 2: High-resolution mapping of homozygous deletions detects DFFB, C1orf174, and LOC100133612 within secondary MDr on 1p36.32.
(A) Chromosome 1 DK analysis of glioblastoma cell line H542 indicates a single homozygous deletion occurring at the 1p36.32 cytoband. (B) High-resolution mapping depicts overlapping homozygous deletions from 3.0Mb to 5.5Mb coordinates on chromosome 1 (indicated by red bars). Copy number data includes tumor samples from our independent analysis plus the TCGA glioblastoma copy number data set. RefSeq gene positions are indicated. Genomic Q-PCR validation analysis (*) of H542, H561, and glioblastoma xenograft X397 confirms homozygous deletion. Q-PCR primer location depicted by black boxes (Supplemental Table S5).
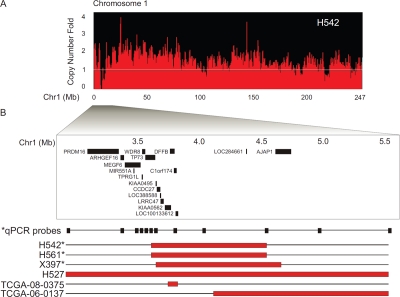
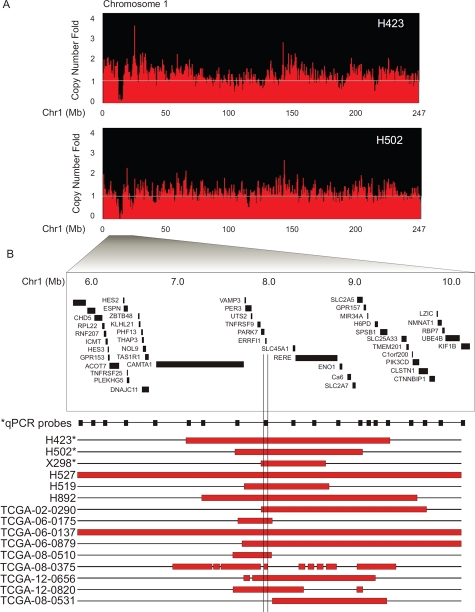
Figure 3: High-resolution mapping of homozygous deletions reveals ERRFI1 within the most frequent MDr on 1p36.23.
(A) DK analysis of chromosome 1 in glioblastoma cell lines H423 and H502 reveals a single homozygous deletion at 1p36.23. (B) High resolution mapping depicts overlapping homozygous deletions from 6.0Mb to 10.0Mb coordinates on chromosome 1 (indicated by red bars). Copy number data include tumor samples from our independent analysis and the TCGA glioblastoma copy number data set. RefSeq gene positions are indicated. Validation by genomic Q-PCR copy number analysis (*) of H423, H502, and glioblastoma xenograft X298 confirms homozygous deletion. Q-PCR primer location depicted by black boxes (Supplemental Table S6).
Glioblastoma cell line H542 presented a homozygous deletion at 1p36.32 (Figure 2A). We mapped the 1p36.32 deletion in H542 by quantitative real-time PCR (Q-PCR) and found that this region contained TP73, KIAA0495, CCDC27, LOC388588, LRRC47, KIAA0562, DFFB, C1orf174, LOC100133612, and LOC284661 (Figure 2B, Supplemental Table 5). Further analysis of additional samples with 1p36.32 deletions revealed an overlapped MDR containing DFFB, C1orf174, and LOC100133612 (Figure 2B).
Cytoband 1p36.23 harbors the most frequently deleted region (Figure 3) on 1p36 in glioblastomas. Two glioblastoma cell lines, H423 and H502, each contained a single focal deletion in 1p36.23 (Figure 3A). We further mapped the 1p36.23 deletions in the two cell lines by Q-PCR. The deleted region contains multiple genes, including CAMTA1, VAMP3, PER3, UTS2, TNFRSF9, PARK7, ERRFI1, SLC45A1, RERE, ENO1, CA6, SLC2A7 and SLC2A5 (Figure 3B, Supplemental Table 6). With the addition of TCGA glioblastoma samples, we found that 15/430 glioblastomas contain homozygous deletions in 1p36.23. Further analysis revealed the most commonly deleted region contains a single gene, ERRFI1 (Figure 3B).
