Abstract
The recent development of analytic high-throughput technologies enables us to take a bird’s view of how metabolism is regulated in real time. We have known for a long time that metabolism is highly regulated at all levels, including transcriptional, posttranslational and allosteric controls. Flux through a metabolic or signaling pathway is determined by the activity of its individual components. Fluxomics aims to define the genes involved in regulation by following the flux. Two technologies are used to monitor fluxes. Pulse labeling of the organism or cell with a tracer, such as 13C, followed by mass spectrometric analysis of the partitioning of label into different compounds provides an efficient tool to study flux and to compare the effect of mutations on flux. The second approach is based on the use of flux sensors, proteins that respond with a conformational change to ligand binding. Fluorescence resonance energy transfer (FRET) detects the conformational change and serves as a proxy for ligand concentration. In contrast to the mass spectrometry assays, FRET nanosensors monitor only a single compound. Both methods provide high time resolution. The major advantages of FRET nanosensors are that they yield data with cellular and subcellular resolution and the method is minimally invasive.
Introduction
Cells and organisms dynamically acclimate their metabolism to changing conditions, such as nutrient availability, temperature or stress. Flux across the plasma membrane and through metabolic pathways is continuously optimized. Sensory systems that are coupled with complex signaling networks adjust the flux and its direction via regulatory circuits. The sensory systems measure the extracellular availability of a given metabolite and other external cues, as well as the intracellular level of the metabolite or subsequent intermediates, and send signals to regulate transporter and enzyme activities by posttranslational modification, protein turnover, or changes in the rate of their biosynthesis. At present, we know little about the receptors that detect the signals, the signaling networks that transmit the information, nor their integration. A new discipline, fluxomics (Box 1), aims to systematically analyze the fluxes occurring within a cell, and at some point even to unravel these networks in a multicellular organism. The availability of large mutant collections, RNA interference (RNAi) or overexpression line libraries, and large chemical libraries now puts us in a position to unravel these networks, provided we have the tools to measure metabolite flux, especially in high throughput. This review discusses two sets of methodologies that have been developed to measure flux: first, mass spectrometric analysis of changes in metabolite levels after pulse labeling, and second, quantitative imaging using fluorescence resonance energy transfer (FRET)-based nanosensors. These recent developments are contrasted to established isotope labeling techniques for metabolic steady-state systems.
Box 1. Glossary.
- Flux
in analogy to electric current, flux is the passage of molecules (moles of a particular metabolite) through a metabolic or transport step per unit cell mass per unit time. Defined by the concentrations of compounds participating in a reaction, enzyme level and enzyme properties.
- Fluxome
commonly referred to as the totality of all fluxes in a system, e.g. a cell. The term fluxome was defined by Sauer et al. [55] as the array of fluxes (reaction rates on a per unit cell volume or per unit cell mass basis) for all of the reactions that occur in the organism. This term is typically used in the context of pulse labeling with 13C-labelled metabolites, followed by mass spectrometry to analyze multiple (typically hundreds of) metabolites in parallel. Such analyses have been expanded to the comparison of fluxes in microorganismal mutants. The term fluxome can also be used to describe the factors (gene networks) that define a specific flux rate.
- Fluxomics
the discipline that analyzes the fluxome as one part of systems biology [56•]. Provides mathematically defined networks of metabolic reactions and their regulation. Fluxomics are defined here as an approach to identify the genes that affect the fluxes which control the steady state of a single metabolite (using FRET nanosensors).
The importance of fluxes
All kinds of physical systems — living cells are no exception — are governed by a dualism between potentials and flows. Potential quantities are related to energy levels whereas flow quantities are related to transport or conversion phenomena. In mechanics, potentials and flows are represented by potential energy and force (a force is a flow of momentum), in hydraulics by pressure and liquid flow, and in electrical systems by voltage and current. For example, voltage can be directly measured by using electrostatic forces, whereas current, that is the flux of electrons, can be measured using a light bulb: the number of collisions per time unit determines the intensity of light emitted. In biological networks, the dualism is given by chemical activities and reaction rates, also called metabolic fluxes. At present, the potential, for example the concentration gradient, can be determined but fluxes must be derived. The precise relation between potentials and flows is described by constitutive laws such as Hooke’s law in mechanics, Ohm’s law for electrical systems, Fick’s law for diffusion, the Nernst equation for electrochemistry, or mass action kinetics in chemistry. However, these are just the simplest examples.
