Abstract
Background
In a human T-cell acute lymphoblastic leukemia (T-ALL) cell line (Molt-4), siRNA-mediated suppression of BCL11B expression was shown to inhibit proliferation and induce apoptosis, functions which may be related to genes involved in apoptosis (such as TNFSF10 and BCL2L1) and TGF-β pathways (such as SPP1and CREBBP).
Methods
The expression levels of the above mentioned genes and their correlation with the BCL11B gene were analyzed in patients with T-ALL using the TaqMan and SYBR Green I real-time polymerase chain reaction technique.
Results
Expression levels of BCL11B, BCL2L1, and CREBBP mRNA in T-ALL patients were significantly higher than those from healthy controls (P <0.05). In T-ALL patients, the BCL11B expression level was negatively correlated with the BCL2L1 expression level (rs = -0.700; P <0.05), and positively correlated with the SPP1 expression level (rs = 0.683; P <0.05). In healthy controls, the BCL11B expression level did not correlate with the TNFSF10, BCL2L1, SPP1, or CREBBP expression levels.
Conclusions
Over-expression of BCL11B might play a role in anti-apoptosis in T-ALL cells through up-regulation of its downstream genes BCL2L1 and CREBBP.
Background
T-cell acute lymphoblastic leukemia (T-ALL) accounts for 15% of newly diagnosed ALL cases in children and 20-25% of ALL cases in adults [1,2]. Overall, these are aggressive malignancies that do not respond well to chemotherapy and have a poorer prognosis than their B-cell counterparts [3]. The development of targeted therapies, including monoclonal antibodies and gene therapy, continues. Small interfering RNA (siRNA) is a promising gene-targeting agent that has shown great potential, particularly in the field of cancer treatment [4-6].
The B-cell chronic lymphocytic leukemia (CLL)/lymphoma 11B (BCL11B) gene plays a crucial role in T-cell development, differentiation, and proliferation [7], and altered expression, mutation, disruption, or rearrangement of BCL11B have been associated with T-cell malignancies [8-11]. BCL11B over-expression has been observed primarily in T-cell malignancies [8,12]. BCL11B has been hypothesized to act as a tumor suppressor gene [9,13], but its precise function remains unclear.
BCL2-like 1 (BCL2L1; Bcl-xL) is similar to Bcl-2 because it restrains the apoptosis induction of multiple stimuli, and is a key factor in the terminal step of apoptosis regulation. Studies have shown that BCL2L1 participates in various protein-protein interactions, playing a role in inhibiting apoptosis. In the endogenous apoptosis pathway, BCL2L1 of the BCL-2 family inhibits apoptosis by blocking the translocation of Bax to the mitochondrial outer membrane [14]. cAMP-response element binding protein (CREBBP) plays a critical role in embryonic development, growth control, and homeostasis by coupling chromatin remodeling to transcription factor recognition. A CREBBP gene rearrangement with chromosomal translocation has been identified in acute myeloid leukemia [15,16] and over-expression of CREBBP was found in Jurkat cells. Additionally, enhancement of apoptotic cell death occurred in the presence of CREB1 siRNA [17]. Tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10; TRAIL) is a tumor necrosis factor superfamily member, and induces apoptosis through its interaction with death receptors. BCL-2 family genes and TNFSF10 probably act together through crosstalk between the intrinsic and death receptor-mediated apoptosis pathways [18]. Secreted phosphoprotein 1 (SPP1) is also known as OPN and its abnormal activation can stimulate tumor growth, invasion, angiogenesis, and immune suppression, with wide-ranging effects on cell proliferation, apoptosis, differentiation, and migration [19,20].
Previous studies [21,22] showed that the inhibition of BCL11B expression by siRNA selectively inhibited proliferation and effectively induced apoptosis in human T-cell acute lymphoblastic leukemia (T-ALL) cell lines (Jurkat, Molt-4). Additionally, global gene expression profiling revealed that BCL11B siRNA-mediated cell apoptosis may be related to BCL-2 family genes of the mitochondrial pathway, and the TRAIL (TNFSF10) gene of the death receptor signaling pathway [22], furthermore, in our previous study, the genes (SPP1 and CREBBP) of the TGF-β pathway (unpublished data). Little is known about the expression pattern of these genes in T-ALL. Thus, analyzing the expression pattern of these genes in malignant T-cells is important because BCL11B disruption and disturbed expression may contribute to the development of T-cell malignancies in humans [8]. In the present study, we further analyzed expression levels of TNFSF10, BCL2L1, SPP1, and CREBBP, and their correlation with BCL11B in male patients with T-ALL, to clarify the role of BCL11B in T-cell malignancies.
