Abstract
Complex traits often involve interactions between different genetic loci. This can lead to sign epistasis, whereby a set of mutations are individually deleterious or neutral but in combination confer a fitness benefit. In order to acquire the beneficial genotype, an asexual population must cross a fitness valley or plateau by first acquiring the deleterious or neutral intermediates. Here, we present a complete, intuitive theoretical description of the valley-crossing process across the full spectrum of possible parameter regimes. We calculate the rate at which a population crosses a fitness valley or plateau of arbitrary width, as a function of the mutation rates, the population size, and the fitnesses of the intermediates. We find that when intermediates are close to neutral, a large population can cross even wide fitness valleys remarkably quickly, so that valley-crossing dynamics may be common even when mutations that directly increase fitness are also possible. Thus the evolutionary dynamics of large populations can be sensitive to the structure of an extended region of the fitness landscape – the population may be likely to pass up directly uphill paths in favor of paths across valleys and plateaus that lead eventually to fitter genotypes. In smaller populations, we find that below a threshold size which depends on the width of the fitness valley and the strength of selection against intermediate genotypes, valley-crossing is much less likely and hence the evolutionary dynamics are less influenced by distant regions of the fitness landscape.
Keywords: Sign Epistasis, Fitness Valleys, Adaptive Evolution
I. INTRODUCTION
Complex traits derive their complexity, in part, from the interactions between multiple genes. This complicates the quantitative description of the evolution of these traits. In some cases, complex phenotypes may evolve through the accumulation of a number of individually beneficial mutations. In others, however, advantageous traits could require multiple mutations in different genes, each of which may be individually neutral or deleterious in the absence of the other mutations. For example, the evolution of a new function in a signal pathway may require mutations in the genes for both a receptor and the corresponding ligand, or in a series of receptor-ligand pairs involved in the pathway (Goh et al., 2000). Other examples include types of cancer that typically occur only after a series of mutations (Knudson, 2001), pathogens that require multiple mutations in order to escape their hosts’ immune response (Levin et al., 2000; McDonald and Linde, 2002; Shih et al., 2007), and the evolution of citrate usage in E. coli (Blount et al., 2008).
In order for a population to acquire an adaptation involving multiple mutations that are individually neutral or deleterious, at least some individuals must first acquire the neutral or deleterious intermediate mutations. In the language of fitness landscapes, there is no directly uphill path from the current genotype to one of higher fitness that corresponds to this adaptation. The population must cross a “fitness valley,” or in the case of neutral intermediates a “fitness plateau,” to reach the higher-fitness state. In this way a population can escape a local peak in fitness space (i.e. a genotype in which no single mutation confers a fitness advantage) by producing a more distantly related higher-fitness genotype (Weinreich and Chao, 2005). Valley-crossing dynamics may also be important when the population is not at a local fitness peak. We want to understand more generally the dynamics in situations where both individually advantageous mutations as well as valley-crossing processes are simultaneously possible.
In general, these evolutionary dynamics depend on the full range of possible pathways by which a population can accumulate mutations to produce nearby higher-fitness genotypes. All of the mutation rates and selective pressures of intermediate genotypes affect the dynamics. We refer to this set of possibilities and relevant parameters as the local structure of the fitness landscape. Unfortunately, very little is known about what fitness landscapes are typical in nature, so it is impossible to say what sorts of evolutionary dynamics are most common. Instead, we aim to lay out the various qualitatively different types of dynamics, and to understand which aspects of the fitness landscape determine the the relative likelihood of different dynamics. As we will see, this provides a new perspective on what could plausibly be typical in evolution. We find, for example, that a population will not necessarily go directly “uphill” in fitness space even if such a change is possible – sometimes valley-crossing will be more likely.
There are two general ways a population can cross a fitness valley. Each of the intermediates can fix in turn through random drift, until eventually the final mutation provides the advantageous effect. We refer to this process as sequential fixation. Alternatively, intermediates can drift at relatively low frequencies, each such intermediate eventually disappearing, until an individual accumulates a combination of mutations that provides a selective advantage. While recombination can bring together such combinations of mutations in a sexual population, in an asexual population they can only occur through multiple mutation events in a single lineage. This latter process in an asexual population has been dubbed “stochastic tunneling” (Iwasa et al., 2004b). Since it is easier for neutral or deleterious mutations to fix through drift in small populations, we expect that in small enough populations sequential fixation will dominate and stochastic tunneling will not occur. In larger populations, on the other hand, neutral and especially deleterious mutations very rarely fix, so we expect that stochastic tunneling will be more important.
The simplest version of the valley-crossing problem in asexuals is when only two mutations, each individually neutral or deleterious, combine to produce a beneficial trait. Kimura (1985) and Carter and Wagner (2002) analyzed this problem in the context of the evolution of pairs of compensatory mutations. Weinreich and Chao (2005) expanded on this work to analyze the valley-crossing problem in both small and large populations for the case of strongly deleterious single-mutant intermediates. This complements the earlier work of Iwasa et al. (2004b), who focused exclusively on the stochastic tunneling process in large populations, but analyzed neutral or arbitrarily deleterious single-mutant intermediates. Durrett and Schmidt (2008) extended this work by also including valley-crossing in small and intermediate-size populations with neutral single-mutants, although without considering the effect of the strength of selection on the double-mutants. For adaptations requiring more than two mutations, Iwasa et al. (2004a) derived the probability of tunneling in large populations with either neutral or strongly deleterious intermediate mutations. Serra and Haccou (2007) extended this work on adaptations requiring more than two mutations to the case of arbitrarily deleterious intermediates. All of this work is also related to the analysis of Barton and Rouhani (1987), who studied a different kind of fitness valley in which there are multiple stable deterministic equilibria.
In this paper, we provide a complete, intuitive description of the valley-crossing problem in asexuals involving any number of intermediates with arbitrary fitness losses. Earlier results are derived as special cases. For the bulk of the paper, we study how an asexual population traverses a particular fitness valley. That is, we imagine that there is one set of mutations that a population must acquire in one specific order to reach one specific beneficial genotype, and that all mutations away from this specific pathway are strongly deleterious. In analyzing this process, we focus primarily on the tunneling process in large populations, but also study the transition to the small-population regime. Our framework allows us to study not only the probability of stochastic tunneling, but also the dynamics of the intermediate mutations, and hence the time required for the beneficial combination of mutations to arise. Our analysis is very much in the spirit of Karlin (1973), as well as Christiansen et al. (1998), though these authors focused on the case where intermediate mutations were also beneficial. In the Discussion, we consider the situation where multiple valley-crossing and possibly directly uphill pathways are possible, leading to the same or different advantageous genotypes. We explain how our analysis of a single pathway can be applied to this more complex situation.
II. MODEL
We consider an asexual population of of N haploid individuals, and study the process by which this population acquires a beneficial trait that requires mutations at K loci. We refer to this as a “K-hit” process. We assume that all combinations of less than K of these mutations are neutral or deleterious relative to the initial genotype, and that only when an individual has acquired all K of them do they confer a benefit. For the bulk of this paper, we analyze the process by which an asexual population traverses a particular path through this genotype space to acquire the K-hit beneficial mutation. That is, we study one specific order in which the intermediate mutations can be acquired, and implicitly assume that any mutation away from this specific pathway is strongly deleterious. Given this order, the fitness of the individual with k mutations is
| (1) |
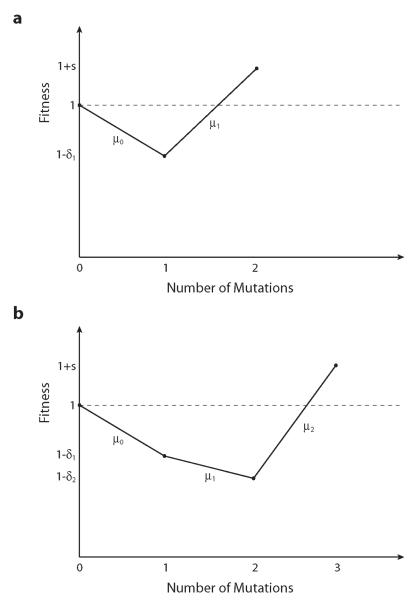
where δk ≥ 0, and we assume that the K-mutant is substantially but not enormously beneficial, 1/N ⪡ s ⪡ 1. We assume that mutations occur from an individual with k mutations to an individual with k + 1 mutations at rate μk. The simplest case, K = 2, is illustrated in Fig. 1a. A case with larger K is illustrated in Fig. 1b. In most realistic situations, there will be a variety of different mutational pathways (e.g. different mutation orders) by which the population can acquire the favored genotype; we show how our analysis describes this more complex situation in the Discussion.
FIG. 1.
An illustration of our model. (a) The simplest case, K = 2. Here the wild-type has fitness 1, and there is a possible double-mutant with fitness 1 + s > 1. The single-mutant, however, has fitness 1 – δ1 < 1. The mutation rate from the wild-type to the single-mutant is μ0, while the mutation rate of the single-mutant to the double-mutant is μ1. (b) A slightly more complex example where K = 3. The wild-type again has fitness 1, single-mutants have fitness 1 – δ1, double-mutants have fitness 1 – δ2, and the triple-mutant is advantageous with fitness 1 + s.