The dualism between substance concentrations, or more precisely chemical potentials, and metabolic fluxes is expressed by the fact that one part is causal for the other. On the one hand, chemical potentials of reactants constitute the driving forces for fluxes. On the other hand, metabolic fluxes change the potentials of metabolite pools. Usually, flows in physical systems are closely related to system functions (i.e. dynamics), whereas potentials rather describe their stock-keeping aspects (i.e. statics). In the field of biochemical networks, in particular, metabolic fluxes are the ultimate manifestation of the cell’s function under certain physiological conditions.
It might appear surprising that flux, such as that across the plasma membrane, is not optimized for a wide spectrum of conditions. The plasma membrane provides a limited compartment [1], however, and a given transporter (at least if not modified), has defined kinetic properties, such as high affinity (typically coupled with low capacity) optimized for importing nutrients present in low abundance or high capacity (typically coupled with low affinity). Thus, when nutrient availability or demand change, either a different set of transporters is required or the properties of the transporter have to be adjusted to allow for optimal flux [2–4]. The same is essentially true for many isozymes. It is thus important to determine both the flux potential and the actual flow, and to compare the system’s behavior under different conditions, for example, different levels of nutrient availability.
In recent years, fluxome analysis under steady-state conditions has become a widely used tool. It is applied to characterize an organism [5,6], to diagnose the effect of genetic manipulations [7], to compare the behavior of one organism under different physiological conditions [8], to detect the presence of certain metabolic pathways [9,10], to compare mutant libraries [11••], or to monitor different growth phases [12]. This tool has been applied to all classes of organisms, including bacteria, unicellular eukaryotes, animal and plant cells, and even whole organs. Some recent reviews overlook the general methodology at the state of the art [13•,14–17]. Particular problems of plant fluxome analysis have been addressed by Ratcliffe and Shachar-Hill [18] and by Schwender et al. [19], and its application in plants is exemplified in a number of papers [20–22].
Methods in fluxomics
Fluxes are usually determined under metabolic steady-state conditions. There is, however, no reason why fluxomics should be restricted to this case. In fact, the rapid redirection of fluxes under dynamic conditions is an important capability of living organisms. Consequently, a new aspect in fluxomics is the consideration of time-dependent fluxes. As we discover below, the experimental methods required to determine these fluxes are quite different from steady-state methods. Interestingly, some current developments are combining methods from dynamic and steady-state approaches to reach new ambitious goals.
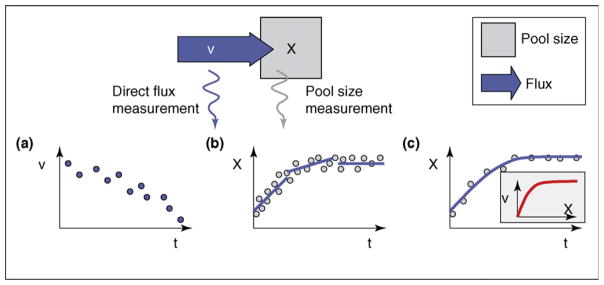
Although, the description of living cells by fluxes seems to be more natural than a description by substance concentrations, cells have predominantly been characterized by their intracellular pool sizes. The reason is rather simple: direct measurements of flows are possible in other physical systems but there is almost no known measurement procedure that directly yields information on metabolic reaction rates (an exotic exception might be the light emission in the luciferase reaction [Figure 1a]). This makes fluxomics a rather singular discipline in the ‘omics’ field.
Figure 1.
Time-dependent flux determination from direct and indirect measurement data. (a) Direct flux measurement, the ideal case. (b) Computation of time derivatives from high-density pool size data. (c) Fitting of a kinetic model to time resolved pool size data.
As an example that illustrates this difference, electrical currents can be measured not only directly by magnetic induction but also indirectly from the voltage change of a capacitor. In the latter case, the signal must be differentiated with respect to time to obtain the time-dependent current. For an electrical system, this is usually not crucial because the measured signal has a high precision. This situation is completely different in the biological case, where measurement noise is significant and a direct differentiation leads to error amplification. To solve this problem, only initial velocities are calculated or signal smoothing algorithms are applied (Figure 1b). However, this helps only in case of a high data density or slowly changing pool sizes. If both approaches are not applicable, a model-based data evaluation is preferred in which a constitutive law is assumed (e.g. Michaelis–Menten kinetics) and only the parameters of this law are estimated (Figure 1c). This strongly reduces the required amount of measured data, but the interpretation of the data is ‘prejudiced’ by the assumed law.
The present review focuses on the comparison and classification of different fluxomics methods for dynamic and static conditions and their interrelations. The flow-potential viewpoint will serve as a guideline. As the dual quantity of fluxes is given by the metabolite pool sizes, methods from metabolomics play an important role here by supplying the raw data for flux determination.