Methods
Samples
Nine newly diagnosed T-ALL patients (male, 6-28 years old; median age, 20 years; white blood cell count (WBC), 1.8-293.5 × 109/L; bone marrow blast percentage: 65-93%; were recruited. The diagnosis of T-ALL was based on cytomorphology, immunohistochemistry, and cytoimmunological analysis. Peripheral blood mononuclear cells (PBMCs) from nine healthy volunteers served as controls (five males and four females, 20-45 years old; median age, 28 years). Peripheral blood was collected by heparin anticoagulation and PBMCs were separated using the Ficoll-Hypaque gradient centrifugation method. The percentage of CD3+cells in PBMCs were detected, there are 75.30 ± 26.77% (range 21.2-97.8%) in PBMCs from T-ALL samples and 59.66 ± 4.75% (range 52.4-65.8%) in PBMCs from healthy control samples.
All procedures were conducted in accordance with the guidelines of the Medical Ethics committees of the health bureau of Guangdong Province, PR China.
RNA extraction and cDNA synthesis
RNA was extracted using the Trizol kit (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into the first-strand cDNA using random hexamer primers and the reverse transcriptase Superscript II Kit (Invitrogen), according to the manufacturer's instructions.
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Quantitative detection of the BCL11B gene expression level in cDNA from PBMCs was performed using TaqMan real-time PCR. PCR was performed as described previously [8]. To precisely determine the copy numbers of BCL11B, a duplex vector, including a fragment of the BCL11B and the β2 microglobulin (β2M) genes was constructed and used as a reference (the duplex vector was a gift from Prof. C.A. Schmidt, Ernst-Moritz-Arndt University Greifswald, Germany). Based on the DNA concentration, measured by spectrophotometry and confirmed by quantitative gel eletrophoresis, standard dilutions of the vector from 107 to 101 copies were prepared [8]. Briefly, PCR was performed in a 25-μL total volume containing 2 μL of cDNA, 25 pmol of each primer (BCL11B-f and BCL11B-b for BCL11B gene amplification; β2Mf and β2Mb for β2M gene amplification), 10 nmol of each dNTP, 1.5 U AmpliTaq Gold (Applied Biosystems, Branchburg, NJ, USA), 5 pmol of 6FAM-TAMRA probe, and PCR buffer containing 4.5 mM MgCl2. After an initial denaturation at 95°C for 5 min, 50 cycles consisting of 95°C for 15 s and 64°C for 1 min were performed. Primers and probes for BCL11B and β2M gene amplification were synthesized by TIB Molbiol Co. (Berlin, Germany; Table 1).
Table 1.
Sequences of primers and probes for real-time PCR (TaqMan method)
| primers/probes | sequence | function |
|---|---|---|
| BCL11Bf | 5'-CACCCCCGACGAAGATGACCAC | forward primer |
| BCL11Bb | 5'-CGGCCCGGGCTCCAGGTAGATG | backward primer |
| BCL11Bp | 5'-6FAM-TCACCCACGAAAGGCATCTGTCCCAAGCA-TAMRA | probe |
| β2Mf | 5'-CTCGCGCTACTCTCTCTTTCT | forward primer |
| β2Mb | 5'-TACATGTCTCGATCCCACTTAACTAT | backward primer |
| β2Mp | 5'-6FAM-CTCACGTCATCCAGCAGAGAATGGAAAGTCA-TAMRA | probe |
The absolute amounts of BCL11B and β2M were measured in two independent assays and BCL11B content per 100,000 β2M copies was calculated using the formula: n = 100000 × BCL11B/β2M.