Our model also applies to asexual diploids, where the fitness of each mutation refers to its fitness given the existing genotype at the homologous portion (e.g. the fitness effect of a mutation from an aa genotype to an Aa genotype is the fitness difference between these two diploid genotypes). In this diploid case, the first mutation of a series could convert a diploid one-locus genotype aa to Aa, and a later mutation to genotype AA; if allele A is a recessive mutation which confers some fitness advantage, mutation in either homologous allele would be individually neutral, but the two mutations together would be advantageous. In this sense our model is related to the earlier work by Karlin and Tavare (1981).
For much of this paper, we will consider the case where the population is large and the frequencies of the intermediate mutations are always small — this is the regime in which stochastic tunneling is important. Under these conditions, we treat the dynamics of the intermediate mutations using a continuous-time branching process, according to which all individuals with k mutations die at rate 1, split into two identical individuals at rate wk, and split into two individuals, one of which has an additional mutation, at rate μk, with all of these processes occurring independently. We assume back-mutations are rare enough to be neglected. This model makes a variety of important quantities easier to calculate analytically, but it is only valid when each individual mutant can be treated independently, which requires that all mutants are at low frequency in the population. We will also consider the case of smaller population sizes and the transition to the regime where the dominant valley-crossing process is sequential fixation of the intermediate genotypes. In this case, we use standard Wright-Fisher dynamics to describe the evolution (Ewens, 2004, pp. 20-21).
III. HEURISTIC ANALYSIS
In this section we lay out a simple intuitive analysis of the valley-crossing problem, which demonstrates the main ideas of our approach. Our analysis follows the general lines of our earlier discussion in Fisher (2007). We first note that when a beneficial mutant arises, it will usually soon go extinct due to random genetic drift. In our haploid model, there is a probability that it will survive this drift, and eventually fix in the population (Ewens, 2004, p. 99). We call the process by which such a lucky beneficial mutant survives drift the establishment of the beneficial mutant; once a beneficial mutation is established (upon reaching a size of order ), its frequency will increase roughly deterministically until the population is dominated by beneficial mutants. We refer to any mutant (beneficial or not) whose descendants will include a beneficial multiple-mutant that will establish as successful. We wish to calculate the time it takes for a beneficial multiple-mutant to first establish.
We begin by considering the simplest case, where a double mutation increases fitness by s (i.e. K = 2), but each of the single mutants is neutral (i.e. δ1 = 0). We refer to this as a two-hit process. We initially consider a population so large that the single mutations essentially never fix. In this case, double-mutants can be produced in two ways. A wild-type individual could acquire both mutations in a single generation. Alternatively, a wild-type could acquire just one mutation, and this lineage could drift neutrally until a second mutation within the lineage produces a beneficial double-mutant. As we will see, this latter process is much more likely, so we begin by considering the rate at which this occurs. To do so, we must calculate the probability, p1, that a single-mutant will be successful (i.e. the probability that an individual in its lineage will acquire a second mutation and establish). The essential property of the single-mutant lineage that determines its probability of producing a double-mutant is its time-integrated population size, ∫n(t)dt, because this is the number of mutational opportunities this lineage presents. We call the value of this integral at time t the “weight” at time t of the single-mutant lineage, and denote it by
| (2) |
The total number of mutational opportunities before the lineage goes extinct is , the total weight of the lineage.
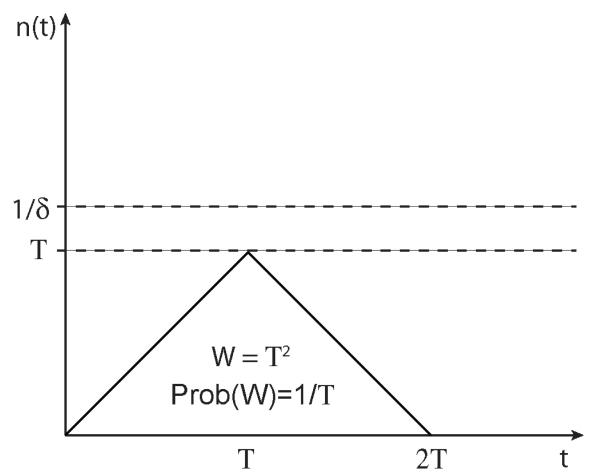
To calculate W, we must understand the dynamics of the single-mutant lineages. In a large population these dynamics are quite simple (Desai and Fisher, 2007). Most of the time the lineage will never reach a substantial size, and will die out within a few generations. Its weight will be of order 1, and the probability that a double mutation occurs and establishes from such a lineage is μ1s times this weight, and is therefore of order μ1s. But with probability of order 1/T, the lineage will survive for more than T generations. If it does, its population size n(T) will be of order T. This will produce a weight of order T2 (a population size of order T for a time of order T). These dynamics are illustrated in Fig. 2, and justified rigorously below and in Appendix B. The probability that such a lineage gives rise to at least one double-mutant that establishes is thus 1 – e−Cμ1sT2, where C is an unknown constant of order 1 and we have used the fact that the occurrence of successful mutations is a Poisson process (the factor C reflects the fact that our estimate of the probability a lineage will reach a given weight is only roughly correct; see Appendix B for a precise calculation.). This means that lineages that survive longer than generations (and hence reach size individuals) are very likely to produce established double-mutants. Thus with probability a single-mutant lives long enough that it is extremely likely to produce a double-mutant that establishes. Since the probability of a single-mutant lineage having weight at least T2 falls off only as 1/T, while the expected number of double-mutants produced by the single-mutant lineage increases as T2 for , the rate at which double-mutants are produced is dominated by these rare lucky single-mutant lineages that reach this size . Thus the overall probability that a single-mutant gives rise to a double-mutant that establishes is simply
| (3) |
FIG. 2.
Sketch of the fate of a mutant lineage which has selective disadvantage δ. Shown is the population size n of the lineage as a function of time t. With probability 1/T, the lineage reaches a size T in roughly T generations, and then drifts to extinction in another T generations. This gives rise to an overall weight W = T2. The overall shape of a “typical” such trajectory is shown (in an actual case, the trajectory would be somewhat “jagged” due to stochastic fluctuations in n(t), but the overall shape would be roughly as shown). This is valid for T < 1/δ; the lineage will almost never reach a population size larger than 1/δ.
If the single-mutant intermediates are deleterious, things are only slightly more complicated. In this case, a single-mutant lineage is still effectively neutral while its population size is small compared to 1/δ1 (Fisher, 2007). On the other hand, it will almost never grow to a size much larger than 1/δ1. Thus if , single-mutant lineages still reach this size with about the same probability as in the neutral case, and all of the neutral results above apply. We have as before . The single-mutant is effectively neutral for the purposes of producing double-mutants — note this can be true even if Nδ1 ⪢ 1 (where the single-mutant is not effectively neutral by conventional definitions). If on the other hand , then the fact that the single-mutant is deleterious matters. In this case, the single-mutant lineage will reach a size of at most order 1/δ1, have a weight of order , and give rise to a double-mutant that establishes with probability of order . Since the probability of this happening is of order δ1, we have
| (4) |
Note that when , this reduces to the neutral result, as it should.
All of our discussion to this point has implicitly assumed that the population size N is large enough that the intermediates can drift to the sizes described above. For the neutral case this means
| (5) |
When this is true, lineages that typically produce double-mutants that establish can do so while staying small compared to N. When Eq. (5) fails, double-mutants establish primarily after a lucky single-mutant has first drifted to fixation. The probability that the neutral mutation fixes is 1/N, after which it will eventually produce a beneficial double-mutant. So for this small-N case we have
| (6) |
Note that this approaches our large-N result at the threshold population size, , as expected. For the case of deleterious mutations, when the condition on N is identical to the neutral case. When , the critical size threshold is instead . For N below this threshold, the single-mutant lineages are always effectively neutral (now because the population size is too small for selection to be felt), and hence the small-population neutral-intermediate results apply: double-mutants establish primarily after a lucky single-mutant has first drifted to fixation, and p1 ~ 1/N. Above this threshold population size, the results above for strongly deleterious intermediates in large populations will generally apply (however, if μ1s is sufficiently small, even strongly deleterious single-mutants are likely to drift to fixation before producing a double-mutant; we discuss this in more detail in our rigorous analysis below).
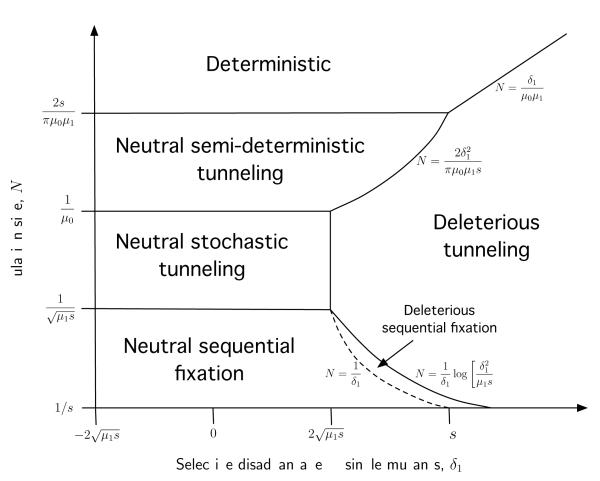
To summarize all of these results, we have the following regimes, which we summarize in Fig. 3. For , the single mutants are effectively neutral; these are the regimes labeled “Neutral” in Fig. 3 (the numerical factors in the boundaries to the regimes follow from our formal analysis below). In this effectively neutral case, for large (but not enormous) N we have (the “neutral stochastic tunneling” regime in Fig. 3), and this transitions to p1 ~ 1/N for smaller populations where (the “neutral sequential fixation” regime in Fig. 3). (Note that none of our analysis thus far has addressed the large-N “neutral semi-deterministic” and “neutral deterministic” regimes shown in the figure; we discuss these in section IV.E below). For larger , we have , as long as μ1s is not too small. This is the “deleterious tunneling” regime in Fig. 3. As μ1s approaches 0, this case becomes slightly more complicated, and is discussed below (this is the “deleterious sequential fixation” regime in Fig. 3). The large-N neutral and deleterious stochastic tunneling regimes were studied earlier by Iwasa et al. (2004b) and Serra and Haccou (2007), while the large-δ1 regimes were analyzed by Weinreich and Chao (2005), although these earlier studies did not explore the full parameter space nor the transitions between regimes.