Flux analysis under highly dynamic conditions
The classical idea of measuring enzyme kinetic data from initial slopes of NAD signals immediately leads to indirect flux analysis methods that are based on time-resolved pool-size signals. A well-known class of experiments to investigate metabolic networks under highly dynamic conditions are represented by stimulus–response experiments ([23•,24; Figure 2a). Here, an external stimulus is imposed on the system by suddenly raising the extracellular concentration of a substance at time zero. The intracellular pathways immediately respond to the stimulus, usually within seconds or minutes. Under these highly dynamic conditions, the most difficult analytical problem is clearly the reliable quantitative measurement of intracellular pool sizes within short time intervals and with minimal interference with cellular functions.
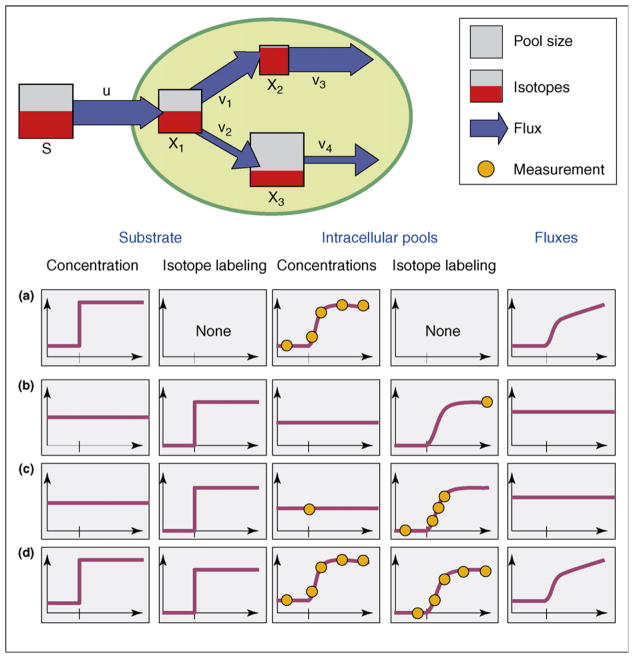
Figure 2.
Conceptionally different approaches to obtain flux information from measured pool size and/or isotope labeling data. (a) Rapid sampling of intracellular pools under highly dynamic conditions of a stimulus response. (b) Standard 13C-labeling experiment under metabolic and isotopic steady-state conditions. (c) Isotopically non-stationary 13C-labeling experiment under metabolic steady-state conditions. (d) Fictive combined experiment under metabolically and isotopically non-stationary conditions.
Two methods have been developed to obtain dynamic flux information from measurements of pool size. In the first, a pulse stimulus in combination with rapid sampling serves to draw samples from a cell culture at high sampling frequency, up to several samples per second [23•,25]. Subsequently, each sample is rapidly inactivated, for example by cold methanol quenching. Cell disruption and separating the intercellular metabolites allows pool sizes to be determined using modern high performance liquid chromatography (LC)-MS instruments [26]. In essence, this is a technical improvement over the method originally introduced to biology by Calvin and Benson [27] to determine the carbon path in photosynthesis. The reliability of this procedure is still a controversy, however, because of the leakage of cell membranes when using methanol quenching. Moreover, it does not provide spatial information when applied to tissues or organs. Neither does it provide subcellular spatial resolution, which is of primary relevance in eukaryotic systems due to their high level of compartmentation. Finally, because of the destructive nature of this approach, the analysis of tissues is limited to parallel sampling. Thus, to date, fluxomics had been restricted to the analysis of unicellular systems in the population average.
An exciting new alternative to this method is given by the direct expression of sensors for the concentration of molecular metabolites in the cell. Certain proteins, specifically chemoreceptors, respond to ligand binding with a conformational change [28]. Protein conformation can be measured directly, using fluorescence resonance energy transfer [29]. FRET reporters in which recognition elements from diverse bacterial chemoreceptors are combined with green fluorescent protein (GFP) variants have permitted the development of genetically encoded flux-sensors for a variety of small molecules, such as calcium, phosphate, carbohydrates (ribose, glucose, maltose and sucrose) and amino acids [30,31••,32•,33•,34,35]. These sensors can detect flux changes in vivo because they monitor steady-state levels of a given ion or metabolite.
A single sensor can already be used to analyze large mutant collections from any organism. The major advantage of these sensors over any other technology lies in the time resolution that they can provide (up to millisecond range as shown for calcium [36,37]). These sensors also provide high spatial resolution; for example, they have successfully been applied to analyze metabolite levels in individual cells within intact organs [38•,39,40••]. On the one hand, a single sensor is able to monitor fewer substances than a MS approach.