Expression levels of TNFSF10, BCL2L1, SPP1, CREBBP, and the reference gene β2-MG were determined by SYBR Green I real-time PCR. Briefly, PCR was performed in a 25-μL total volume containing 1 μL of cDNA, 9 μL of 2.5× SYBR Green mix (Tiangen, Beijing, PR China), and 10 μmol/L primer pairs. The following cycling conditions were used: initial denaturation at 95°C for 2 min, followed by 44 cycles at 95°C for 15 s, and 81°C (TNFSF10, SPP1, CREBBP, and β-2-MG) or 84°C (BCL2L1) for 1 min. The relative amounts of the genes of interest and the β2M reference gene were measured in two independent assays. The 2(-ΔΔCT) method was used to present the data of the genes of interest relative to an internal control gene [23,24]. The efficiencies of real-time PCR for expression analysis of different genes were evaluated using diluted Molt-4 cDNA (1, 5-1, 5-2, 5-3, 5-4) as templates to construct relative standard curves. Additionally, the specific amplification of PCR products was analyzed by melting curve analysis and agarose electrophoresis. Primers used in the SYBR Green I real-time PCR for all four gene amplifications were synthesized by Shanghai Biological Engineering Technology Services Co., Ltd. (Table 2).
Table 2.
Sequences of primers for real-time PCR (SYB Green I method)
| primers | sequence | function |
|---|---|---|
| TNFSF10 | 5'-GAGTATGAACAGCCCCT-3' | forward primer |
| TNFSF10 | 5'-GTTGCTTCTTCCTCTGGT-3' | backward primer |
| BCL2L1 | 5'-AAACTGGGTCGCATTGTGG-3' | forward primer |
| BCL2L1 | 5'-TCTCGGCTGCTGCATTGTTC-3' | backward primer |
| SPP1 | 5'-ACAGCCAGGACTCCATTGA-3' | forward primer |
| SPP1 | 5'-TCAGGTCTGCGAAACTTCTTAG-3' | backward primer |
| CREBBP | 5'-CGGTTTCTCGGCGAATGAC-3' | forward primer |
| CREBBP | 5'-CATTTCCTATTCCTGGGTTGAT-3' | backward primer |
RT-PCR for TNFSF10, BCL2L1, SPP1, and CREBBP genes was performed using the same primers as described above, and the PCR products were sent to Shanghai Invitrogen Biotechnology Co. for DNA sequence analysis.
Statistical analyses
Independent-sample t -test analysis was used for the BCL11B gene mRNA levels in different samples, while the Mann-Whitney U test and Spearman's rank correlation analyses were used for non-normally distributed data using the SPSS 13.0 statistical software. Differences were considered statistically significant at P < 0.05.
Results
Over-expression of BCL11B gene in T-ALL
The expression level of BCL11B mRNA in PBMCs from patients with T-ALL (1821.81 ± 1896.58 copies/105 β2M copies) was significantly higher than that from healthy controls (259.71 ± 182.72 copies/105 β2M copies; t = 2.46; P = 0.039; Figure 1). PCR products from β2M and BCL11B genes were confirmed by 2.5% gel electrophoresis (Figure 2D, E).
Figure 1.
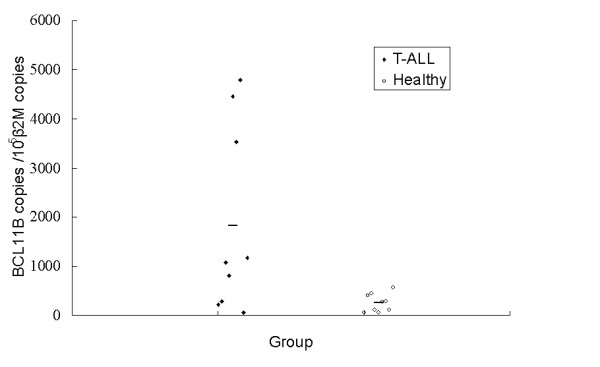
Expression levels of the BCL11B gene in PBMCs from T-ALL and healthy controls.
Figure 2.