FIG. 3.
Schematic of the different parameter regimes for the case K = 2. In the sequential fixation regime, the population usually crosses the valley by first fixing the single-mutants. For N∣δ1∣ < 1 (the neutral sequential fixation regime), the single-mutants are approximately neutral, and the expected time for the beneficial double-mutants to establish is just τ ≈ 1/μ0. For larger Nδ1 (the deleterious sequential fixation regime, between the dotted and solid lines), the process is slowed by selection against single-mutants, and τ ≈ 1/Nμ0ρ1, where ρ1 is given by Eq. (29). For larger values of N or δ1, the valley-crossing dynamics are dominated by tunneling. In the neutral stochastic tunneling regime, τ is given by the neutral limit in Eq. (28), while in the deleterious tunneling regime, τ is given by the strongly-deleterious limit of Eq. (28). In the neutral semi-deterministic tunneling regime, the initial increase of the single-mutant class is approximately deterministic, and τ is given by the first line of Eq. (35). In the deterministic regime, the dynamics are fully deterministic, and τ is given by Eq. (37). Note that the transitions between regimes are not sharp, but represent smooth crossovers. Note also that the parameter regimes describing weakly deleterious intermediates also apply to weakly beneficial intermediates (the region of the figure where δ1 < 0).
Thus far we have only considered the probability that a single-mutant lineage will give rise to a double-mutant that establishes. We must also understand the rate at which the single-mutant lineages arise in the first place, and the amount of time that it takes such a lineage to produce a double-mutant, given that it is destined to do so. The former is simple: single-mutants arise as a Poisson process at rate Nμ0. Since each single-mutant has a probability p1 of being successful, the expected time until the first successful single-mutant is produced is generations. In a large population with neutral or weakly deleterious intermediates, this is . We have seen that in this regime the successful single-mutant lineage will typically survive of order generations, and produce the first successful double-mutant after this time. Thus the total expected time for the successful double-mutant to be produced is . Similar calculations apply to the other regimes described above. This expression is only valid for Nμ0 < 1; if single-mutants are produced more frequently, it is likely that a second successful single-mutant lineage will arise while the first is still drifting, so we can no longer assume that only the first successful lineage matters. We analyze this more carefully in section IV.E below.
We can understand more complex multi-hit processes by iterating the above analysis. For example, consider a three-hit beneficial mutation (K = 3) with neutral intermediates. A double-mutant will produce a successful beneficial triple-mutant with probability . Thus in a large enough population a single-mutant will produce a double-mutant destined to produce a successful triple-mutant with probability
| (7) |
The population size must be large for this to obtain: we need . For slightly deleterious mutations the result is the same, although now the single-mutant must be very close to neutral for this to hold. When these conditions are met, however, this three-hit process is only a factor of (μ2/s)1/4 more improbable than the two-hit one, rather than the naive guess that it would be a factor of μ2 more improbable. We describe these more complex processes in more detail in our rigorous analysis below.
Finally, we note that although we have drawn a clear line between one-hit and multi-hit processes, the actual distinction is much less sharp. Take for example the case of two-hit processes, K = 2. We showed above that for weakly deleterious intermediates, the behavior is identical to the case of neutral intermediates. In fact, the argument we made there also applies to the case where the intermediates are weakly beneficial. If single-mutants have some advantage σ1, then when , the fact that the intermediate is advantageous is not felt before the double-mutant arises. Thus our neutral result still applies, and even though each mutation is independently beneficial, the dynamics are still those of a two-hit process with neutral intermediates. This is reflected in the region where δ1 < 0 in Fig. 3. Similar behavior holds for the more complex multi-hit situation, although the intermediates have to be closer to precisely neutral for the neutral results to apply.
IV. FORMAL ANALYSIS
We now turn to formal analysis, and rigorously derive and extend the results described heuristically above. We first focus on describing the fate of a given k-mutant lineage, using Laplace transforms to calculate the probability that this lineage will be successful for arbitrary selective coefficients and mutation rates. We then calculate the expected time that a successful k-mutant lineage will drift before producing the first successful (k + 1)-mutant. We next consider the entire trajectory of evolution, from the initial wild-type to the eventual fixation of the beneficial mutants, paying special attention to the case of beneficial double-mutants. In doing so, we describe the population sizes for which the beneficial mutants are more likely to establish via a tunneling process than via the sequential fixation through drift of the intermediate mutants.
A. The probability a mutation in a large population is successful
We begin by rigorously calculating pk, the probability that a k-mutant will be successful. In this section we focus on the case of a large population, where the frequencies of all intermediate mutations are always small compared to the total population size. We also assume that the absolute number of mutants is small enough that we can ignore competition between multiple potentially successful lineages; we will examine this assumption in section IV.E below. We use an argument based on the weights of mutant lineages; for an alternative derivation see Appendix C.
By definition, for k < K, a k-mutant individual will be successful if and only if one of its descendants is a successful (k + 1)-mutant. Thus pk will depend on pk+1, the probability that a (k + 1)-mutant individual will be successful. pk+1 will in turn depend on pk+2, and so on. We can therefore determine all the probabilities of success recursively, starting with the probability that a (beneficial) K-mutant individual will be successful, .
For a given k-mutant individual, let n(t) be the number of its k-mutant descendants in the population at time t (note that descendants that have accumulated additional mutations are not included in n). Each of these descendants has a probability μkdt of producing a (k + 1)-mutant in a time dt, and each of these mutants has a probability pk+1 of being successful. The probability that the chosen lineage will produce a successful mutant in the interval [t, t + dt) is therefore μkpk+1n(t)dt. This is a Poisson process, so the total probability that the lineage will produce a successful mutant before going extinct, given n(t), is 1–e−∫μkpk+1n(t)dt = 1–e−μkpk+1W, where W is the weight as defined above, W ≡∫n(t)dt. To obtain pk, we must average this over all possible values of W, giving
| (8) |
where P(W = w)dw is the probability that the weight is between w and w + dw.
We see that pk depends only on the expectation of the exponential of W. This makes it convenient to consider the Laplace transform φ of the probability density function, defined as
| (9) |
With this definition, we can rewrite Eq. (8) as
| (10) |
We calculate φ in Appendix A and find
| (11) |
Combining Eqs. (10) and (11), we find that pk is given by
| (12) |
The derivation of Eq. (11) assumes that N is large enough that the probability that k-mutants will drift to a high frequency is very small, so that such lineages contribute negligibly to the integral in Eq. (9). This requirement becomes more stringent as y approaches 0, so for very small μkpk+1 the population must be very large for Eq. (12) to hold. We will derive the explicit condition on N in section IV.D below.
For biological values of μk and pk+1, we will typically have μkpk+1 ⪡ 1, in which case Eq. (12) is well-approximated for any value of δk < 1 by
| (13) |
In the limits of neutral or strongly deleterious mutations, Eq. (13) simplifies further to
| (14) |
which agrees with the heuristic calculation given above.
So far, we have written pk in terms of pk+1, but we can apply Eq. (12) recursively to get an expression for pk involving only the selection coefficients and the mutation rates. Most important is p1, the probability that a given single-mutant will eventually give rise to the successful beneficial K-mutant. The general expression that this gives is not illuminating, but in the cases where the intermediate mutants are either all close to neutral or all strongly deleterious, we find the simple expressions
| (15) |
When K is large and all the intermediate mutants are close to neutral, note that the probability p1 that a single-mutant will be successful depends only weakly on the ultimate selective advantage s of the eventual beneficial mutants. This is because p1 is essentially the probability that a single-mutant lineage will drift to a very large size and generate many mutants, and it is relatively likely that the first such large lineage will drift to an enormous size, large enough so that at least a few descendants are likely to acquire a very large number of mutations. In contrast, when the intermediate mutants are strongly deleterious, the successful single-mutant lineage is likely to be small and produce just a few lucky mutant offspring, and the same is true for all the other successful intermediate mutant lineages. Since each k-mutant lineage must be roughly equally lucky, the selection coefficients and mutation rates at the end of the valley are just as important to p1 as δ1 and μ1.
Note that these results are only valid when the population size is large enough that intermediate mutants are always at low frequency in the total population. This condition is not particularly restrictive when the intermediate mutations are strongly deleterious. However, it can easily fail when the intermediates are close to neutral; in particular when K is large the population sizes required are often enormous. We calculate the population sizes needed for these results to hold, and describe what happens when they do not, in section IV.D below. These critical population sizes are also illustrated in Fig. 3, as the boundary between the “sequential fixation” and “tunneling” regimes. Further, our results above also require that population sizes not be too large. When they exceed a critical value, we must consider competition between different mutant lineages. This situation is the “neutral semi-deterministic tunneling” and “deterministic” regimes in Fig. 3; we analyze these in section IV.E below.