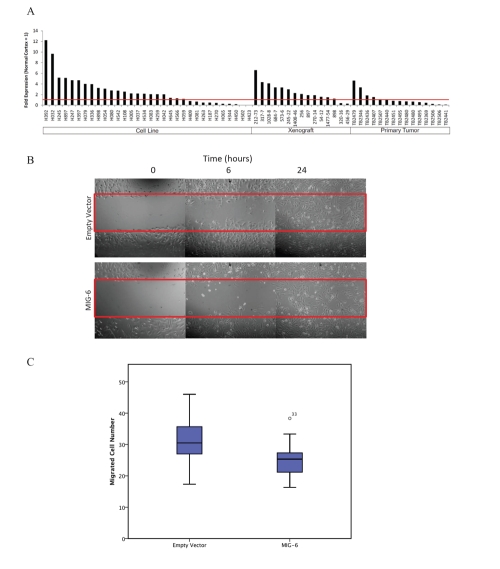
ERRFI1 is a candidate tumor suppressor which functions in normal cells as a negative regulator of EGFR and the ErbB family [24-26]. We measured ERRFI1 mRNA levels in a panel of 62 glioblastoma samples and detected downregulation of ERRFI1 expression in 34% of the samples tested (Figure 4A).
Figure 4: ERRFI1 is silenced in glioblastomas and reduces cell migration in H423 glioblastoma cells.
(A) Quantification of ERRFI1 mRNA levels in glioblastomas (32 cell lines, 15 xenografts, and 15 primary tumors) by Q-PCR. Results presented as fold expression relative to normal cortex control (indicated by red line). Restoration of ERRFI1 expression in H423 cells results in decreased cell migration as measured by (B) wound healing assay and (C) trans-well migration assay (T-test, p < 0.001).
We further investigated the possible pathogenic function of ERRFI1 in glioblastoma cells. Glioblastoma cell line H423 harbors a homozygous deletion of ERRFI1 and did not express endogenous ERRFI1 (Figure 4A). We transfected H423 cells with pCMV-6-entry-ERRFI1 and performed wound-healing experiments on the transfected cells. Compared to cells transfected with a control vector, H423 cells with ERRFI1 expression had impaired wound healing (Figure 4B). Furthermore, we conducted trans-well assays and demonstrated that H423 cells with ERRFI1 expression had a lower rate of trans-well migration than control cells (Figure 4C).
TACC3 on 4p16.3 is overexpressed and correlates with the expression of Aurora kinases in glioblastomas
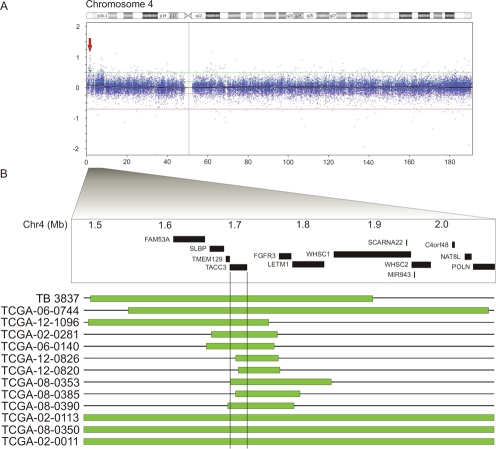
Focal gains on 4p16.3 were also frequently identified in a subset of glioblastomas (Figure 5A). We mapped the 4p16 duplications in a data set of 430 glioblastomas. The gains clustered in a minimal gained region centered on 4p16.3 from 1.5 to 2.0Mb (Figure 5B), containing SLBP, TMEM129, TACC3, FGFR3 and LETM1, of which TACC3 most commonly displayed gain of copy number. A Q-PCR analysis of an independent panel of glioblastoma samples detected genomic duplications of TACC3 in 5 out of 101 samples. In addition, compared with its adjacent genes, SLBP, TMEM129 and FGFR3 at 4p16.3, we found that TACC3 displayed a predominant overexpression pattern in glioblastomas (Figure 6A, S1 D).
