Abstract
To improve transfection efficiency and reduce the cytotoxicity of polymeric gene vectors, reducible polycations (RPC) were synthesized from low molecular weight (MW) branched polyethyleneimine (bPEI) via thiolation and oxidation. RPC (RPC-bPEI0.8kDa) possessed a MW of 5 kDa~80 kDa, and 50%~70% of the original proton buffering capacity of bPEI0.8kDa was preserved in the final product. The cytotoxicity of RPC-bPEI0.8kDa was 8~19 times less than that of the gold standard of polymeric transfection reagents, bPEI25kDa. Although bPEI0.8kDa exhibited poor gene condensing capacities (~2 µm at a weight ratio (WR) of 40), RPC-bPEI0.8kDa effectively condensed plasmid DNA (pDNA) at a WR of 2. Moreover, RPC-bPEI0.8kDa/pDNA (WR ≥ 2) formed 100~200 nm-sized particles with positively charged surfaces (20~35 mV). In addition, the results of the present study indicated that thiol/polyanions triggered the release of pDNA from RPC-bPEI0.8kDa/pDNA via the fragmentation of RPC-bPEI0.8kDa and ion-exchange. With negligible polyplex-mediated cytotoxicity, the transfection efficiencies of RPC-bPEI0.8kDa/pDNA were approximately 1200~1500-fold greater than that of bPEI0.8kDa/pDNA and were equivalent or superior (~7-fold) to that of bPEI25kDa/pDNA. Interestingly, the distribution of high MW RPC-bPEI0.8kDa/pDNA in the nucleus of the cell was higher than that of low MW RPC-bPEI0.8kDa/pDNA. Thus, the results of the present study suggest that RPC-bPEI0.8kDa has the potential to effectively deliver genetic materials with lower levels of toxicity.
1. Introduction
Increasing interest in the cytosolic release of genetic materials has prompted the development of degradable polymeric carriers with low cytotoxicity. Their degradation can be triggered by cellular stimuli [1]. Among stimuli, intracellular pH [1, 2] and reduction-oxidation (redox) potential [3, 4] are the most commonly investigated triggers. The acidity of endosomes/lysosomes can trigger polymeric degradation [1]; however, the exposure of the released genetic materials to the endolysosomes reduces their bioavailability to target sites such as the cytosol and the nucleus. Redox potential is an attractive stimulus because of the strong gradient between the reducing subcellular compartments (i.e., cytosol and nucleus) and the oxidizing extracellular environments [3]. Disulfide bonds (~S-S~) [3–10] and diselenide bonds (~Se-Se~) [11] are promising chemical bonds for redox-induced polymeric degradation. Unlike pH-induced degradation, redox potential can trigger the release of biotherapeutics into the cytosol or the nucleus after the polymeric drug carriers escape from the endolysosomal compartment [7, 12].
Most reducible polycations (RPC) have been prepared from a range of cationic compounds that possess disulfide bonds in the middle of their chemical structures [4, 6, 8–10] or thiol groups at their terminal ends [4, 12–16]. Polycations with disulfide bonds have been prepared from dithiobis(succinimidyl propionate) (DSP) [9], N,N’-cystamine bisacrylamide (CBA) [6, 8, 10], and dimethyl-3,3’-dithiobis(propionimidate) (DTBP) [9], which react with amines. Upon reaction with primary amines, DSP is converted into amide linkages; thus, primary amines that are useful for gene condensation are lost during the reaction [9]. CBA generates new secondary or tertiary amines with proton buffering activity by consuming primary or secondary amines, respectively (called the Michael addition) [6, 8, 10]. However, CBA-mediated reactions produce low molecular weight (LMW) products (lower than 10 kDa) [6–8, 10] and produce unregulated polymeric architectures (branched or crosslinked) when primary and secondary amines coexist. When DTBP reacts with primary amines, the number of positive charges in the molecule is maintained due to the formation of amidine linkages; however, due to its rapid hydrolysis in aqueous solution, excess DTBP (~10 equivalents) has been frequently used. As a result, the MWs of the products are difficult to control.
One method for the synthesis of reducible ‘S-S’ polycations is the oxidation of thiolated compounds with positive charges. Using mono-, di-, tri-, or multi-thiolated molecules, well-designed polymer architectures (linear, branched, grafted, or crosslinked structures) can be obtained. However, thiolated species are often prepared through long and complicated reaction sequences [12, 13], and expensive peptides containing cysteine residues are required [14–16].