B. Time to the generation of the next mutant
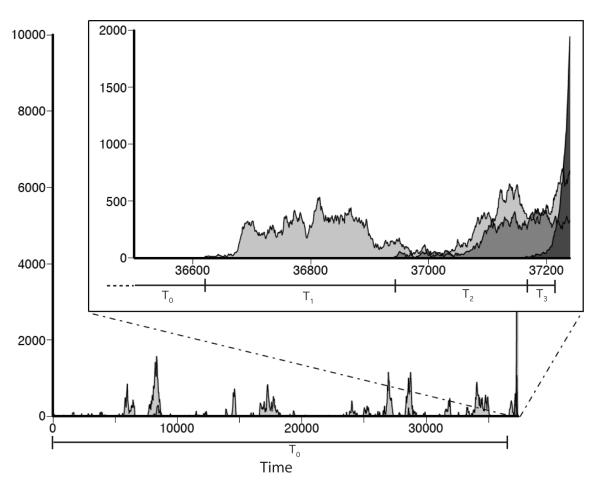
Thus far we have focused on the probability that a given mutation will be successful. What we are ultimately interested in, however, is the time Te that it takes for the beneficial multiple mutant to establish. We will focus on calculating the expected time, τ ≡ E[Te]. To do this, it will be helpful to write Te as , where T0 is the time for the first successful single-mutant to occur, Tk is the time that the first successful k-mutant drifts before producing the first successful k + 1-mutant, and TK is the establishment time of the first beneficial mutant, once it arises. An example of the dynamics and this division of Te for the case K = 3 is illustrated in Fig. 4. Defining τk ≡ E[Tk], we can write the total expected time as , and find τ by calculating τ0, τK, and τk. Note, however, that this division of Te is invalid at large population sizes because sometimes the first successful mutant will be overtaken by a later one; we describe this situation in section IV.E below.
FIG. 4.
A typical example of the dynamics by which a beneficial triple-mutant (K = 3) is acquired, from an individual-based computer simulation. Shown in light gray is the population size of single-mutants, in darker gray is the population size of double-mutants, and the darkest gray represents triple mutants. Inset is a magnified view of the last few hundred generations in this simulated dynamics. The waiting time T0 for the first successful single-mutant to arise is shown along with the time T1 this single-mutant drifts before giving rise to the first successful double-mutant, the time T2 this double-mutant drifts before giving rise to the first successful triple-mutant, and the time T3 for this triple-mutant to establish. We see that the total time until the beneficial mutant establishes (Te) is dominated by T0. The total population size is N = 105, the fitness parameters are δ1 = δ2 = 10−3, s = 0.1, and the mutation rates are μ0 = 10−6, μ1 = μ2 = 10−4, which corresponds to a neutral stochastic tunneling regime.
We begin by calculating τ0. Making the assumption that the population is dominated by wild-type individuals until the beneficial mutation arises, new single-mutant lineages are generated at a roughly constant rate Nμ0 (we discuss the situation when this assumption is invalid in section IV.D below). Since each of these lineages has a probability p1 of being successful, T0 will be exponentially distributed with expectation .
The expected time τK for a successful beneficial mutant lineage to establish has been analyzed by Barton (1998) and Desai and Fisher (2007), who found , where γ = 0.577 … is Euler’s constant.
We now turn to calculating τk, the expected time for which a successful k-mutant lineage will drift before producing the successful (k + 1)-mutant. To do this, we calculate the probability pk(t) that the lineage will be successful by time t (i.e., the probability that a k-mutant lineage will produce a successful (k + 1)-mutant after drifting for no more than time t). Just as pk depends on the expectation of the exponential of the weight W of the lineage, pk(t) depends on the expectation of the exponential of the weight accumulated by time t, defined as . The probability of success by time t is then
| (16) |
Again, we see that it is natural to use a Laplace transform for W(t) to calculate pk(t). We thus define the Laplace transform ϕ(y, t) ≡ E[e−yW(t). In Appendix A, we find that ϕ(y, t) is given by
| (17) |
where the dependence on y is contained in a±, which are defined as
| (18) |
Combining this with Eq. (16) gives an explicit expression for pk(t):
| (19) |
with y = μkpk+1 in a±. Note that as t increases, W(t) approaches W, and ϕ and pk(t) approach the values from the previous section: limt→∞ ϕ(y, t) = φ(y) and limt→∞ pk(t) = pk.
From pk(t), it is straightforward to find τk. The cumulative distribution function for Tk, the time for a successful k-mutant lineage to produce a successful (k + 1)-mutant, is just pk(t)/pk. Since Tk is a continuous positive random variable, its expectation is given by the integral of the complement of this cumulative distribution function:
| (20) |
Inserting the values from Eq. (12) and Eq. (19) into Eq. (20) and performing the integration gives
| (21) |
For biological parameter values (μkpk+1 ⪡ 1), this is
| (22) |
When all the mutations are either nearly neutral or strongly deleterious, this simplifies to
| (23) |
Note that Eq. (23) agrees with our earlier heuristic arguments that a successful neutral k-mutant lineage should drift for approximately generations, and that a successful deleterious k-mutant lineage is likely to have survived for approximately 1/δk generations.
Putting these results together, we find that the total expected time until the beneficial mutants establish is
| (24) |
where τk is given by Eq. (21). As we will describe in section IV.E below, Eq. (24) is only valid when τ is dominated by τ0, the expected waiting time until the first successful single-mutant occurs. This is the situation considered by Weinreich and Chao (2005) and Iwasa et al. (2004b). As can be seen from our expressions above, a sufficient condition for this to be true is Nμ0 ⪡ 1. Below we will derive alternative expressions for τ valid in larger populations, Nμ0 ⪢ 1.
C. The case K = 2, beneficial double mutants
Many of the above complex expressions are easier to understand in the simplest possible case, K = 2. In this case, p2 = s, and the chance that a single mutant individual will be successful is (from Eq. (13))
| (25) |
| (26) |
Eq. (25) agrees with the result (10) in Iwasa et al. (2004b), , to leading order in δ1 and μ1s. The small- and large-δ1 approximations thus agree as well.
The expected time until the spread of the beneficial mutants is (from Eqs. (23) and (24))
| (27) |
| (28) |
In Eq. (28), we have used the fact that τ1 and can be neglected for Nμ0 ⪡ 1. Note that the strongly-deleterious approximation reduces to Eq. (2) in Weinreich and Chao (2005), (where we have adjusted for a haploid population) when the mutation rates are equal, μ0 = μ1 ≡ μ.
D. Smaller population sizes
So far we have assumed that N is large enough that intermediate mutations never become a substantial fraction of the total population before the beneficial mutants start to spread. This was implicit in our calculation of τ0, as well as in the branching process approximation we used to calculate the rest of the τk. For smaller N, we can no longer treat mutant lineages as independent, so the branching process approximation breaks down. In addition, it becomes likely that intermediate mutants will drift to fixation before the beneficial mutants are generated; the dynamics shift from being dominated by stochastic tunneling to being dominated by sequential fixation. In this section, we calculate the value of N that separates these two regimes, and find the rate at which beneficial mutations are produced in the small-population, sequential-fixation regime.
For simplicity, first consider the case where double-mutants are beneficial, so that the probability p1 that a single-mutant individual is successful via tunneling is given by Eq. (25). A given single-mutant individual has probability
| (29) |
of giving rise to a lineage that will drift to fixation. If p1 ⪢ ρ1, then stochastic tunneling will dominate the dynamics, and the assumption we made in deriving Eq. (12) that lineages which drift to fixation make only a negligible contribution to p1 will necessarily be valid. If ρ1 ⪢ p1, then the single-mutant genotype is likely to dominate the population before the first successful beneficial mutant is produced. The transition p1 = ρ1 occurs at the population size N×, where
| (30) |
| (31) |
This threshold population size N× is the boundary between the tunneling and sequential fixation regimes shown in Fig. 3. Note that the second expression for N× in Eq. (31) is always smaller than the first, so is always a sufficient condition for tunneling to be more likely than fixation of single mutants. Again, this agrees with our intuition that if a lineage drifts to size ~ , it will achieve a weight ~ 1/μ1s, and will therefore be likely to have generated a successful double mutant. If δ1 ⪡ 1, the second expression in Eq. (31) reduces to (this is the boundary between the deleterious sequential fixation and deleterious tunneling regimes in Fig. 3), in agreement with the result (4) of Weinreich and Chao (2005), adjusted for a haploid population. Intuitively, we can understand this result by noting that if N < 1/δ1, the single-mutant is effectively neutral and can drift to fixation, and that even if N ~ 1/δ1, so that the single-mutant is slightly deleterious, it can still fix before generating a successful double-mutant individual if the rate of producing successful double mutants, μ1s, is small enough (this is the “deleterious sequential fixation” regime in Fig. 3).
The expected waiting time τseq for the beneficial mutants to establish via fixation of the intermediate mutants is approximately the sum of the expected time for a single-mutant lineage destined for fixation to be produced and the expected time for a successful double-mutant lineage to arise on a background of single-mutants:
| (32) |
Note this equation assumes no back-mutations which would take the population back to the original wild-type, though it would be straightforward to include these (this would increase τseq by a factor of roughly two if the forward and back mutation rates were the same). In practice, τseq will be dominated by either the first or second term; when the two are comparable we will typically be in a parameter regime where Eq. (32) does not apply. Note that since τseq is only relevant in small populations, the time for the single-mutants to drift to fixation after being produced and the time for the successful double mutants to establish after being produced will generally be much smaller than the terms we have included in Eq. (32). It is straightforward to show that as the population size approaches the threshold value N×, the expected waiting time τseq approaches the value τ found for the parameter regime where tunneling is more likely, as long as we assume N×μ0 ⪡ 1 so that Eq. (27) for τ is valid. Thus, the two parameter regimes join smoothly together.