Figure 5: High resolution mapping identifies TACC3 as the glioblastoma-targeted gene on 4p16.3.
(A) Illumina Bead-Chip analysis of chromosome 4 in glioblastoma primary tumor TB3837 reveals a single focal gain at 4p16.3. (B) High resolution mapping depicts overlapping gain of copy number from 1.5Mb to 2.1Mb coordinates on chromosome 4 (indicated by green bars). Copy number data includes tumor samples from our independent analysis and the TCGA glioblastoma copy number data set. RefSeq gene positions are indicated.
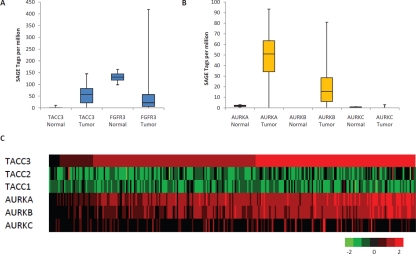
Figure 6: TACC3 is the predominant gene upregulated in 4p16.3 and correlates with Aurora kinase expression.
SAGE of (A) TACC3 and FGFR3 and (B) Aurora kinases, AURKA, AURKB, and AURKC in normal brain (n = 2) and glioblastoma (n = 16) samples (data adapted from [5]). (C) Gene expression of TACC and Aurora kinase family members using the TCGA glioblastoma expression data set (n = 266) indicates correlation between TACC3 and AURKA (correlation coefficient = 0.76) and between TACC3 and AURKB (correlation coefficient = 0.70). Red indicates high gene expression level and green indicates low gene expression level.
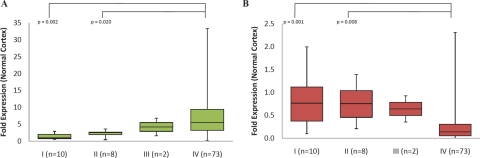
Dysregulation of the human Transforming Acidic Coiled Coil (TACC) genes is thought to be important in the development of several cancers. The three human TACC proteins, TACC1, TACC2, and TACC3, are core components of the centrosome and have non-overlapping functions in the normal cell and the cancer cell [27,28]. TACC1 overexpression promotes cellular transformation in vitro [29] and in vivo [30]. TACC2 (AZU-1) is a putative tumor suppressor in breast cancer [31]. TACC3 expression is upregulated and associated with shorter median survival in patients with non-small-cell lung cancer [32]. However, TACC3 expression is reduced in ovarian and thyroid cancer [33,34]. We therefore assessed expression of the TACC genes in a panel of gliomas (WHO Grades I-IV) by Q-PCR (Figure 7). We found significant overexpression of TACC3 in Grade IV gliomas. Conversely, TACC2 transcript levels were significantly downregulated in Grade IV gliomas.
Fig 7. Grade-specific TACC3 upregulation and TACC2 downregulation in gliomas.
(A) TACC3 and (B) TACC2 mRNA levels in a panel of 93 glioma samples were assessed by Q-PCR. Results presented as fold expression relative to normal cortex control. I (WHO classification glioma Grade I), II (WHO classification glioma Grade I), III (WHO classification glioma Grade III), IV (WHO classification glioma Grade IV).
TACC3 can be phosphorylated by Aurora-A kinase and plays an important role in mitosis [35-37]. Therefore, to determine the correlation of TACC3 and Aurora kinase expression, we used Serial Analysis of Gene Expression (SAGE) [5] and TCGA expression data sets to evaluate TACC3 and Aurora kinase gene expression in glioblastomas (Figure 6B, 6C). We found a significant expression correlation between TACC3 and Aurora A (correlation coefficient = 0.76), as well as between TACC3 and Aurora B (correlation coefficient = 0.70) (Figure 6C).
DISCUSSION
While many of the most common copy number gains and deletions have been characterized in glioblastomas, the significance of many other genomic events remains unknown. Utilizing an independent set of glioblastomas and publicly available data, in this study, we focused on the identification and characterization of genes on two frequently altered regions, 1p36 and 4p16. We found that ERRFI1 is a potential glioblastoma-targeted tumor suppressor gene and TACC3 is a potential oncogene.