Since these FRET sensors are genetically encoded, they provide subcellular resolution. This subcellular resolution is not achieved by optical methods, but genetically by targeting. The addition of targeting signals has successfully been used to monitor glucose flux across the membrane of the endoplasmic reticulum (ER) or nuclear fluxes [31••,41. Thus, local fluxes can be monitored by anchoring the sensors in specific membranes, for example at the cell surface [32••], and it is conceivable that even flux in localized domains, such as rafts, can be monitored this way. Finally, and most importantly, the sensor approach is non-destructive, making it possible to monitor an intact organ (or organism) with minimal invasion. Obviously, the expression of the sensors with a cell adds a new buffer for the analyte of interest that may affect metabolic flux.
The computer evaluation of dynamic experiments to estimate the time-dependent intracellular fluxes between the metabolite pools must currently rely on the assumption of a reaction kinetic mechanism (i.e. constitutive laws) for all of the involved transport and reaction steps. This gives rise to a differential equation model that describes the dynamics of intracellular pool sizes. By fitting this model to the measured data, the in vivo parameters of the enzyme kinetics are estimated ([42•]; Figure 3). Finally, the time-dependent metabolic fluxes (as a function of time) can be directly computed from the fitted model [43,44•].
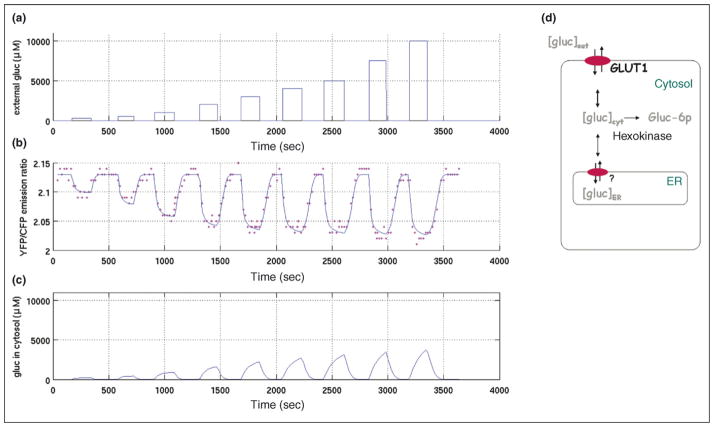
Figure 3.
Changes in glucose concentration in HepG2 cells stably expressing FLIPglu600 μ. (a) External perfused glucose concentration. (b) Measured yellow fluorescent protein (YFP)/cyan fluorescent protein (CFP) emission ratios and simulation curve based on a kinetic model. (c) Cytosolic glucose concentration from simulation. (d) Underlying compartment model for analysis of glucose homeostasis in HepG2 cells (modified from Fehr et al. [31••]). Glucose is transported reversibly across the plasma and ER membranes and is phosphorylated irreversibly in the cytosol.
13C metabolic flux analysis in steady state
13C metabolic flux analysis (13C-MFA) is currently the best-established fluxomics technology. In contrast to flux analysis under highly dynamic conditions, the 13C-MFA method always requires the assumption of metabolic steady state for the entire duration of the experiment. This limits the possible applications, but on the other hand, no assumptions on reaction kinetics are needed to derive the fluxes from the available measurements. Clearly, in this situation, knowledge of the intracellular metabolite pool sizes does not help to determine the fluxes.
This major source of information for computing the fluxes is the isotope labeling of substrates that are fed into the system (usually with 13C but other isotopes are discussed [45•]). After switching the feed to a labeled substrate, the isotopes are distributed over the intercellular network by metabolic activities (Figure 2b). After some time, both the metabolic fluxes and the fluxes and fractions of labeled material in the metabolite pools can be shown to have reached a steady state [44•]. This fractional information can be obtained using NMR or MS. Interestingly, once again, the intracellular fluxes have to be determined from a concentration-like quantity.
The measured fractions of isotopic labeling in intracellular pools form the basis of a rather complex mathematical model that describes the distribution of labeled material over cellular metabolism and relates the unknown fluxes with the given measurements. This is not a real biological model but rather a physical model that provides probabilistic rules describing the distribution of isotope label. Thus, the validity of this complex model is non-critical, in contrast to mechanistic reaction kinetic models for biochemical networks in a dynamic metabolic state.
By fitting the isotope distribution model to the measured data, intracellular fluxes can finally be determined [17]. Additional information, given by the knowledge of forward and backward fluxes in bidirectional reaction steps, is generated by this method. This information can also be obtained from dynamic models that are based on reaction kinetics, if all reversible reaction steps are consequently modeled with reversible reaction kinetic formulae. This, in turn, increases the number of parameters to be estimated from the rapid-sampling data.