For larger values of K, Eq. (30) and Eq. (31) for the threshold population size are still valid, provided we replace s by p2. However, for K > 2, N× only describes the population size above which the first successful double-mutant individual is likely to be produced before single-mutant individuals dominate the population, while the dominant valley-crossing dynamics may involve a combination of some intermediate mutants fixing and others succeeding via tunneling. Thus even for N > N×, it may still be likely that intermediate mutants reach a high frequency or even fix before the first successful K-mutant individual is produced, and even if N < N× stochastic tunneling may still play an important part in the valley-crossing process.
We will explicitly characterize the dynamics of the simplest case, in which all intermediate mutants are neutral. In this case, a single k-mutant has a probability ρk = 1/N of producing a lineage which drifts to fixation. If pk > 1/N for a given k < K, then the successful k-mutant lineage is likely to produce the successful k + 1-mutant lineage before it drifts to fixation, and our derivation of Eq. (12) is valid. Thus there is a sequence of K – 2 threshold population sizes for 1 < k < K, with . For N between and , the first k mutations are likely to drift to fixation, after which it is likely that the beneficial mutants will establish via stochastic tunneling before the k + 1-mutants can drift to fixation. The total expected time for populations with size between and to cross the valley is approximately the sum of the expected times for each of the first k successful mutant lineages to be produced once the preceding genotype is fixed in the population, along with the expected time for the successful k + 1-mutant lineage to be produced after the k-mutants drift to fixation:
| (33) |
Here we are ignoring the relatively small times for successful mutant lineages to drift to fixation or produce the next successful mutant lineage by tunneling. The generalization to arbitrarily deleterious intermediate mutants is straightforward, although the effect of the fixation of deleterious mutants on the mean fitness must be taken into account, as well as the possibility that there may be population sizes for which the dominant dynamics involve tunneling through lower-order mutants with higher-order mutants drifting to fixation.
E. Larger population sizes
Eq. (24) for the total expected time for the establishment of the beneficial mutants implicitly assumes that the first successful mutant lineage will be the one that eventually dominates the population. But if the population size is sufficiently large, multiple lineages may compete against each other. In particular, the first lineage that would have been successful in the absence of competition may be superseded by a later lineage that happens to produce beneficial mutants unusually quickly (i.e., in much less than the mean time ). This will occur with an appreciable frequency if the expected time for a successful lineage to drift before the beneficial mutation establishes is larger than the expected time for a successful lineage to arise: . In this section, we explore the values of N for which our approximation is valid, and describe what happens when it is not. We restrict our analysis to the case K = 2 for simplicity.
For Nμ0 ⪡ 1, we have seen above that , so no more than a few mutant lineages will typically be present in the population at a given time. Thus the first successful lineage will produce beneficial mutants that establish before another successful lineage can arise and overtake it. Our approximation above is valid, and the dynamics of the system are characterized by the time τ derived above.
We now consider what happens for larger values of N. For Nμ0 ⪢ 1, the total number of single mutants at time t, which we will denote n1(t), is well-approximated by its expectation (Fisher, 2007). Thus we can solve the deterministic differential equation for n1(t) to obtain
| (34) |
where the first line of Eq. (34) corresponds to effectively neutral single-mutants, and the second to deleterious single-mutants in mutation-selection balance. Double-mutants will be produced at the (time-dependent) rate R = n1(t)μ1.
If this rate R remains less than 1 until after the first double-mutant lineage establishes, then typically that first established lineage will dominate the population, and we can find the expected time, τsd, for this lineage to arise using an analysis similar to that used for single-mutant lineages above. That is, we define W1(t) to be the total weight of single-mutants by time t, . Then the probability that a successful double-mutant lineage will have been produced by time t is p2(t) = 1 – e−μ1sW1(t). The expected time is therefore given by
| (35) |
where we have used Eq. (34) to derive the approximations in Eq. (35). Note that the expected establishment time in the case of deleterious intermediates is the same as found for Nμ0 ⪡ 1 in Eq. (28). However, the expected time for neutral intermediates is different, and the threshold value of δ1 below which single-mutants are effectively neutral is larger by a factor proportional to . We refer to this large-N neutral regime as the “neutral semi-deterministic tunneling” regime in Fig. 3 (the subscript in τsd refers to “semi-deterministic”). There is no corresponding deleterious semi-deterministic tunneling regime, since as we have just seen in the deleterious case the establishment time is the same as it is for Nμ0 ⪡ 1.
As mentioned above, Eq. (35) is only valid when double-mutants are produced rarely enough that the first successful double-mutant lineage will dominate the population (i.e. R < 1). From the time when this first successful lineage arises, it takes ~ γ/s generations to establish, after which it has a doubling time of ~ log 2/s, while new successful double-mutant lineages are being produced at a rate ~ n1(τsd)μ1s. So a second double-mutant lineage will be likely to establish and make a significant contribution to the total double-mutant population if . Note that this is the same as the condition for being able to treat the double-mutant population deterministically, R = n1(τsd)μ1 ⪢ 1. Using our expression for τsd above, we see that Eq. (35) is valid for N < Ndet, where
| (36) |
This Ndet is the boundary between the “deterministic” and “neutral semi-deterministic/deleterious tunneling” regimes in Fig. 3.
In the deterministic regime, where N > Ndet, multiple lineages of double-mutants are produced very quickly, and the total number of double-mutants is well-approximated by its mean. By solving the deterministic differential equation for the number of double-mutants, we find that this mean is . We can use this to calculate the time for the beneficial mutants to establish, τd (here the subscript refers to “deterministic”), which is the time at which . We find
| (37) |
Note that τd < 0 for sufficiently large Nμ0, reflecting the fact that in extremely large populations there will be even more double-mutants at long times than would be expected from a single successful lineage arising immediately at t = 0 (see Desai and Fisher (2007) for a discussion of these subtleties in the definition of the establishment time).
Combining this expression for τd with Eq. (28) for τ, Eq. (32) for τseq, and Eq. (35) for τsd, we now have a complete description of the typical trajectories of populations with K = 2 for all biological values of N, μ0, μ1, δ1, and s. This is shown in Fig. 3. As a function of N, there are four regimes. For very small values of N, N < N× (as defined in Eq. (30)), the mutations fix sequentially and the beneficial mutant establishes after a time τseq, as given by Eq. (32). These are the neutral and deleterious sequential fixation regimes in Fig. 3. For N× < N < 1/μ0, our main analysis applies and the beneficial mutants establish after a time τ as given by Eq. (24). These are the neutral stochastic tunneling and deleterious tunneling regimes in Fig. 3. For 1/μ0 < N < Ndet, we can treat the single-mutants deterministically but still require a stochastic analysis of the beneficial mutants; this is the neutral semi-deterministic tunneling regime in Fig. 3, as well as the large-N part of the deleterious tunneling regime. In this semi-deterministic regime, the beneficial mutants establish after a time τsd, as given by Eq. (35). Finally, for N > Ndet, the analysis is fully deterministic and the beneficial mutants establish after a time τd, as given by Eq. (37). This is the deterministic regime in Fig. 3.
For larger values of K, there are yet more possible regimes. In these cases, when Nμ0 ⪡ 1, the time for the beneficial mutants to establish is given by Eq. (24), the extension of Eq. (32), or Eq. (33) (or its generalization for deleterious intermediate mutants), as described above. When N is larger than this, however, an analysis in the spirit of this section is necessary. In general, there can be a regime where single-mutants are treated deterministically but stochastic analysis is required for the rest, a regime where single and double-mutants are treated deterministically but stochastic analysis is needed for the rest, and so on. Note that there may be some regimes where population is large enough that some mutant classes must be treated deterministically, but also small enough that some intermediate mutants are likely to fix. We do not enumerate all the possibilities here, but these cases can all be analyzed using the approach we have developed above.
V. SIMULATIONS
To complement the analytical results described above, we performed stochastic individual-based computer simulations of our model. We focused on the cases K = 2 and K = 3, and verified our results for the time τ it takes for the population to acquire the K-hit adaptation, across a range of population sizes, mutation rates, and fitnesses of the intermediates.
To implement our simulations, we evolved a simulated population using time steps of dt = 10−2 generations. At the beginning of each time step, the mean fitness of the population was calculated, after which each k-mutant individual divided into two k-mutants with probability , produced a (k + 1)-mutant with probability μkdt, and died with probability dt. These three events occurred simultaneously and independently for each individual, for a total of 3N independent events per time step. If the population size N* at the end of a time step was different from N, then the population size was normalized by multiplying the size of each mutant class by N/N*, rounding to the nearest integer, and then adjusting the number of individuals in the largest class to bring the total number of individuals to exactly N. Each simulation run continued until the population consisted entirely of K-mutants. At the end of the run, the last time at which there were no K-mutants was recorded, and taken to be the time of the production of the first successful K-mutant. We ran 500 independent simulations for each set of parameters, and averaged these results to produce each data point shown in Fig. 5 and Fig. 6.