The objective of the present study was to enhance beneficial transfection characteristics (gene condensation with small amounts of polycations, proton buffering-mediated endosomal disruption, and high transfection efficiency) and to reduce cellular toxicity by mimicking high molecular weight (HMW) branched polyethyleneimine (bPEI) polymers (HMW bPEIs with a Mw of 25 kDa; bPEI25kDa), which are the gold standard of polymeric transfection reagents. Because bPEI0.8kDa possesses both primary amines, and secondary and tertiary amines, which can be used to condense genes and to enhance the proton buffering capacity of the polymers, respectively, the RPCs were synthesized from LMW bPEI (Mw 0.8 kDa; bPEI0.8kDa). bPEI0.8kDa molecules were linked via thiolation with 2-iminothiolane, followed by oxidation in aqueous solution. A procedure based on the use of 2-iminothiolane was employed in order to avoid problems associated with the use of DTBP. Moreover, by converting primary amines to amidine groups, the number of positive charges in the final product was maintained. The chemical, physical, and biological characteristics of bPEI0.8kDa-based RPC (RPC-bPEI0.8kDa) were investigated, and the cytotoxicity, proton buffering capacity, gene condensation ability, and transfection efficiency of RPC-bPEI0.8kDa was compared to that of non-degradable bPEI.
2. Materials and methods
2.1. Materials
Branched PEIs (bPEI0.8kDa (Mw 0.8 kDa, Mn 0.6 kDa), bPEI25kDa (Mw 25 kDa, Mn 10 kDa), and bPEI750kDa (Mw 750 kDa, Mn 60 kDa)), 2-iminothiolane, l-cysteine hydrochloride monohydrate, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), RPMI 1640 medium, Ca2+-free and Mg2+-free Dulbecco’s phosphate buffered saline (DPBS), triethylamine (TEA), dimethyl sulfoxide (DMSO), 4-(2-hydroxy-ethyl)-1-piperazine (HEPES), d-glucose, sodium bicarbonate, recombinant human insulin, ethidium bromide (EtBr), dithiothreitol (DTT), heparin, Hoechst 33342, and paraformaldehyde (PFA) were purchased from Sigma-Aldrich (St. Louis, MO). bPEI1.8kDa (Mr 1.8 kDa) was purchased from Polysciences, Inc (Warrington, PA), and plasmid DNA (pDNA) encoding firefly luciferase (gWiz-Luc or pLuc) was purchased from Aldevron, Inc. (Fargo, ND). Fetal bovine serum (FBS), penicillin-streptomycin antibiotics, trypsin-EDTA solution, LysoTracker®Red, and YOYO-1 were purchased from Invitrogen, Inc. (Carslbard, CA). Luciferase assay kit and BCA™ protein assay kit were purchased from Promega Corporation (Madison, WI) and Pierce Biotechnology, Inc. (Rockford, IL), respectively.
2.2. Cells and cell culture
HEK293 cells (human embryonic kidney cell line) and MCF7 cells (human breast adenocarcinoma cell line) were cultured in DMEM and RPMI 1640, respectively. The culture medium was supplemented with glucose (4.5 g/L for HEK293 cells and 2 g/L for MCF7 cells) and 10% FBS, and the cells were grown under humidified air containing 5% CO2 at 37°C. Additionally, insulin (4 mg/L) was added to the medium of MCF7 cells (RPMI 1640).