13C metabolic flux analysis on the ultra-short time scale
The standard 13C-MFA method takes rather long experimental durations, approximately 2–3 times the doubling times of the microorganism, because the label must first accumulate in the biomass before being measured. In many cases, it is not possible to keep a living system in a metabolic steady state for such a long time. Examples in which this has been achieved, however, include cells in transient growth phases, cells in industrial fermentation conditions (fed batch) or genetically unstable recombinant cells. Application to slowly growing or non-growing cells is of particular interest for plant physiology. An application that is unique to plants is given by studies on C1-metabolism in photoautotrophic plants because, in this case, the isotopic steady state contains no information on metabolic fluxes if the substrate carries only a single carbon atom [46].
Nevertheless, most physiological states of a cellular system can be considered to be in at least a quasi metabolic steady state, which means that, for short time durations (minutes to hours), metabolism is approximately in a steady state. In this case, a novel 13C-flux analysis method can be applied that does not rely on the steady-state assumption for label enrichment in metabolite pools. By contrast, the enrichment of labeled material in the metabolite pools is now observed under isotopically transient conditions ([47]; Figure 2c).
The new isotopically non-stationary method represents an interesting fusion of the methods for MFA under highly dynamic conditions that are based on rapid sampling and 13C-MFA under steady-state conditions. Metabolite pools are kept constant but must be measured because they represent the system’s capacity for labeled material.
This isotopically non-stationary method (INST-MFA) became possible only recently because mass spectrometers are now able to determine reliable fractional label information from low-concentration pools of intermediate metabolites [48]. The processes for the computational evaluation of such transient data are substantially more difficult than those for the evaluation of data describing the isotopic steady state because the accumulation of labeled materials in the intracellular pools is now described by a dynamic model. Thus, the system of equations that describes the distribution of label over the network has to be replaced by differential equations [49•].
A first application of INST-MFA to Escherichia coli proved that information on all intracellular fluxes could be obtained in just 15 s, during which about 20 samples were drawn from the culture [50••]. This makes isotopically non-stationary metabolic flux analysis a promising candidate for the analysis of slowly growing plant or animal cells. Several teams are currently working to establish the new method from experimental and theoretical view-points [45•,49•,51,52].
Conclusions and new horizons
A still-speculative extension of the INST-MFA method may be the use of experiments under both metabolic and isotopic steady states (Figure 2d). An exploratory simulation study [24] has already shown that INST-MFA would significantly improve the available information on reaction kinetic parameters in vivo and, ultimately, on the dynamic fluxes.
Another challenging development is the model-free determination of flux information from pool-size data by smoothing and differentiation of high-density data (Figure 1b). It should be noticed that this is already possible in other chemical disciplines, as has been demonstrated with the determination of diffusion flows [53].
Altogether, a detailed theoretical basis has been developed for fluxome analysis using 13C-labeling. These mathematical tools are currently adapted for fluxome studies using the FRET flux sensors. FRET sensors can provide direct information on steady-state levels and fluxes in a specific subcellular compartment. Moreover, these sensors provide information on flux in individual cells and thus significantly expand the potential of metabolomics. The combination of FRET sensors with microfluidics and modeling promises new insights into the regulation of metabolism in response to a changing environment, and into the underlying signaling networks not only in multicellular, eukaryotic systems but also in microorganisms from E. coli to yeast. Both 13C and nano-sensor MFA technologies will rapidly be applied to fluxomics, as exemplified in the case of yeast mutants that are affected in sugar signaling [54].