FIG. 5.
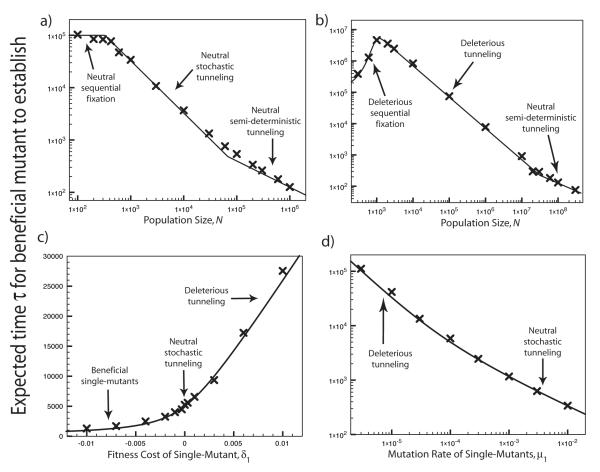
Comparison of theoretical predictions for τ, the expected time for the beneficial mutants to establish, to simulation results. These plots are for the case K = 2, with double-mutants having selective advantage s = 0.1. The crosses are the simulation results, while the solid curves show the theoretical predictions. The different parameter regimes are indicated on the plots. In (a) and (b), population size N is varied while the other parameters are held constant. In (a), μ0 = 10−5, μ1 = 10−4, and δ1 = 2×10−4, so that the single-mutants are effectively neutral. In (b), μ0 = 10−5, μ1 = 10−6, and δ1 = 7 × 10−3, so that the single-mutants are strongly deleterious for intermediate population sizes. In (c), the selective disadvantage δ1 of the single-mutants is varied, with N = 105, μ0 = 10−6, and μ1 = 4 × 10−5. Note that the tunneling-based theoretical prediction Eq. (27) is accurate even for negative values of δ1, corresponding to beneficial single-mutants. In (d), the rate of mutation from single- to double-mutants, μ1, is varied, with N = 105, μ0 = 10−6, and δ1 = 3 × 10−3.
FIG. 6.
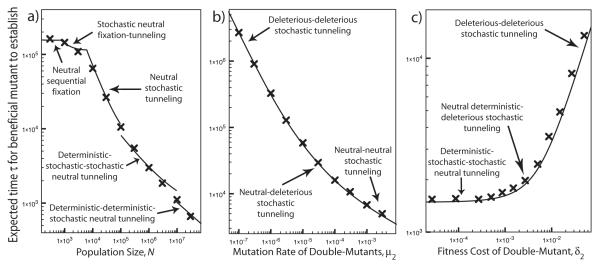
Comparison of theoretical predictions for τ, the expected time for the beneficial mutants to establish, to simulation results. These plots are for the case K = 3, with triple-mutants having selective advantage s = 0.1. The crosses are the simulation results, while the solid curves show the theoretical predictions. The different parameter regimes are indicated on the plots. In (a), population size N is varied while the other parameters are held constant at μ0 = 10−5 and μ1 = μ2 = δ1 = δ2 = 2 × 10−5, so that single- and double-mutants are effectively neutral. In the “stochastic neutral fixation-tunneling” regime, the most likely way for the population to cross the fitness valley is for single-mutants to drift to fixation, after which the triple-mutants establish through stochastic tunneling. In the “deterministic-stochastic-stochastic neutral tunneling” regime, the single-mutant class is treated deterministically and the higher mutant classes are stochastic. In the “deterministic-deterministic-stochastic neutral tunneling” regime, both single- and double-mutants are taken to be deterministic, with stochastic effects important only for the triple-mutants. The jumps in the theoretical prediction reflect points at which the different regimes do not fuse smoothly. In (b), the rate of mutation from double- to triple-mutants is varied, with N = 105, μ0 = 10−6, μ1 = 4 × 10−4, δ1 = 2 × 10−4, and δ2 = 5 × 10−3. The “neutral-deleterious stochastic tunneling” regime refers to a region of parameter space in which single-mutants are effectively neutral while double-mutants are strongly deleterious; the other parameter regimes are named analogously. In (c), the fitness cost of the double-mutant is varied, with N = 5 × ×105, μ0 = 5 × 10−5, μ1 = 3 × 10−5, μ2 = 5 × 10–, and δ1 = 8 × 10−4. The “neutral deterministic-deleterious stochastic tunneling” regime refers to a region of parameter space in which single-mutants are effectively neutral and increase in number roughly deterministically while double-mutants are strongly deleterious.
In Fig. 5, we compare our theoretical predictions for the time, τ, until the advantageous genotype establishes to our simulation results for the case K = 2. Our theoretical results are in excellent agreement with the simulations across a range of population sizes N, fitness costs of the single-mutant δ1, and mutation rate of the single mutant μ1. Note in particular that our theory accurately describes the transitions between the sequential fixation, tunneling, and semi-deterministic regimes as a function of N (Fig. 5a-b), and the transition between the neutral tunneling and deleterious tunneling regimes as a function of δ1 and μ1 (Fig. 5c-d). However, right at the transition Nμ0 ≈ 1 between the neutral stochastic tunneling regime and the semi-deterministic regime, our predictions are only accurate to about 30% (Fig. 5a). This is presumably because in this regime, both stochastic fluctuations in the number of single-mutants and competition between mutants are important.
In Fig. 6, we compare our theoretical predictions to the results of our simulations for the case K = 3. Once again, our theoretical results are in good agreement with the simulations, and accurately describe the transitions between the various regimes described by our analysis.
VI. DISCUSSION
Our analysis has provided a complete description of the rate at which an asexual population traverses a specific path through genotype space, involving fitness valleys or plateaus, to a particular fitter genotype. In general, however, there can be several different possible paths to the same final genotype. More interestingly, there could be many different fitter genotypes that are several mutations away from the original wild-type, with different paths leading to each.
In each such complex situation, the probability that evolution proceeds along any particular pathway or finds any particular beneficial genotype depends on the mutation rates of selective pressures involved — that is, on the local structure of the fitness landscape. Since very little is known about this structure, we cannot say which of the various modes of evolution (possibly involving various degrees of valley-crossing) are most important in nature. Instead, we will aim to describe a range of qualitatively different types of evolutionary behavior that are possible, and to understand which aspects of the structure of fitness landscapes are key to determining which of these different modes are important in nature. As we will see, this leads to some surprising insights — for example, that valley-crossing processes could be quite routine even when directly uphill pathways are also possible.
In order to address these questions, we must extend our analysis of simple evolutionary trajectories, in which only there is only one possible pathway to one possible beneficial genotype, to describe more complex situations with several possible pathways to several possible beneficial genotypes. Fortunately, each of these more complex possibilities can be broken down into multiple possible simpler evolutionary trajectories, of the type we have analyzed above. Thus our analysis provides a toolbox for studying these more complex situations. Note that in principle our earlier results also allow us to consider the case where one of the possible mutations increases the mutation rates at the other loci, as for example is common for some forms of cancer, although we will not explicitly discuss this situation here (Lengauer et al., 1998).
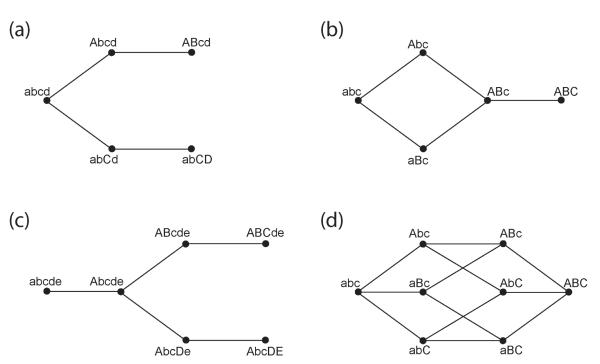
It may initially seem that the rate at which a population acquires a particular favorable genotype is simply the sum of the rates at which the population traverses all the possible pathways to that genotype. However, this is not the case. Instead, the overall rate involves a complicated combination of the probabilities of each possible path and the fitnesses of all the different intermediates. The basic types of situations that can arise are illustrated in Fig. 7. The essential rule is that whenever two pathways are entirely disjoint, the overall rate at which the population acquires an adaptation is the sum of the rates for each pathway. The same is true for pathways that overlap (i.e. share intermediate genotypes) without branching, or that branch (i.e. an intermediate genotype can mutate into two or more different further intermediate genotypes) at genotypes that are strongly deleterious.
FIG. 7.
Characteristic situations where there are several different pathways to a final advantageous genotype or genotypes. In each panel, the initial genotype is at the left, the final advantageous genotype or genotypes at the right, and all intermediate genotypes are assumed to be neutral or deleterious. In (a), the population initially has genotype abcd, and there are two entirely disjoint pathways by which evolution can proceed: either mutation A and then mutation B can occur, or mutation C and then mutation D. The overall rate at which a population acquires one of the two fitter genotypes equals the sum of the rates of acquiring each separately, regardless of the fitness of the intermediates. The probability the population acquires genotype AB before genotype CD is given by their relative rates. In (b), the population can acquire mutations A or B in either order, and then mutation C. Again, the overall rate equals the sum of the rates for the two possible pathways independently. In (c), the potential evolutionary trajectory branches after the population acquires mutation A. The population can then acquire mutations B and C, or mutations D and E, to reach an advantageous genotype. If genotype Abcde is strongly deleterious, the overall rate is the sum of the rates for each pathway. If Abcde is effectively neutral, the overall rate is lower than the sum of the rates for each pathway. In (d), the three mutations A, B, and C can be acquired in any order to reach advantageous genotype ABC. Because there are intermediate branching points, the overall rate is lower than the sum of the rates for each possible pathway if the single-mutants are effectively neutral.