Deletions in 1p36 have been previously reported in glioblastoma genomic studies, indicating that multiple tumor suppressors exist in this locus [19,38-42]. In our analysis, we observed several regions of deletions across 1p36. However, the most striking and frequent deletions occur between the 5 and 10Mb coordinates on 1p36.23, centering on the candidate tumor suppressor ERRFI1. Deletion of ERRFI1 has been reported to activate EGFR and sustain MAPK signaling, resulting in tumor phenotypes in numerous tissues in ERRFI1 knockout mice [43-45]. ERRFI1 is also frequently deleted, mutated, or down-regulated in breast and lung cancers, as well as in glioblastomas[38,42,46]. In a recent study, overexpression of ERRFI1 was shown to decrease proliferation in glioblastoma cells, binding EGFR with STX8, and driving internalized EGFR to late endosomes for degradation, whereas knockdown of ERRFI1 expression resulted in increased tumor invasion [47]. Our biological data are consistent with these previous findings, showing that restoring ERRFI1 expression in an ERRFI1-deficient glioblastoma cell line decreases glioblastoma cell migration. Our genetic and biological data, as well as data from other studies, suggests that ERRFI1 is another key component in the EGFR signaling pathway involved in glioblastoma development.
Genomic alterations of 4p16.3 have been reported in several cancers, indicating the presence of one or more oncogenes [48-51]. Using an integrative genomic strategy, we have analyzed genes altered in the 4p16.3 region in glioblastomas and revealed that TACC3 is the primary glioblastoma-targeted gene in this region. Furthermore, a TACC3 somatic mutation (p.E680K) was reported in one of 22 glioblastomas [5]. TACC3 has a conserved function to promote centrosomal microtubule assembly, a process which is often altered in cancer cells [27,52]. Consequently, genetic silencing of TACC3 results in destabilized microtubules, defects in chromosome alignment, and mitotic defects [52-54].
The Aurora family of serine-threonine kinases is comprised of three members (A, B, and C) that cooperate with many other proteins, including the TACC family, to direct chromosome assembly and segregation during mitosis [27,55-57]. Dysfunction of Aurora kinases can disrupt genomic integrity and lead to aneuploidy, mitotic arrest, and cell death. Aurora A and B are of particular interest since they have been shown to be overexpressed in a broad range of human tumors and are often associated with poor outcome. Thus, regulators of the mitotic spindle apparatus, including Aurora kinases and its substrates, are attractive targets for small-molecule therapeutics [35,58-60]. In our study, we found a strong correlation between Aurora kinase and TACC3 expression, indicating that in the Aurora kinase/TACC pathway, the dysregulated kinase and its substrate, may contribute synergetically to glioblastoma pathogenesis and could serve as targets for molecular-based intervention.
MATERIALS AND METHODS
Tumor samples
DNA samples were obtained from brain tumor cell lines, xenografts and primary brain tumors. Brain tumor tissue samples were obtained from the Preston Robert Tisch Brain Tumor Center Biorepository at Duke University Medical Center by an IRB-approved protocol. All samples were obtained in accordance with the Health Insurance Portability and Accountability Act. Frozen sections were made from each tumor sample and examined by light microscopy by a board-certified neuropathologist to ensure that more than 95% of each section consisted of tumor cells. Normal patient DNA samples were obtained from peripheral blood. Normal adult and fetal brain genomic DNA was obtained from BioChain Institute, Inc. (Hayward, CA). Samples were defined as pediatric if taken from a patient between the ages of 0 and 19.