Acknowledgments
This work was supported by grants from Bundesministerium für Bildung und Forschung (BMBF) (SysMAP Project) to WW and from the US National Institute of Health (Roadmap Initiative ‘Metabolomics technology development’ [R33DK070272]) and US Department of Energy (DE-FG02-04ER15542) to WBF.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hennaut C, Hilger F, Grenson M. Space limitation for permease insertion in the cytoplasmic membrane of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1970;39:666–671. doi: 10.1016/0006-291x(70)90257-3. [DOI] [PubMed] [Google Scholar]
- 2.Marini AM, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu KH, Tsay YF. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Springael JY, Andre B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dauner M, Storni T, Sauer U. Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J Bacteriol. 2001;183:7308–7317. doi: 10.1128/JB.183.24.7308-7317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Winden WA, van Gulik WM, Schipper D, Verheijen PJT, Krabben P, Vinke JL, Heijnen JJ. Metabolic flux and metabolic network analysis of Penicillium chrysogenum using 2D [13C, 1H] COSY NMR measurements and cumulative bondomer simulation. Biotechn Bioeng. 2003;83:75–92. doi: 10.1002/bit.10648. [DOI] [PubMed] [Google Scholar]
- 7.Emmerling M, Dauner M, Ponti A, Fiaux J, Hochuli M, Szyperski T, Wüthrich K, Bailey JE, Sauer U. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J Bacteriol. 2002;184:152–164. doi: 10.1128/JB.184.1.152-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiefer P, Heinzle E, Zelder O, Wittmann C. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. Appl Environ Microbiol. 2004;70:229–239. doi: 10.1128/AEM.70.1.229-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer E, Sauer U. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J Biol Chem. 2003;278:46446–46451. doi: 10.1074/jbc.M307968200. [DOI] [PubMed] [Google Scholar]
- 10.Petersen S, de Graaf AA, Eggeling L, Möllney M, Wiechert W, Sahm H. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J Biol Chem. 2000;275:35932–35941. doi: 10.1074/jbc.M908728199. [DOI] [PubMed] [Google Scholar]
- 11••.Fischer E, Sauer U. Large-scale in vivo fluxes reveal rigidity and suboptimal performance of B. subtilis metabolism. Nat Genet. 2005;37:636–640. doi: 10.1038/ng1555. One of the first real high-throughput applications of metabolic flux analysis under steady-state conditions. [DOI] [PubMed] [Google Scholar]
- 12.Wahl A, El Massaoudi M, Schipper D, Wiechert W, Takors R. Serial 13C-based flux analysis of an L-phenylalanine-producing E. coli strain using the sensor reactor. Biotechnol Prog. 2004;20:706–714. doi: 10.1021/bp0342755. [DOI] [PubMed] [Google Scholar]
- 13•.Fernie AR, Geigenberger P, Stitt M. Flux an important, but neglected, component of functional genomics. Curr Opin Plant Biol. 2005;8:174–182. doi: 10.1016/j.pbi.2005.01.008. An excellent introduction to flux analysis and an update regarding its relation to genomics. [DOI] [PubMed] [Google Scholar]
- 14.Sauer U. Metabolic networks in motion. 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellerstein MK. In vivo measurement of fluxes through metabolic pathways: the missing link in functional genomics and pharmaceuticel research. Annu Rev Nutr. 2003;23:379–402. doi: 10.1146/annurev.nutr.23.011702.073045. [DOI] [PubMed] [Google Scholar]
- 16.Kelleher JK. Probing metabolic pathways with isotopic tracers: insights from mammalian metabolic physiology. Metab Eng. 2004;6:1–5. doi: 10.1016/j.ymben.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001;3:195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 18.Ratcliffe RG, Shachar-Hill Y. Measuring multiple fluxes through plant metabolic networks. Plant J. 2006;45:490–511. doi: 10.1111/j.1365-313X.2005.02649.x. [DOI] [PubMed] [Google Scholar]
- 19.Schwender J, Ohlrogge J, Shachar-Hill Y. Understanding flux in plant metabolic networks. Curr Opin Plant Biol. 2004;7:309–317. doi: 10.1016/j.pbi.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Roscher A, Kruger NJ, Ratcliffe RG. Strategies for metabolic flux analysis in plants using isotope labelling. J Biotechnol. 2000;77:81–102. doi: 10.1016/s0168-1656(99)00209-6. [DOI] [PubMed] [Google Scholar]
- 21.Sriram G, Fulton DB, Iyer VV, Peterson JM, Zhou R, Westgate ME, Spalding MH, Shanks JV. Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractions: 13C labeling, two dimensional [13C, 1H] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant Physiol. 2004;136:3043–3057. doi: 10.1104/pp.104.050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinzle E, Matsuda F, Miyagawa H, Wkasa K, Nishioka T. Estimation of metabolic fluxes, expression levels and metabolite dynamics of a secondary metabolic pathway in potato using label pulse feeding experiments combined with kinetic network modeling and simulation. Plant J. 2007 doi: 10.1111/j.1365-313X.2007.03037.x. in press. [DOI] [PubMed] [Google Scholar]