On the other hand, when a branching point is effectively neutral, the behavior is more complex. Imagine for example a given intermediate genotype A, where mutations to genotypes B and C are possible, with rates μB and μC respectively. Each of these further mutations has some probability of eventually being successful, pB and pC respectively. If only mutation B were possible, the intermediate genotype would have to drift to a size to be likely to give rise to a successful mutant. This would occur with probability . Since both mutations are possible, the intermediate genotype only has to drift to a size , which occurs with probability . Note that the difference between the probability of reaching size nBC and that of reaching size nB is smaller than the difference in the sizes themselves. Essentially, the intermediate A can drift to a much smaller size and still be successful, but this is only increased in probability by the square root of the ratio of the sizes. Thus the rate of the overall process is only the square root of the sum of the rates of the two processes.
Several specific examples help illustrate the behavior. We begin by considering the situation where there is only one advantageous genotype possible, separated from the original wild-type by K mutational steps. Because the mutations can occur in different orders, there are multiple paths to acquire this fitter genotype — since K mutations are needed, there are a total of K! possibilities. It will prove useful to compare in each case the rate at which the population acquires the advantageous genotype to the rate if only one of the K! pathways were possible (i.e. the mutations had to be acquired in one specific order).
The simplest case is when all the K! possible intermediates are deleterious with the same sufficiently large δ, and each possible mutation occurs with the same rate μ. Then our earlier analysis applies, with δi = δ for all i between 1 and K — 1, and the mutation rate μk = (K – k)μ. The result (from Eq. (15)) is that the rate at which the fitter genotype is acquired is increased by a factor of K! relative to the case where each of these mutations has to be acquired in one specific order. In other words, the overall rate is just the sum of the rate of each pathway. More generally, as long as all possible intermediates are sufficiently deleterious, the rate at which the fitter genotype is acquired is the sum over that for all the possible paths, even if the fitnesses of the intermediates (and possibly the mutation rates) depend on the order of the mutations.
If some of the intermediates are effectively neutral, the behavior is more subtle. For example, if all the intermediates are neutral with each mutation having the same rate, μ, one again can use our earlier analysis to include the effects of all the possible orderings, as this is equivalent to having μ0 = Kμ, μ1 = (K – 1)μ, …, μK–1 = μ. This yields an overall enhancement of the fixation probability by a factor of K(K – 1)1/2(K – 2)1/4 … 21/2K–2 (again from Eq. (15)), relative to the case where the mutations can only be acquired in one particular order. This factor is no larger than K(K – 1), and hence much less than the K! we would expect if the rates for each of the possible pathways could be simply added together.
Although the scenario of a single beneficial genotype with moderately large K may indeed be relevant in many situations involving high mutation rates, more interesting are situations in which many different beneficial genotypes are within reach. If each of these involves completely distinct, non-overlapping sets of mutational changes, then the probabilities of any of them reaching fixation are independent, under the conditions on the population size for which our primary results obtain. The relative likelihood of each possible result is thus given by the ratio of the rates for each given by our earlier analysis. On the other hand, if the mutational paths to the beneficial genotypes overlap, the behavior is more complicated.
Consider first a simple example for which a particular neutral or deleterious first mutation enables many, say M, different conditionally beneficial second mutations. If each of the second mutation rates are the same, μ1, and the fitnesses of the beneficial double-mutants are also the same, the rate at which the population acquires one of the beneficial genotypes is straightforward: in our earlier calculations this is equivalent to replacing μ1 by Mμ1. When the intermediate is sufficiently deleterious, the rate at which the population acquires one of the M possible beneficial genotypes is simply M times that for each alone. But for a neutral or weakly deleterious intermediate, this combinatoric factor is smaller — only . In this neutral case, if the second mutations have different rates μ1,j and selective advantages sj with j = 1 … M, then the probability a single mutant is successful at generating an established beneficial genotype is : note that the sum appears inside the square root because the neutral intermediate only needs to last long enough for one of the many possible second mutations to become established.
When more than two mutations are needed to reach the beneficial genotypes, general results can be obtained iteratively, but are very messy. The important features are encompassed by a few illustrative cases. If all the intermediates are sufficiently deleterious, then again the rates can be summed over all the paths (and over the final advantageous genotypes). But if the intermediates are close to neutral, this is not the case. Consider a situation where the possible evolutionary pathways form a branching tree. In this scenario, each mutation can give rise to M possible next mutations, each with rate μ. Each of these in turn give rise to M possibilities for the subsequent mutation, and so on until a beneficial genotype is reached. Whether or not this situation is broadly relevant depends on the structure of fitness landscapes, but it is quite plausible that in many situations each genotype has a roughly equal number of “promising” future directions that may lead to beneficial combinations of alleles. If we restrict the analysis to all the beneficial genotypes requiring K mutations from the initial state, there are MK possible paths from the initial genotype to a beneficial K-mutant. If all the intermediates have the same fitness, this is equivalent to the simple K-step process we have analyzed, with each μ replaced by Mμ. For sufficiently deleterious intermediates, this gives rise to an enhancement of the overall rate at which one of these beneficial genotypes is found by a factor of MK. For the neutral case, the overall fixation probability is N(Mμ)2(s/Mμ)1/2K–1 which is larger than the single path rate by a factor which increases only gradually with K: from M for K = 1 to M2 in the limit of large K.
In situations where multiple advantageous genotypes are possible, we can also ask which is most likely to be acquired first. This is straightforward: the probability that a particular genotype is acquired first is proportional to the rate at which it is established. Thus this seemingly abstract discussion of relative rates has broad implications for the way in which populations adapt. For example, the conventional assumption is that adaptation is far more likely to occur by single mutations which each increase the fitness, since double mutations or more complex processes are far less probable. Yet consider for example a situation in which one adaptation requires two mutations, while another requires only one to confer a benefit. It might intuitively seem that the latter is highly unlikely to fix before the former. Yet our analysis shows that this is not necessarily true. If a single-mutant is beneficial (K = 1, the one-hit process), it has a probability of establishing given that it occurs. In a large population, if a double-mutant is beneficial with weakly deleterious intermediates (K = 2), we have seen that the double-mutant has a probability of arising given that the initial mutation occurs. Thus the ratio of the probabilities of these events is of order , rather than the much smaller mutation rate for the second mutation, μ1. In other words, the two-hit process is not necessarily much more unlikely than the one-hit. In addition, it is crucial to consider the number of possibilities. The total number of possible double (and higher) mutations is enormously larger than the number of possible single mutations. Thus we might expect that there are more available beneficial two-mutation combinations than there are beneficial one-hit mutations, particularly if we are near a local fitness peak. Given this, it is entirely plausible that beneficial two-hit mutations arise faster than beneficial one-hit mutations, and hence populations could tend to acquire these more complex adaptations even when simpler one-hit adaptations are also possible. Shih et al. (2007) have found that this indeed seems to be case for influenza A hemagglutinin evolution.
Unfortunately, very little is known about what fraction of possible double, triple, and higher-hit mutations are likely to be beneficial, and hence what the differences in initial mutation rates are for these different types of events (or in the language of our previous discussion, what the value of M is). As we have seen, which types of events dominate evolution depends on this number of combinatoric possibilities and various other parameters in a complex way — for example, we have just seen that when intermediates are close to neutral, multiple-hit processes are not much less likely than single-hit ones, but at the same time the overall rate of these events does not increase linearly with the number of possibilities M, but only as . Since we have very little understanding of any of these parameters, it is not at all clear which types of events dominate evolution. What we have learned instead is what aspects of the structure of fitness landscapes determine the relative likelihoods of different types of evolutionary behavior. Better information about these aspects of the structure of genome space is sorely needed in order to understand how organisms, particularly microbes, adapt; we have discussed some of the open questions along these lines in Fisher (2007).
Given a particular structure of genome space, our results give some insight into how different populations will explore this space. We have seen that in small enough populations (in the sequential fixation or deleterious tunneling regimes), one-hit processes will typically be much faster than multiple-hit ones, even if there are many possible multiple-hit processes. Thus a small population will adapt by “choosing” among the possible single mutations that directly increase fitness. It will choose at random among these mutations (weighted by their establishment probabilities) if it is sufficiently small; if it is somewhat larger, clonal interference processes may allow it to tend to “choose” one of the best possible single mutations. Adaptation will progress by this series of individually beneficial steps, even if by doing so the population “misses” more-fit genotypes separated by neutral or slightly deleterious intermediates.
A larger population, because it is in a stochastic tunneling regime, can “see” further away in genotype space. In such a population, two-hit processes are easily found, and hence single mutations that offer small increases in fitness (or lead to genetic dead ends) will tend not to be fixed, in favor of double mutations that offer larger increases. Still larger populations can explore three-hit mutations, and so on. Thus the threshold population sizes we have calculated for transitions between regimes can be thought of as the characteristic sizes above which a population can “see” a step further in genotype space. Populations in different regimes will adapt and explore genome space in qualitatively different ways.