2.3. Synthesis and characterization of reducible polycations
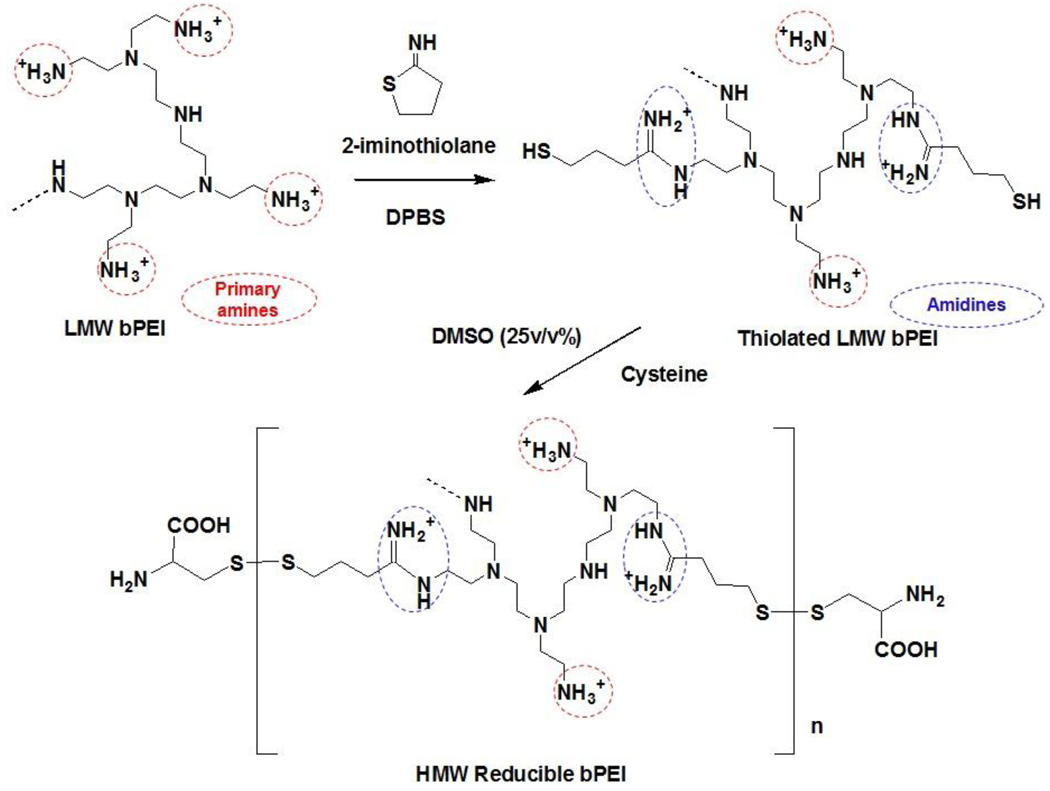
RPCs were synthesized from LMW bPEI0.8kDa via thiolation and oxidation, as shown in Fig. 1. The primary amines of bPEI0.8kDa (550 mg; 687.5 µmol of bPEI0.8kDa based on a Mw of 800; 3.2 mmol of amines based on the theoretical ratio (i.e., 1°:2°:3°=25%:50%:25%) of amines in bPEI) were thiolated with 2-iminothiolane (1.2~2 equivalents of bPEI0.8kDa) in DPBS (55 mL; pH 7.0~7.4) for 12 hr at room temperature (RT). Upon completion, DMSO (18.33 mL; one third DPBS by volume) and l-cysteine hydrochloride monohydrate (0.2~0.24 mmol) were added to the solution, and DMSO-induced oxidative polymerization of thiolated bPEI0.8kDa was conducted for 24 hr at RT. To remove DMSO and excess reactants, the polymer solution was dialyzed for 24 hr in deionized water using a dialysis membrane (MWCO 3500 Da). Finally, bPEI-based RPCs (RPC-bPEI0.8kDas) were lyophilized. The conditions used in the synthesis of RPC-bPEI0.8kDas are summarized in Table 1. The structure of RPC-bPEI0.8kDa was confirmed by 1H-NMR spectroscopy in D2O, and the MWs of RPC-bPEI0.8kDas were estimated by measuring the viscosities of the polymer solution and were compared to the Mns of commercial bPEIs [9, 17].
Fig. 1.
The synthesis of bPEI-based reducible polycations (RPC-bPEI).
Table 1.
Synthetic conditions of RPC-bPEI0.8kDa
| bPEI0.8kDa (µmol)1 |
Iminothiolane (µmol) |
Mole ratios of iminothiolane per bPEI0.8kDa |
l-Cysteine (µmol) |
|
|---|---|---|---|---|
| RPC-bPEI0.8kDa1 | 687.5 | 1375 | 2 | 200 |
| RPC-bPEI0.8kDa2 | 687.5 | 1375 | 2 | 240 |
| RPC-bPEI0.8kDa3 | 687.5 | 1100 | 1.6 | 240 |
| RPC-bPEI0.8kDa4 | 687.5 | 825 | 1.2 | 240 |
calculated based on Mw.
Using HEK293 and MCF7 cells, the cytotoxicity of the RPC-bPEI0.8kDas was measured with a MTT-based cell viability assay [18] and was compared to that of non-degradable bPEI25kDa and bPEI0.8kDa. HEK293 and MCF7 cells were seeded in 96-well plates at a density of 2×103 cells/well and 5×103 cells/well, respectively, and were cultured for 24 hr prior to the addition of RPC-bPEI0.8kDas and non-degradable bPEIs. In the assay, the cells were exposed to different concentrations of the polymer solution for 24 hr. Upon treating the cells (0.1 mL of culture medium) with a solution of MTT (10 µL; 5 mg/mL), surviving cells transform MTT into formazan crystals. After 4 hr of incubation, the culture medium was aspirated and DMSO was added to dissolve the crystals. Finally, the absorbance of the solution was measured at 570 nm with a microplate reader.