- 23•.Theobald U, Mailinger W, Baltes M, Rizzi M, Reu M. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: I. Experimental observations. Biotechnol Bioeng. 1997;55:305–316. doi: 10.1002/(SICI)1097-0290(19970720)55:2<305::AID-BIT8>3.0.CO;2-M. Pioneering work in the field of metabolic high-throughput rapid sampling techniques. [DOI] [PubMed] [Google Scholar]
- 24.Wahl A. Metabolic Engineering VI: From recDNA Towards Engineering Biological Systems. Leeuwenhorst, The Netherlands: Engineering Conference International; 2006. Metabolic and isotopic instationary experiments: an exploratory simulation study. [Google Scholar]
- 25.Lange HC, Eman M, van Zuijlen G, Visser D, van Dam JC, Frank J, de Mattos MJT, Heijnen JJ. Improved rapid sampling for in vivo kinetics of intracellular metabolites in Saccharomyces cerevisiae. Biotechnol Bioeng. 2001;75:406–415. doi: 10.1002/bit.10048. [DOI] [PubMed] [Google Scholar]
- 26.Mashego MR, Wu L, van Dam JC, Ras C, Vinkel JL, van Winden W, van Gulik WM, Heijnen JJ. MIRACLE: mass isotopomer ratio analysis of U-[13]C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol Bioeng. 2004;85:620–628. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- 27.Calvin M, Benson AA. The path of carbon in photosynthesis. Science. 1948;107:476–480. doi: 10.1126/science.107.2784.476. [DOI] [PubMed] [Google Scholar]
- 28.Shilton BH, Flocco MM, Nilsson M, Mowbray SL. Conformational changes of three periplasmic receptors for bacterial chemotaxis and transport: the maltose-, glucose/galactose-and ribose-binding proteins. J Mol Biol. 1996;264:350–363. doi: 10.1006/jmbi.1996.0645. [DOI] [PubMed] [Google Scholar]
- 29.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehr M, Frommer WB, Lalonde S. Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc Natl Acad Sci USA. 2002;99:9846–9851. doi: 10.1073/pnas.142089199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Fehr M, Takanaga H, Ehrhardt DW, Frommer WB. Evidence for high-capacity bidirectional glucose transport across the endoplasmic reticulum membrane by genetically encoded fluorescence resonance energy transfer nanosensors. Mol Cell Biol. 2005;25:11102–11112. doi: 10.1128/MCB.25.24.11102-11112.2005. The authors use a FRET sensor for glucose to measure flux across the ER membrane. The sensor was targeted to the ER lumen using an ER signal sequence at the amino-terminus and an ER retention signal. The data derived from out-of-equilibrium measurements were used to develop a simple flux model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc Natl Acad Sci USA. 2005;102:8740–8745. doi: 10.1073/pnas.0503274102. A FRET sensor for glutamate that was anchored on the exterior surface of the plasma membrane is used to measure efflux of the neurotransmitter from neuronal cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Gu H, Lalonde S, Okumoto S, Looger LL, Scharff-Poulsen AM, Grossman AR, Kossmann J, Jakobsen I, Frommer WB. A novel analytical method for in vivo phosphate tracking. FEBS Lett. 2006;580:5885–5893. doi: 10.1016/j.febslet.2006.09.048. The authors develop a FRET sensor for inorganic phosphate using the same strategy as used for sugar and amino-acid nanosensors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lager I, Fehr M, Frommer WB, Lalonde S. Development of a fluorescent nanosensor for ribose. FEBS Lett. 2003;553:85–89. doi: 10.1016/s0014-5793(03)00976-1. [DOI] [PubMed] [Google Scholar]
- 35.Lager I, Looger LL, Hilpert M, Lalonde S, Frommer WB. Conversion of a putative Agrobacterium sugar-binding protein into a FRET sensor with high selectivity for sucrose. J Biol Chem. 2006;281:30875–30883. doi: 10.1074/jbc.M605257200. [DOI] [PubMed] [Google Scholar]
- 36.Iwano M, Shiba H, Miwa T, Che FS, Takayama S, Nagai T, Miyawaki A, Isogai A. Ca2+ dynamics in a pollen grain and papilla cell during pollination of Arabidopsis. Plant Physiol. 2004;136:3562–3571. doi: 10.1104/pp.104.046961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen GJ, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF, et al. Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;289:2338–2342. doi: 10.1126/science.289.5488.2338. [DOI] [PubMed] [Google Scholar]
- 38•.Kerr RA, Schafer WR. Intracellular Ca2+ imaging in C. elegans. Methods Mol Biol. 2006;351:253–264. doi: 10.1385/1-59745-151-7:253. This study demonstrates the applicability of nanosensors for monitoring flux (in this case of calcium) in intact organisms. [DOI] [PubMed] [Google Scholar]