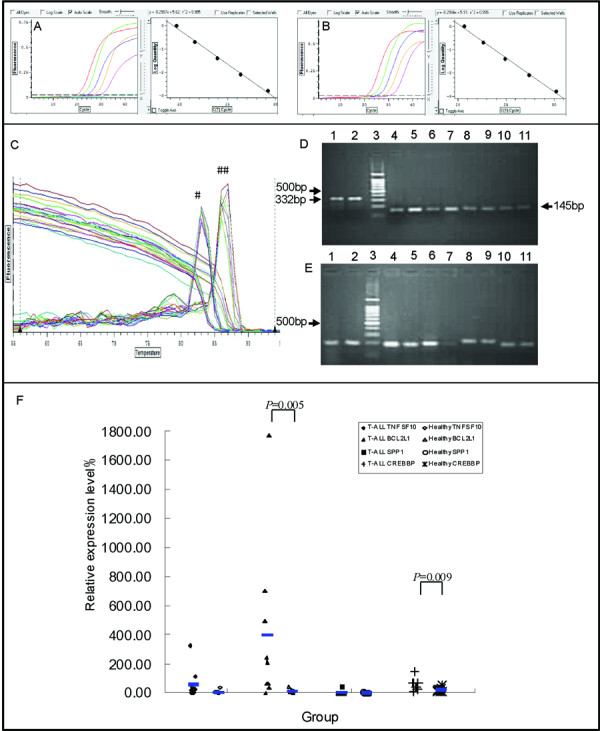
Features of the expression of TNFSF10, BCL2L1, SPP1, and CREBBP genes in T-ALL and healthy groups. A, B: Accurate standard curve graphs of BCL2L1 and the β2M control gene are shown using diluted Molt-4 cDNA as the template. The amplification efficiency of BCL2L1-related genes was more than 95%, and consistent with the high amplification efficiency of the β2M reference gene. C: Melting curves of the BCL2L1 and β2M genes from nine patients. #: Specific peak of the β2M reference gene begins at 81°C. ##: Specific peak of the BCL2L1 gene begins at 84°C. D: PCR products of the β2M gene by 2.5% agarose gel electrophoresis analysis. The size of the PCR products of the β2M gene used for the BCL11B reference is 332 bp (line 1, 2) and that used for the four genes of interest is 145 bp (line 4-11). Line 3: DNA ladder. E: PCR products analyzed by 2.5% agarose gel electrophoresis. Line 1-2: BCL11B (193bp), line 3: DNA ladder, line 4-5: BCL2L1 (202 bp), line 6-7: CREBBP (206 bp), line 8-9: SPP1 (241 bp), line 10-11: TNFSF10 (190 bp). F: Relative expression levels of the four genes of interest in T-ALL and healthy groups.
Expression of TNFSF10, BCL2L1, SPP1, and CREBBP genes in T-ALL
The high amplification efficiency of the four genes of interest (TNFSF10, BCL2L1, SPP1, and CREBBP) was consistent with that of the β2M reference gene. For example, the accurate standard curve graphs of BCL2L1 and β2M control gene amplification are illustrated in Figure 2A and 2B (r2 = 0.995). The amplification efficiencies of BCL2L1 and the β2M control gene were 95.30% and 95.16%, respectively, and the melting curves are shown in Figure 2C. PCR products from the β2M control gene and genes of interest were confirmed using 2.5% gel electrophoresis (Figure 2D, E), followed by sequence confirmation (data not shown).
Relative expression levels of BCL2L1 mRNA (397.82 ± 565.98%) and CREBBP mRNA (53.28 ± 39.21%) in patients with T-ALL were significantly higher than those from healthy controls (BCL2L1: 10.83 ± 11.18%; CREBBP: 20.80 ± 13.50%; P < 0.05), whereas the relative expression levels of TNFSF10 and SPP1 mRNA showed no significant difference between T-ALL and healthy groups (Figure 2F).
In T-ALL patients, Spearman's rank correlation analyses revealed that the BCL11B expression level was negatively correlated with the BCL2L1 relative expression level (rs = -0.700; P = 0.036; Figure 3A), and positively correlated with the SPP1 relative expression level (rs = 0.683; P = 0.042; Figure 3B). The BCL11B expression level did not exhibit an obvious correlation with TNFSF10 or CREBBP relative expression levels. No significant correlation was found between the BCL11B gene and the other four genes of interest in the healthy controls.
Figure 3.
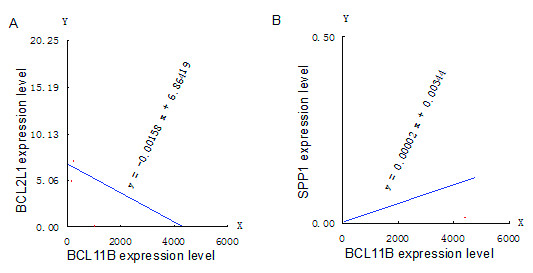
Linear correlation analyses of the BCL11B and BCL2L1 genes (A) and SPP1 gene (B) in T-ALL samples.
