Abstract
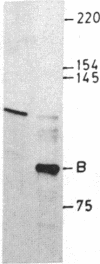
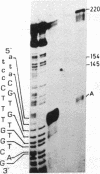
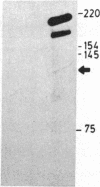
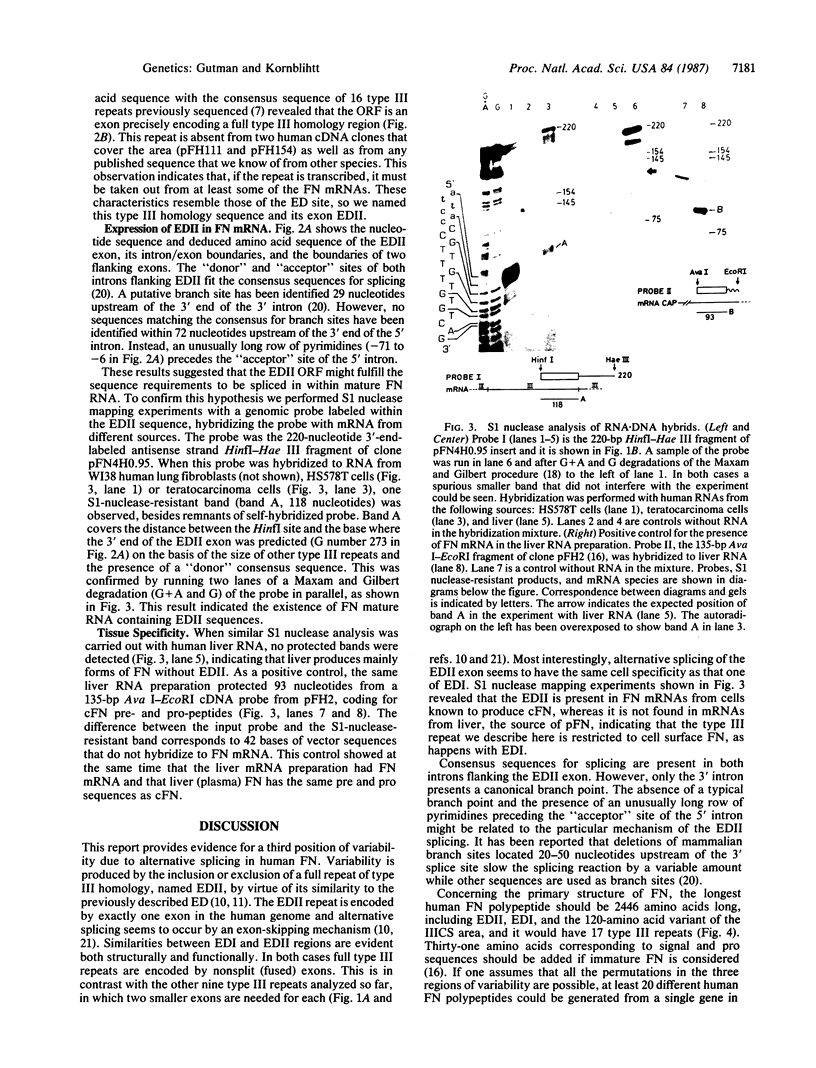
We describe here a third region of variability in human fibronectin (FN) due to alternative RNA splicing. Two other positions of alternative splicing have been reported previously (ED and IIICS). The third region involves a 273-nucleotide exon encoding exactly one 91-amino acid repeat of type III homology, located between the DNA- and the cell-binding domains of FN, which is either included in or excluded from FN mRNA. The two mRNA variants arising by an exon-skipping mechanism are present in cells known to synthesize the cellular form of FN. However, liver cells, which are the source of plasma FN, produce only messengers without the extra type III sequence. Therefore, the region described here resembles, both structurally and functionally, the previously described ED (for extra domain) region, located toward the C terminus of the molecule, between the cell- and heparin- (hep 2) binding domains. We conclude that both the extra type III repeat (named EDII) and ED represent sequences restricted to cellular FN. Combination of all the possible patterns of splicing in the three regions described to date may generate up to 20 distinct FN polypeptides from a single gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anson D. S., Choo K. H., Rees D. J., Giannelli F., Gould K., Huddleston J. A., Brownlee G. G. The gene structure of human anti-haemophilic factor IX. EMBO J. 1984 May;3(5):1053–1060. doi: 10.1002/j.1460-2075.1984.tb01926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Nguyen H. T., Medford R. M., Destree A. T., Mahdavi V., Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985 May;41(1):67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cossu G., Andrews P. W., Warren L. Covalent binding of lactosaminoglycans and heparan sulphate to fibronectin synthesized by a human teratocarcinoma cell line. Biochem Biophys Res Commun. 1983 Mar 29;111(3):952–957. doi: 10.1016/0006-291x(83)91392-x. [DOI] [PubMed] [Google Scholar]
- Gutman A., Yamada K. M., Kornblihtt A. Human fibronectin is synthesized as a pre-propolypeptide. FEBS Lett. 1986 Oct 20;207(1):145–148. doi: 10.1016/0014-5793(86)80029-1. [DOI] [PubMed] [Google Scholar]