Digital Karyotyping
We generated 18 glioblastoma DK libraries. Additionally, we included 1 primary and 8 glioblastoma cell line DK libraries from the Cancer Genome Anatomy Project [19] (http://cgap.nci.nih.gov/SAGE/DKViewHome). Protocols for performing DK and software for the extraction and analysis of genomic tags are available at www.digitalkaryotyping.org [61]. Experimental tag sequences were compared to predicted human genome virtual tags and were visualized by using SageGenie DKView (http://cgap.nci.nih.gov/SAGE/DKViewHome). Homozygous deletions were screened by using a sliding window size of 150 virtual tags (~600 kb in size). Putative homozygous deletions were defined as events with a tag density ratio (observed tags/expected tags in window) of < 0.05. Amplifications were identified using a sliding window size of 50 virtual tags (~200 kb in size). Putative amplifications were defined as events with a tag density ratio of > 6.
High density SNP arrays
Eighty-four glioblastoma samples were analyzed for copy number variation by utilizing HumanHap550-Duo and Human610-Quad BeadChips with the Illumina Infinium Whole Genome Genotyping Assay. Data was pre-processed to generate logR intensity ratios using Illumina BeadStudio. Data was converted into copy number calls and visualized by using Nexus Copy Number software (BioDiscovery, Inc., El Segundo, CA).
TCGA Data
Genome-wide Level 3 TCGA copy number data was downloaded with the TCGA Data Portal Data Access Matrix (http://cancergenome.nih.gov/dataportal) from Hudson-Alpha Cancer Genome Characterization Center, Memorial Sloan Kettering Cancer Center, and Harvard Medical School-Dana Farber Cancer Institute. Candidate deletions were defined as events with seg mean ≤ −1. Candidate gains were defined as events with seg mean ≥ 0.4. Events less than 30kb and events occurring in two or more tumors with identical start and stop positions were removed, because they were likely artifact or copy number polymorphisms. Candidate gene TCGA expression data from the University of North Carolina and Broad Institute was downloaded with the TCGA Data Portal Data Browser. Gene expression values from the TCGA database represent the ratio of tumor expression to normal expression. The expression value for a given gene for a given patient is the log2 ratio of the tumor expression of the gene in the patient to a synthetic normal sample.
Bioinformatic Analysis
Nexus Copy Number software (BioDiscovery, Inc) was used to visualize copy number data. The following values were used for screening of genetic events of homozygous loss and amplification: Significance Threshold, 1×10−8; Min probes per segment, 2; Max contiguous probe spacing (Kbp), 1000; Gain, 0.5; Loss, −0.7. For identification of areas of duplication, the Nexus default settings were as follows: Significance threshold, 1×10−6, Min probes per segment, 5; Max contiguous probe spacing (Kbp), 1000; Gain, 0.2.
Quantitative Real-Time PCR
The genomic DNA content and mRNA expression levels of genes of interest within tumor and normal cells were quantified by quantitative real-time polymerase chain reaction (Q-PCR). Genomic DNA from normal blood cells served as controls, and genomic DNA content was normalized to that of Line-1. For mRNA expression measurement, cDNA from normal human adult cortex was used as the control and cDNA content was normalized to that of GAPDH. Primers for genomic mapping of 1p36.32 and 1p36.23 are listed in Supplemental Table S5 and Supplemental Table S6.
Wound Healing Assay
H423 glioblastoma cells were maintained in ZO+ media (Gibco, Carlsbad, CA) with 10% fetal calf serum (FCS). Cells were transfected with pCMV6-entry or pCMV6-ERRFI1. Cells were plated at 100% confluence in 6-well plates. The cell monolayer was wounded with a p1000 pipette tip and wound healing was documented at 0, 6 and 24 hours by photographs.
Transwell Migration Assay
H423 cells were transfected with pCMV6-entry or pCMV6-ERRFI1, washed, and resuspended in serum-free media, then seeded into transwell membranes at 10,000 cells per well with 5% FCS as a chemoattractant. After 24 hours, cells were removed from the upper chamber then fixed in 100% methanol on the lower surface of the membrane and stained with crystal violet (2% solution in ethanol) and counted (experiment performed in triplicate wells, 12 fields counted per well). Statistical analyses were conducted with PASW (SPSS) 18.0 (IBM, Armonk, North Castle, NY).