Discussion
Increasing numbers of translocations involving the BCL11B locus [8,10,11] or high levels of BCL11B mRNA expression in most T-ALL cases [8,12] have been reported; however, the mechanism of BCL11B-mediated oncogenesis remains unknown. To clarify the role of BCL11B in T-cell malignancies, we further analyzed the expression levels of TNFSF10, BCL2L1, SPP1, and CREBBP genes and their correlations with BCL11B in patients with T-ALL and controls. Over-expression of the BCL11B gene, as well as BCL2L1 and CREBBP mRNA, were characteristic features of T-ALL.
Recent evidence has suggested that multiple mechanisms may regulate the release of mitochondrial factors, some of which depend on the action of caspases. BCL2L1 may inactivate caspase-8 by decreasing death-inducing signaling complex (DISC) formation in the plasma membrane, nucleus, and Golgi complex while diverting DISC formation to the mitochondria. The inhibitory effects of BCL2L1 on DISC formation may play a significant role in protecting endothelial cells from hypoxia/reoxygenation (H/R)-induced cell death [25]. Thus, over-expression of the BCL2L1 gene suggests that it might be related to the occurrence of T-ALL by defective regulation of apoptosis. During the process of T-ALL, over-expressed BCL2L1 is thought to suppress the activity of caspase-8; thus, as a kind of protection mechanism, the TNFSF10 gene of some patients is highly expressed, promoting caspase-8 activity in response to this abnormal cell proliferation. However, the low expression level of SPP1 in untreated Molt-4 cells differed from the high expression levels found in mostly solid tumors [26]. Additionally, our findings indicated no significant difference in SPP1 gene expression in the T-ALL group. Comprehensive analysis revealed that T-ALL occurred in the presence of BCL11B, BCL2L1, and CREBBP gene over-expression, which was closely related to blocking apoptosis of malignant T cell, whereas the TNFSF10 gene was also highly expressed in some patients, which may partly correct the imbalance.
Correlation analysis of BCL11B in the T-ALL group revealed that the BCL11B expression level was negatively correlated with that of BCL2L1 (Bcl-xL), although over-expression of both genes was found in T-ALL samples. This suggested that BCL2L1 was affected by the BCL11B gene in transcriptional regulation, and both participated in the same protein-protein interactions, acting as apoptosis regulators along with a competitive target protein downstream. In BCL11B-knockdown T-cell lines, when exposed to growth stimuli, T cells exhibit apoptosis in S phase with concomitant decreases in the cell-cycle inhibitor p27 and the anti-apoptotic protein Bcl-xL, due to transcriptional repression [13]. However, BCL11B and BCL2L1 protein levels in the T-ALL group still remain to be validated. Correlation analysis of BCL11B in the T-ALL group revealed that the BCL11B expression level was positively correlated with the relative SPP1 expression level. The expression of SPP1 was significantly down-regulated with BCL11B silencing by RNA interference, suggesting that the SPP1 gene may be a target of the BCL11B gene in transcriptional regulation (unpublished data). SPP1 gene silencing in vitro significantly increased mitochondrial cytochrome c release, and the inhibitory action of the Wnt target gene osteopontin (SPP1) on mitochondrial cytochrome c release determines renal ischemic resistance [27]. Thus, the SPP1 gene may play a consistent role in anti-apoptotic effects with the BCL11B gene, by decreasing mitochondrial cytochrome c release. The hypothetical regulatory network of apoptosis in BCL11B and related genes is shown in Figure 4. However, the role of the SPP1 gene in T-cell malignancies is unclear, because low expression of SPP1 was detected in T-ALL.
Figure 4.