- 39.Gordon S, Dickinson MH. Role of calcium in the regulation of mechanical power in insect flight. Proc Natl Acad Sci USA. 2006;103:4311–4315. doi: 10.1073/pnas.0510109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell. 2006;18:2314–2325. doi: 10.1105/tpc.106.044073. The authors analyze a set of glucose nanosensors that have differing affinities in intact roots and in leaf slices, a first step towards comparative analysis of flux in intact organs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fehr M, Lalonde S, Ehrhardt DW, Frommer WB. Live imaging of glucose homeostasis in nuclei of COS-7 cells. J Fluoresc. 2004;14:603–609. doi: 10.1023/b:jofl.0000039347.94943.99. [DOI] [PubMed] [Google Scholar]
- 42•.Wahl SA, Haunschild MD, Oldiges M, Wiechert W. Unravelling the regulatory structure of biochemical networks using stimulus response experiments and large scale model selection. IEE Proc Syst Biol. 2006;153:275–286. doi: 10.1049/ip-syb:20050089. State of the art in the field of computational methods for metabolic stimulus response experiments with rapid sampling data. [DOI] [PubMed] [Google Scholar]
- 43.Oldiges M, Noack S, Wahl A, Qeli E, Freisleben B, Wiechert W. From enzyme kinetics to metabolic network modelling – visualization tool for enhanced kinetic analysis of biochemical network models. Eng Life Sci. 2006;6:155–162. [Google Scholar]
- 44•.Wiechert W, Wurzel M. Metabolic isotopomer labeling systems. Part I: global dynamic behaviour. Math Biosci. 2001;169:173–205. doi: 10.1016/s0025-5564(00)00059-6. The theory behind mathematical modeling and simulation of 13C-labelling experiments. [DOI] [PubMed] [Google Scholar]
- 45•.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. State of the art in the field of computational methods for isotope labeling experiments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan JA. Metabolic Engineering VI: From recDNA Towards Engineering Biological Systems. Leeuwenhorst, The Netherlands: Engineering Conference International; 2006. Development of 13C metabolic flux analysis for photoautotrophic metabolism? [Google Scholar]
- 47.Wiechert W, Nöh K. From stationary to instationary metabolic flux analysis. In: Kragl U, editor. Technology Transfer in Biotechnology. Springer; 2005. pp. 145–172. [DOI] [PubMed] [Google Scholar]
- 48.Luo B, Grönke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and TCA cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 49•.Nöh K, Wahl A, Wiechert W. Computational tools for isotopically instationary 13C labelling experiments under metabolic steady state conditions. Metab Eng. 2006;8:554–577. doi: 10.1016/j.ymben.2006.05.006. The first implementation of a computational framework for fully instationary 13C-labelling experiments. [DOI] [PubMed] [Google Scholar]
- 50••.Nöh K, Grönke K, Luo B, Takors R, Oldiges M, Wiechert W. Metabolic flux analysis at ultra short time scale: isotopically non stationary 13C labeling experiments. J Biotechnol. 2007 doi: 10.1016/j.jbiotec.2006.11.015. in press. The first fully evaluated experiment proving the concept of isotopically non stationary 13C labelling experiments. [DOI] [PubMed] [Google Scholar]
- 51.Grotkjaer T, Akesson M, Christensen B, Gombert AK, Nielsen J. Impact of transamination reactions and protein turnover on labeling dynamics in 13C labeling experiments. Biotechnol Bioeng. 2004;86:209–216. doi: 10.1002/bit.20036. [DOI] [PubMed] [Google Scholar]
- 52.van Winden W, van Dam JC, Ras C, Kleijn RJ, van Gulik WM, Heijnen JJ. Metabolic-flux analysis of Saccharomyces cerevisiae CEN. PK113-7D based on mass isotopomer measurements of 13C-labeled primary metabolites. FEM Yeast Res. 2005;5:559–568. doi: 10.1016/j.femsyr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 53.Bardow A, Göke V, Koβ HJ, Lucas K, Marquardt W. Concentration-dependent diffusion coefficients from a single experiment using model-based Raman spectroscopy. Fluid Phase Equilibria. 2005;228–229:357–366. [Google Scholar]
- 54.Raghevendran V, Gombert AK, Christensen B, Kotter P, Nielsen J. Phenotypic characterization of glucose repression mutants of Saccharomyces cerevisiae using experiments with 13C-labelled glucose. Yeast. 2004;21:769–779. doi: 10.1002/yea.1136. [DOI] [PubMed] [Google Scholar]
- 55.Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wuthrich K, Bailey JE. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol. 1999;181:6679–6688. doi: 10.1128/jb.181.21.6679-6688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Bornholdt S. Systems biology. Less is more in modeling large genetic networks. Science. 2005;310:449–451. doi: 10.1126/science.1119959. This manuscript provides a theoretical framework, describing the different layers of systems biology and the role of fluxomics. [DOI] [PubMed] [Google Scholar]