SUPPLEMENTAL FIGURE AND TABLES
Acknowledgments
We thank Melissa J. Ehinger and Diane L. Satterfield for glioblastoma sample processing. We thank B. Ahmed Rasheed and David Lister for glioblastoma DNA purification. This work was supported by The Pediatric Brain Tumor Foundation Institute at Duke, by the NIH grants NCI R01CA118822, and by a Cancer Institute NSW Research Scholars Award (C.A.P).
Abbreviations used:
- TACC3
transforming, acidic coiled-coil containing protein 3
- FGFR3
fibroblast growth factor receptor 3
- AURKA
aurora kinase A
- AURKB
aurora kinase B
- AURKC
aurora kinase C
- ERRFI1
ERBB receptor feedback inhibitor 1
- EGFR
epidermal growth factor receptor
- ERBB
epidermal growth factor receptor family
- TCGA
The Cancer Genome Atlas
- CNV
copy number variation
- SAGE
Serial Analysis of Gene Expression
- Q-PCR
quantitative polymerase chain reaction
- FCS
fetal calf serum.
Footnotes
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
REFERENCES
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006;24:1253–65. doi: 10.1200/JCO.2005.04.5302. [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008 doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis M, Olson JJ, Mikkelsen T, Lehman N, Aldape K, Alfred Yung WK, Bogler O, Vandenberg S, Berger M, Prados M, Muzny D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eley GD, Reiter JL, Pandita A, Park S, Jenkins RB, Maihle NJ, James CD. A chromosomal region 7p11.2 transcript map: its development and application to the study of EGFR amplicons in glioblastoma. Neuro Oncol. 2002;4:86–94. doi: 10.1093/neuonc/4.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jen J, Harper JW, Bigner SH, Bigner DD, Papadopoulos N, Markowitz S, Willson JK, Kinzler KW, Vogelstein B. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994;54:6353–8. [PubMed] [Google Scholar]
- 10.Freire P, Vilela M, Deus H, Kim YW, Koul D, Colman H, Aldape KD, Bogler O, Yung WK, Coombes K, Mills GB, Vasconcelos AT, Almeida JS. Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS ONE. 2008;3:e4076. doi: 10.1371/journal.pone.0004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Tayrac M, Etcheverry A, Aubry M, Saikali S, Hamlat A, Quillien V, Le Treut A, Galibert MD, Mosser J. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer. 2009;48:55–68. doi: 10.1002/gcc.20618. [DOI] [PubMed] [Google Scholar]
- 12.Lo KC, Rossi MR, LaDuca J, Hicks DG, Turpaz Y, Hawthorn L, Cowell JK. Candidate glioblastoma development gene identification using concordance between copy number abnormalities and gene expression level changes. Genes Chromosomes Cancer. 2007;46:875–94. doi: 10.1002/gcc.20474. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–41. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 15.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16:559–66. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 16.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Jr., Marks JR, Dressman HK, West M, Nevins JR. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 17.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, Harpole D, Marks J, Berchuck A, Ginsburg GS, Febbo P, Lancaster J, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 18.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, Lengauer C, Cummins JM, Velculescu VE, Fults DW, McLendon RE, Bigner DD, Yan H. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–24. [PubMed] [Google Scholar]
- 19.Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 96:169–79. doi: 10.1007/s11060-009-9959-4. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z, Zhou L, Killela P, Rasheed AB, Di C, Poe WE, McLendon RE, Bigner DD, Nicchitta C, Yan H. Glioblastoma proto-oncogene SEC61gamma is required for tumor cell survival and response to endoplasmic reticulum stress. Cancer Res. 2009;69:9105–11. doi: 10.1158/0008-5472.CAN-09-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Vinci A, Infusini E, Nigro S, Monaco R, Giaretti W. Intratumor distribution of 1p deletions in human colorectal adenocarcinoma is Commonly homogeneous. Cancer. 1998;83:415–22. [PubMed] [Google Scholar]