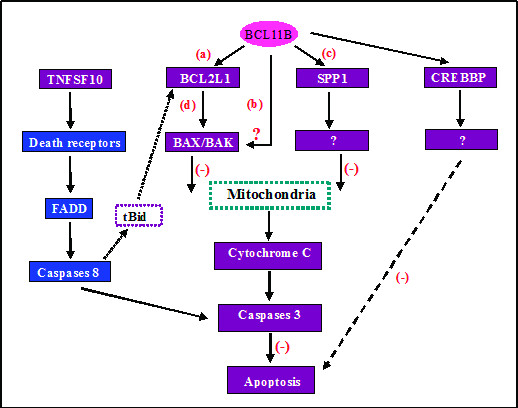
Schematic representation of the regulatory network of apoptosis in BCL11B and its related genes. (a) BCL2L1 is affected by the BCL11B gene in transcriptional regulation. (b, d) BCL11B and BCL2L1 participate in the same protein-protein interactions, along with competitive downstream target proteins. BCL2L1 (Bcl-xL) normally interferes with the mitochondrial programmed cell death pathway by sequestering proapoptotic proteins such as BCL2-associated × protein (BAX) and BCL2-antagonist/killer 1 (BAK1; BAK), suggesting that BAX/BAK may be competitive target proteins downstream of BCL11B. (c) The SPP1 gene may be a target of the BCL11B gene in transcriptional regulation: it plays a consistent role in anti-apoptotic effects with the BCL11B gene by decreasing mitochondrial cytochrome c release.
Conclusions
The expression pattern of the BCL11B gene and four of its related genes (TNFSF10, BCL2L1, SPP1, and CREBBP) was characterized in T-ALL. Over-expression of BCL11B may play a role in anti-apoptosis in T-ALL cells through up-regulation of its downstream genes BCL2L1 and CREBBP.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YQL made contributions to conception and design laboratory study. XH, SHC, QS, LJY, and BL performed the laboratory technique process and the laboratory analyses. LYZ, SXG and XD were responsible of the patient's treatment and carried out acquisition of clinical data. YQL and XH coordinated the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Xin Huang, Email: huangmingxin2001@sina.com.
Shaohua Chen, Email: jnshaohuachen@163.com.
Qi Shen, Email: 84441287@qq.com.
Lijian Yang, Email: jnyanglijian@163.com.
Bo Li, Email: libo517@hotmail.com.
Liye Zhong, Email: zhongliye99@163.com.
Suxia Geng, Email: gsx76@126.com.
Xin Du, Email: miyadu@hotmail.com.
Yangqiu Li, Email: yangqiuli@hotmail.com.
Acknowledgements
The project was sponsored by grants from National Natural Science Foundation of China (No. 30771980), the Fundamental Research Funds for the Central Universities (No. 21610604) and the Guangdong Science & Technology Project (No. 2007B030703008; and 2009B050700029).
References
- Rivera GK, Crist WM. In: Principles and Practice of Hematology. Blood. Handin RI, Stossel TP, Lux SE, editor. 1995. Acute lymphoblastic leukemia; pp. 743–759. [Google Scholar]
- Uckun FM, Sensel MG, Sun L, Steinherz PG, Trigg ME, Heerema NA. Biology and treatment of childhood T-lineage acute lymphoblastic leukemia. Blood. 1998;91:735–746. [PubMed] [Google Scholar]
- Morris JC, Waldmann TA, Janik JE. Receptor-Directed Therapy of T-Cell Leukemias and Lymphomas. J Immunotoxicol. 2008;5:235–248. doi: 10.1080/15476910802129661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- Devi RS. siRNA-based approaches in cancer therapy. Cancer Gene Therapy. 2006;13:819–829. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- Przybylski GK, Dik WA, Wanzeck J, Grabarczyk P, Majunke S, Martin-Subero JI, Siebert R, Dölken G, Ludwig WD, Verhaaf B, van Dongen JJ, Schmidt CA, Langerak AW. Disruption of the BCL11B gene through inv 14 q11.2q32.31 results in the expression of BCL11B-TRDC fusion transcripts and is associated with the absence of wild-type BCL11B transcripts in T-ALL. Leukemia. 2005;19:201–208. doi: 10.1038/sj.leu.2403619. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Nordigården A, Jönsson JI, Söderkvist P. Bcl11b mutations identified in murine lymphomas increase the proliferation rate of hematopoietic progenitor cells. BMC Cancer. 2007;7:195. doi: 10.1186/1471-2407-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl S, Konig M, Spath K. Juxtaposition of the BCL11B gene to a novel region at 17q by a t(14;17) (q32; Q21) in childhood T-Cell lymphoblastic lymphoma [abstract] Blood. 2007;110:101B. [Google Scholar]