- Hackett A. J., Smith H. S., Springer E. L., Owens R. B., Nelson-Rees W. A., Riggs J. L., Gardner M. B. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J Natl Cancer Inst. 1977 Jun;58(6):1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- Hynes R. Molecular biology of fibronectin. Annu Rev Cell Biol. 1985;1:67–90. doi: 10.1146/annurev.cb.01.110185.000435. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Umezawa K., Vibe-Pedersen K., Baralle F. E. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985 Jul;4(7):1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A. R., Vibe-Pedersen K., Baralle F. E. Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. EMBO J. 1984 Jan;3(1):221–226. doi: 10.1002/j.1460-2075.1984.tb01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt A. R., Vibe-Pedersen K., Baralle F. E. Isolation and characterization of cDNA clones for human and bovine fibronectins. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3218–3222. doi: 10.1073/pnas.80.11.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Odermatt E., Tamkun J. W., Hynes R. O. Repeating modular structure of the fibronectin gene: relationship to protein structure and subunit variation. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6571–6575. doi: 10.1073/pnas.82.19.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Paul J. I., Schwarzbauer J. E., Tamkun J. W., Hynes R. O. Cell-type-specific fibronectin subunits generated by alternative splicing. J Biol Chem. 1986 Sep 15;261(26):12258–12265. [PubMed] [Google Scholar]
- Peters J. H., Ginsberg M. H., Bohl B. P., Sklar L. A., Cochrane C. G. Intravascular release of intact cellular fibronectin during oxidant-induced injury of the in vitro perfused rabbit lung. J Clin Invest. 1986 Dec;78(6):1596–1603. doi: 10.1172/JCI112752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. E., Thøgersen H. C., Skorstengaard K., Vibe-Pedersen K., Sahl P., Sottrup-Jensen L., Magnusson S. Partial primary structure of bovine plasma fibronectin: three types of internal homology. Proc Natl Acad Sci U S A. 1983 Jan;80(1):137–141. doi: 10.1073/pnas.80.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J. W., Hynes R. O. Plasma fibronectin is synthesized and secreted by hepatocytes. J Biol Chem. 1983 Apr 10;258(7):4641–4647. [PubMed] [Google Scholar]
- Tamkun J. W., Schwarzbauer J. E., Hynes R. O. A single rat fibronectin gene generates three different mRNAs by alternative splicing of a complex exon. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5140–5144. doi: 10.1073/pnas.81.16.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibe-Pedersen K., Kornblihtt A. R., Baralle F. E. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J. 1984 Nov;3(11):2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. M. Cell surface interactions with extracellular materials. Annu Rev Biochem. 1983;52:761–799. doi: 10.1146/annurev.bi.52.070183.003553. [DOI] [PubMed] [Google Scholar]