- 22.Bièche I, Khodja a, Lidereau R. Deletion mapping of chromosomal region 1p32-pter in primary breast cancer. Genes, chromosomes & cancer. 1999;24:255–63. doi: 10.1002/(sici)1098-2264(199903)24:3<255::aid-gcc11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Ragnarsson G, Eiriksdottir G, Johannsdottir JT, Jonasson JG, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. British journal of cancer. 1999;79:1468–74. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, Castellani L, Alemà S, Alimandi M, Segatto O. Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene. 2003;22:4221–34. doi: 10.1038/sj.onc.1206516. [DOI] [PubMed] [Google Scholar]
- 25.Ballarò C, Ceccarelli S, Tiveron C, Tatangelo L, Salvatore AM, Segatto O, Alemà S. Targeted expression of RALT in mouse skin inhibits epidermal growth factor receptor signalling and generates a Waved-like phenotype. EMBO reports. 2005;6:755–61. doi: 10.1038/sj.embor.7400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Pickin Ka, Bose R, Jura N, Cole Pa, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–4. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–88. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci U S A. 2000;97:14352–7. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58:165–70. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- 30.Cully M, Shiu J, Piekorz RP, Muller WJ, Done SJ, Mak TW. Transforming acidic coiled coil 1 promotes transformation and mammary tumorigenesis. Cancer Res. 2005;65:10363–70. doi: 10.1158/0008-5472.CAN-05-1633. [DOI] [PubMed] [Google Scholar]
- 31.Chen HM, Schmeichel KL, Mian IS, Lelievre S, Petersen OW, Bissell MJ. AZU-1: a candidate breast tumor suppressor and biomarker for tumor progression. Mol Biol Cell. 2000;11:1357–67. doi: 10.1091/mbc.11.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung CK, Jung JH, Park GS, Lee A, Kang CS, Lee KY. Expression of transforming acidic coiled-coil containing protein 3 is a novel independent prognostic marker in non-small cell lung cancer. Pathol Int. 2006;56:503–9. doi: 10.1111/j.1440-1827.2006.01998.x. [DOI] [PubMed] [Google Scholar]
- 33.Ulisse S, Baldini E, Toller M, Delcros JG, Gueho A, Curcio F, De Antoni E, Giacomelli L, Ambesi-Impiombato FS, Bocchini S, D’Armiento M, Arlot-Bonnemains Y. Transforming acidic coiled-coil 3 and Aurora-A interact in human thyrocytes and their expression is deregulated in thyroid cancer tissues. Endocr Relat Cancer. 2007;14:827–37. doi: 10.1677/ERC-07-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauffart B, Vaughan MM, Eddy R, Chervinsky D, DiCioccio RA, Black JD, Still IH. Aberrations of TACC1 and TACC3 are associated with ovarian cancer. BMC Womens Health. 2005;5:8. doi: 10.1186/1472-6874-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeRoy PJ, Hunter JJ, Hoar KM, Burke KE, Shinde V, Ruan J, Bowman D, Galvin K, Ecsedy JA. Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 2007;67:5362–70. doi: 10.1158/0008-5472.CAN-07-0122. [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol. 2005;170:1047–55. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barros TP, Kinoshita K, Hyman AA, Raff JW. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol. 2005;170:1039–46. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichimura K, Vogazianou AP, Liu L, Pearson DM, Backlund LM, Plant K, Baird K, Langford CF, Gregory SG, Collins VP. 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene. 2008;27:2097–108. doi: 10.1038/sj.onc.1210848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law ME, Templeton KL, Kitange G, Smith J, Misra A, Feuerstein BG, Jenkins RB. Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet Cytogenet. 2005;160:1–14. doi: 10.1016/j.cancergencyto.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Yin D, Ogawa S, Kawamata N, Tunici P, Finocchiaro G, Eoli M, Ruckert C, Huynh T, Liu G, Kato M, Sanada M, Jauch A, Dugas M, Black KL, Koeffler HP. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res. 2009;7:665–77. doi: 10.1158/1541-7786.MCR-08-0270. [DOI] [PubMed] [Google Scholar]
- 41.Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–28. [PubMed] [Google Scholar]