- Su XY, Della-Valle V, Andre-Schmutz I, Lemercier C, Radford-Weiss I, Ballerini P, Lessard M, Lafage-Pochitaloff M, Mugneret F, Berger R, Romana SP, Bernard OA, Penard-Lacronique V. HOX11L2/TLX3 is transcriptionally activated through T-cell regulatory elements downstream of BCL11B as a result of the t(5;14) (q35;q32) Blood. 2006;108:4198–4201. doi: 10.1182/blood-2006-07-032953. [DOI] [PubMed] [Google Scholar]
- Oshiro A, Tagawa H, Ohshima K, Karube K, Uike N, Tashiro Y, Utsunomiya A, Masuda M, Takasu N, Nakamura S, Morishima Y, Seto M. Identification of subtype-specific genomic alterations in aggressive adult T-cell leukemia/lymphoma. Blood. 2006;107:4500–4507. doi: 10.1182/blood-2005-09-3801. [DOI] [PubMed] [Google Scholar]
- Kamimura K, Mishima Y, Obata M. Lack of Bcl11b tumor suppressor results in vulnerability to DNA replication stress and damages. Oncogene. 2007;26:5840–5850. doi: 10.1038/sj.onc.1210388. [DOI] [PubMed] [Google Scholar]
- Breckenridge DG, Xue D. Regulation of mitochondrial membrane permeabilization by BCL-2 family proteins and caspases. Curr Opin Cell Biol. 2004;16:647–652. doi: 10.1016/j.ceb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Borrow J, Stanton VP Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, Civin CI, Disteche C, Dubé I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. The translocation t(8;16)(p11;13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- Giles RH, Dauwerse JG, Higgins C, Petrij F, Wessels JW, Beverstock GC, Döhner H, Jotterand-Bellomo M, Falkenburg JH, Slater RM, van Ommen GJ, Hagemeijer A, van der Reijden BA, Breuning MH. Detection of CBP rearrangements in acute myelogenous leukemia with t(8;16) Leukemia. 1997;11:2087–2096. doi: 10.1038/sj.leu.2400882. [DOI] [PubMed] [Google Scholar]
- Caravatta L, Sancilio S, di Giacomo V, Rana R, Cataldi A, Di Pietro R. PI3-K/Akt-dependent activation of cAMP-response element-binding (CREB) protein in Jurkat T leukemia cells treated with TRAIL. J Cell Physiol. 2008;214:192–200. doi: 10.1002/jcp.21186. [DOI] [PubMed] [Google Scholar]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy Bcl-2 apoptotic switch in cancer. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standal T, Borset M, Sundan A. Role of osteopontin in adhesion, migration, cell survival, and bone remodeling. Exp Oncol. 2004;26:179–184. [PubMed] [Google Scholar]
- Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006;16:79–87. doi: 10.1016/j.tcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Huang X, Chen S, Yang LJ, Chen SH, Zhou YB, Schmidt CA, Li YQ. Effects of down-regulating BCL11B expression on the proliferation, apoptosis and global gene expression profiling of Molt-4 cells [Abstract] Blood. 2009;114:4505. doi: 10.1182/blood-2009-02-206219. [DOI] [Google Scholar]
- Grabarczyk P, Przybylski GK, Depke M, Völker U, Bahr J, Assmus K, Bröker BM, Walther R, Schmidt CA. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26:3797–3810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang J, Kim HP. Bcl-XL disrupts death-inducing signal complex formation in plasma membrane induced by hypoxia/reoxygenation. FASEB J. 2004;18:1826–1833. doi: 10.1096/fj.04-2047com. [DOI] [PubMed] [Google Scholar]
- Rodrigues LR, Teixeira JA, Schmitt FL, Paulsson M, Lindmark-Mänsson H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1087–1097. doi: 10.1158/1055-9965.EPI-06-1008. [DOI] [PubMed] [Google Scholar]
- Viñas JL, Sola A, Jung M, Mastora C, Vinuesa E, Pi F, Hotter G. Inhibitory action of Wnt target gene osteopontin on mitochondrial cytochrome c release determines renal ischemic resistance. Am J Physiol Renal Physiol. 2010;299:F234–242. doi: 10.1152/ajprenal.00687.2009. [DOI] [PubMed] [Google Scholar]