While this intuition is useful and may provide a basis for further work, it glosses over important subtleties. Most importantly, it envisions a population as inhabiting a “point” in genome space, moving from there to another point, and so on. In actuality, a large population can contain significant genetic diversity, and can spread out substantially among nearby neutral genotypes. This will be particularly true if there are a large number of paths leading to different potential adaptations, with Mμ0 larger than the rate at which beneficial mutations establish. Understanding these dynamics remains an important challenge, and will be necessary if we are to form a more complete understanding of how asexual populations adapt and explore genome space.
Acknowledgements
This research was supported in part by NIH Grant P50GM071508 to the Lewis-Sigler Institute, by NIH grant GM28016, by an NSF Graduate Research Fellowship, and by a Stanford Graduate Fellowship.
Appendix A: Laplace transforms
We wish to derive an expression for the Laplace transform of the probability density function of . As mentioned in the main text, φ, the Laplace transform of the probability density function of W, can then be found from ϕ by taking the limit as t goes to infinity. However, calculating ϕ(y, t) is difficult to do directly because W(t) is not a Markov random variable. An easier approach is to instead consider the two-dimensional (Markov) random variable (n(t), W(t)), and calculate . Once we have found φ, we can evaluate it at x = 0 to average over all values of n(t) and recover the Laplace transform for the weight: ϕ(y, t) = φ(0, y, t).
To derive φ, we will follow a procedure similar to that used by Kendall (1948). We first need to find an equation for the time evolution of the joint probability density of the lineage size n(t) and the weight W(t), which we denote by pt(n, w). For an infinitesimal time dt, pt satisfies
| (38) |
where the first term on the right-hand side is the probability that a death occurred in [t, t + dt), the second term is the probability of a birth, and the third term is the probability that neither a birth nor a death occurred. Rearranging terms, and using , we can rewrite Eq. (38) as a partial differential equation for pt:
| (39) |
By definition, , where we have defined pt(n, w) ≡ 0 for all n, w < 0. Differentiating both sides of this equation with respect to time, and using Eq. (39) to replace , we can find a partial differential equation for φ:
| (40) |
In deriving the last term in Eq. (40), we have used integration by parts:
since pt(n,±∞) = 0 for n ≠ 0.
We can solve Eq. (40) using the method of characteristics. If we write the characteristics as x(y, t), then they must satisfy
| (41) |
Solving this differential equation, we find that φ must depend on x and t only through , where a± are the roots in e−x of the right-hand side of Eq. (41):
| (42) |
Note that 0 < a− < 1 < a+. Since the lineage starts at time t = 0 with one individual (p0(n, w) = δn,1 δ(w)), φ must satisfy boundary condition φ(x, y, 0) = e−x, which gives
| (43) |
From this, the simpler Laplace transform of the probability density of W(t) follows immediately:
| (44) |
The Laplace transform of the probability density function of W follows from this:
| (45) |
Appendix B: Probability density of W
Although Eq. (45) is all we need to confirm the results of our original intuitive calculation, it does not by itself show that our argument (using the weights of lineages) underlying that calculation was correct. In order to do this, we must show first that the cumulative probability of a lineage achieving a weight of at least w goes like ~ for w ⪡ 1/δ2, and falls off rapidly for larger w. Equivalently, we wish to show that the probability density for the weight of a lineage, P(w), goes like P(w) ~ w−3/2 before falling off at 1/δ2. From this, we will also be able to show that, given a probability μσ per individual per unit time of producing a successful mutant, the typical weight for a successful lineage is ~ 1/(μσ) for , and ~ 1/δ2 for . (Here σ, the probability of success for a mutant individual, could represent an actual selective advantage s, or it could be the probability that the individual’s descendants will fix after acquiring additional mutations necessary for a selective advantage.)
We can find P(w) by taking the inverse Laplace transform of φ(y). Applying standard identities of Laplace transforms (see Arfken and Weber (1995), Tables 15.1 and 15.2) to Eq. (45), we obtain
| (46) |
where I1 is a modified Bessel function of the first kind. This exact result is valid for both positive and negative δ, although for δ < 0 (beneficial mutants), there will be a positive probability that the lineage achieves infinite weight, corresponding to fixation.
For w ⪢ 1, which includes all weights large enough to be relevant for δ ⪡ 1, Eq. (46) is well-approximated by the asymptotic expansion (see Arfken and Weber (1995), Table 11.2)
| (47) |
Assuming that δ ⪡ 1, we can Taylor expand the argument of the exponential to obtain
| (48) |
which exhibits exactly the behavior that we predicted, behaving like w−3/2 until falling off rapidly at w ~ 1/δ2.
To find the typical weight of a successful lineage, we note that the probability density of the weight of a successful lineage is
where the probability of success p is given by Eq. (12). Plugging in our approximate expression for P(w), we see that
| (49) |
This expression can then be integrated to give the cumulative distribution function. Although the exact expression is not illuminating, the asymptotics are exactly as predicted by our heuristic argument: for μσ ⪢ δ2/4, the cumulative distribution is dominated by weights w ≲ 1/μσ, while for μσ ⪡ δ2/4, it is dominated by w ≲ 1/δ2.
Appendix C: Alternative derivation of pk
Eq. (12) for pk can easily be derived without referring to weights by using a first-step analysis, although this derivation does not provide the same intuitive understanding, and does not provide an expression for τ. To perform the first-step analysis, consider a k-mutant individual, having a probability pk of success. There are four possibilities for the first event that will occur to this individual: with rate 1, it will die; with rate 1–δk, it will divide into two k-mutant individuals; with rate μkpk+1, it will produce a successful (k+1)-mutant; and with rate μk(1–pk+1), it will produce an unsuccessful (k+1)-mutant. In the first case (death), the individual has zero probability of being successful. In the second case (reproduction), each of the offspring has probability pk of being successful, for a total probability of 1 – (1 – pk)2 of success. In the third case (reproduction producing a successful mutant), the probability of success is, by definition, 1. In the final case, (reproduction producing an unsuccessful mutant), we can ignore the unsuccessful (k + 1)-mutant, leaving us in the original situation of a single k-mutant individual with a probability pk of success.
Equating the original probability of success to the probability of success summed over the four possible first events yields a quadratic equation for pk:
| (50) |
(The denominator of the right-hand side is the sum of the rates of the different possible first events.) Solving Eq. (50) for pk gives
the same expression derived via a more complicated calculation in Appendix A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Arfken GB, Weber HJ. Mathematical methods for physicists. fourth edition Academic Press; San Diego: 1995. [Google Scholar]
- Barton N. The effect of hitch-hiking on neutral genealogies. Genetical Research. 1998;72:123–133. [Google Scholar]
- Barton NH, Rouhani S. The frequency of shifts between alternative equilibria. Journal of Theoretical Biology. 1987;125:397–418. doi: 10.1016/s0022-5193(87)80210-2. [DOI] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. PNAS. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AJR, Wagner GP. Evolution of functionally conserved enhancers can be accelerated in large populations: a population-genetic model. Proceedings of the Royal Society B. 2002;269:953–960. doi: 10.1098/rspb.2002.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen FB, Otto SP, Bergman A, Feldman MW. Waiting with and without recombination: The time to production of a double mutant. Theoretical Population Biology. 1998;53:199–215. doi: 10.1006/tpbi.1997.1358. [DOI] [PubMed] [Google Scholar]
- Desai MM, Fisher DS. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett R, Schmidt D. Waiting for two mutations: With applications to regulatory sequence evolution and the limits of darwinian evolution. Genetics. 2008;180:1501–1509. doi: 10.1534/genetics.107.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W. Mathematical population genetics. second edition Springer-Verlag; New York: 2004. [Google Scholar]
- Fisher DS. Evolutionary dynamics. In: Bouchaud JP, Mezard M, Dalibard J, editors. Complex Systems. Volume LXXXV. Springer-Verlag; Amsterdam: 2007. (Lecture Notes of the Les Houches Summer School). [Google Scholar]
- Goh C-S, Bogan AA, Joachimiak M, Walther D, Cohen FE. Co-evolution of proteins with their interaction partners. Journal of Molecular Biology. 2000;299:283–293. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Michor F, Nowak MA. Evolutionary dynamics of invasion and escape. Journal of Theoretical Biology. 2004a;226:205–214. doi: 10.1016/j.jtbi.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Michor F, Nowak MA. Stochastic tunnels in evolutionary dynamics. Genetics. 2004b;166:1571–1579. doi: 10.1534/genetics.166.3.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. Sex and infinity: A mathematical analysis of the advantages and disadvantages of genetic recombination. In: Hiorns RW, Hiorns MSB, editors. The Mathematical Theory of the Dynamics of Biological Populations. Academic Press; New York: 1973. pp. 155–194. [Google Scholar]
- Karlin S, Tavare S. The detection of a recessive visible gene in finite populations. Genetical Research Cambridge. 1981;37:33–46. [Google Scholar]
- Kendall DG. On the generalized ”birth-and-death” process. Annals of Mathematical Statistics. 1948;19:1–15. [Google Scholar]
- Kimura M. The role of compensatory neutral mutations in molecular evolution. Journal of Genetics. 1985;64:7–19. [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nature Reviews Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BA, Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- Serra MC, Haccou P. Dynamics of escape mutants. Theoretical Population Biology. 2007;72:167–168. doi: 10.1016/j.tpb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Shih AC-C, Hsiao T-C, Ho M-S, Li W-H. Simultaneous amino acid substitutions at antigenic sites drive influenza a hemagglutinin evolution. PNAS. 2007;104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Chao L. Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution. 2005;59:1175–1182. [PubMed] [Google Scholar]