- 42.Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, Huang J, Dhakal S, Ivanova E, Xiao Y, Zhang H, Hu J, Stommel JM, Lee MA, Chen AJ, Paik JH, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci U S A. 107:6912–7. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferby I, Reschke M, Kudlacek O, Knyazev P, Pantè G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fässler R, Klein R. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nature medicine. 2006;12:568–73. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 44.Jin N, Gilbert JL, Broaddus RR, DeMayo FJ, Jeong JW. Generation of a Mig-6 conditional null allele. Developmental Genetics. 2007;45:716–21. doi: 10.1002/dvg.20348. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y-W, Staal B, Su Y, Swiatek P, Zhao P, Cao B, Resau J, Sigler R, Bronson R, Vande Woude GF. Evidence that MIG-6 is a tumor-suppressor gene. Oncogene. 2007;26:269–76. doi: 10.1038/sj.onc.1209790. [DOI] [PubMed] [Google Scholar]
- 46.Anastasi S, Sala G, Huiping C, Caprini E, Russo G, Iacovelli S, Lucini F, Ingvarsson S, Segatto O. Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene. 2005;24:4540–8. doi: 10.1038/sj.onc.1208658. [DOI] [PubMed] [Google Scholar]
- 47.Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, Huang J, Dhakal S, Ivanova E, Xiao Y, Zhang H, Hu J, Stommel JM, Lee Ma, Chen A-J, Paik J-H, et al. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010:1–6. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiemeney LA, Sulem P, Besenbacher S, Vermeulen SH, Sigurdsson A, Thorleifsson G, Gudbjartsson DF, Stacey SN, Gudmundsson J, Zanon C, Kostic J, Masson G, Bjarnason H, Palsson ST, Skarphedinsson OB, Gudjonsson SA, et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet. 42:415–9. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM, Bergsagel PL. Frequent translocation t(4;14) (p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–4. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart JP, Thompson A, Santra M, Barlogie B, Lappin TR, Shaughnessy J., Jr Correlation of TACC3, FGFR3, MMSET and p21 expression with the t(4;14)(p16.3;q32) in multiple myeloma. Br J Haematol. 2004;126:72–6. doi: 10.1111/j.1365-2141.2004.04996.x. [DOI] [PubMed] [Google Scholar]
- 51.Castro P, Creighton CJ, Ozen M, Berel D, Mims MP, Ittmann M. Genomic profiling of prostate cancers from African American men. Neoplasia. 2009;11:305–12. doi: 10.1593/neo.81530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–41. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider L, Essmann F, Kletke A, Rio P, Hanenberg H, Wetzel W, Schulze-Osthoff K, Nurnberg B, Piekorz RP. The transforming acidic coiled coil 3 protein is essential for spindle-dependent chromosome alignment and mitotic survival. J Biol Chem. 2007;282:29273–83. doi: 10.1074/jbc.M704151200. [DOI] [PubMed] [Google Scholar]
- 54.Piekorz RP, Hoffmeyer A, Duntsch CD, McKay C, Nakajima H, Sexl V, Snyder L, Rehg J, Ihle JN. The centrosomal protein TACC3 is essential for hematopoietic stem cell function and genetically interfaces with p53-regulated apoptosis. Embo J. 2002;21:653–64. doi: 10.1093/emboj/21.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–50. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 57.Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Jr., Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14:1639–48. doi: 10.1158/1078-0432.CCR-07-2179. [DOI] [PubMed] [Google Scholar]
- 59.Warner SL, Bearss DJ, Han H, Von Hoff DD. Targeting Aurora-2 kinase in cancer. Mol Cancer Ther. 2003;2:589–95. [PubMed] [Google Scholar]
- 60.Schneider L, Essmann F, Kletke A, Rio P, Hanenberg H, Schulze-Osthoff K, Nurnberg B, Piekorz RP. TACC3 depletion sensitizes to paclitaxel-induced cell death and overrides p21WAF-mediated cell cycle arrest. Oncogene. 2008;27:116–25. doi: 10.1038/sj.onc.1210628. [DOI] [PubMed] [Google Scholar]
- 61.Leary RJ, Cummins J, Wang TL, Velculescu VE. Digital karyotyping. Nat Protoc. 2007;2:1973–86. doi: 10.1038/nprot.2007.276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.