To measure the proton buffering capacities of the polymers, the RPC-bPEI0.8kDas were titrated via traditional acid-base titration methods [18]. RPC-bPEI0.8kDa and bPEI0.8kDa (10 mg) were dissolved in 150 mM NaCl (aq., 10 mL) and 1 N NaOH (aq., 100 µL). The polymer solution (1 mg/mL; 3 mL) was titrated with 0.1 N HCl at RT, and changes in the pH of the polymer solution were monitored. The proton buffering capacities of RPC-bPEI0.8kDas and bPEI0.8kDa were compared at a pH of 7.4~5.1 to determine the behavior of the polymers at the endolysosomal pH. To calculate the buffering capacity, the equation proposed by Ou et al. [6] was modified because the moles of protonable amines in the polymers are difficult to determine:
Here, ΔVHCl is the volume of HCl solution (0.1 N) required to increase the pH of the polymer solution from pH 7.4 to 5.1, CHCl is the concentration of HCl solution (0.1 N), and mPolymer is the mass (3 mg) of the polymer. The estimated proton buffering capacities of RPC-bPEI0.8kDas were compared with that of bPEI0.8kDa.
2.4. Preparation and physicochemical characteristics of polyplexes
As previously reported [19, 20], polycations (RPC-bPEI0.8kDas and bPEIs) were combined with pDNA in HBG (pH 7.4, 20 mM HEPES, 5 wt/vol% glucose). After incubation (20 µL per 1 µg of pDNA) for 30 min at RT, the resultant polyplexes were used in further studies. In the present investigation, either the weight ratio (WR; polycation (wt) per pDNA (wt)) or the N/P ratio (amines (N) of polycation per phosphate groups (P) of pDNA) was varied.
Condensation of pDNA with RPC-bPEI0.8kDa was monitored with a gel electrophoresis assay [21, 22]. RPC-bPEI0.8kDa/pDNA complexes with different complexation ratios were loaded into the wells of a 0.8 wt/vol% agarose gel supplemented with EtBr. A constant voltage (100 V) was applied to the polyplex-loaded gel in 0.5X TBE buffer for 60 min. Uncomplexed pDNA or exposed pDNA in the polyplexes was detected with a UV illuminator. In addition, to evaluate thiol-triggering pDNA exposure or release from RPC-bPEI0.8kDa/pDNA complexes, the polyplexes were exposed to heparin and/or a solution of DTT in 150 mM NaCl for 1 hr and then subjected to electrophoresis.
The surface charge and particle size of RPC-bPEI0.8kDa/pDNA complexes were measured with a Zetasizer 3000HS (Malvern Instrument, Inc, Worcestershire, UK) at a wavelength of 677 nm and a constant angle of 90° at RT. The concentration of pDNA in the polyplex solution was set to 2.5 µg/mL and 5 µg/mL, respectively.
2.5. Transfection-mediated biological characteristics of polyplexes
As previously reported [20, 21, 23], the transfection experiments were conducted in 6-well plates. HEK293 and MCF7 cells were seeded at a density of 1×105 cells/well and 5×105 cells/well, respectively, and the cells were cultured for 24 hr. One hour before transfection, the complete culture medium was replaced with serum-free and insulin-free medium. The polyplexes (20 µL per µg of pDNA) were added, and the cells were transfected for 4 hr. Upon completion, the cells were incubated for an additional 44 hr in the complete culture medium. When the transfection experiments were complete, the cells were rinsed twice with DPBS and lysed in a lysis buffer. The relative luminescence unit (RLU) and protein content of the transfected cells were measured according to the protocols of the manufacturer.
To induce the cellular uptake of the polyplexes, the cells were prepared in 6-well plates, and polyplexes (20 µL per 1 µg of pDNA) prepared with YOYO-1-intercalated pDNA were added into the wells. After 4 hr of incubation, the cells were detached from the plates and fixed with a 4% solution of PFA. Cells containing fluorescent polyplexes were monitored via flow cytometry (FACScan Analyzer, Becton-Dickinson, Franklin Lakes, NJ), and a primary argon laser (488 nm) and a fluorescence detector (530±15 nm) were used to detect YOYO-1. The uptake of the polyplexes into the cells was analyzed with a gated population of at least 5,000 cells.
HEK293 and MCF7 cells were seeded in 12-well plates at a density of 5×104 cells/well and 2.5×105 cells/well, respectively, and the MTT-based cytotoxicity of the polyplex-transfected cells was assessed. The experimental procedure used to evaluate the cytotoxicity of transfected cells was similar to that of the in vitro transfection studies; however, the concentration of the polyplexes was set to 10 µL per 0.5 µg of pDNA. After 48 hr of transfection, a solution of MTT (0.1 mL; 5 mg/mL) was applied to the cells (1 mL of culture medium), and the cytotoxicity of the polyplexes was measured after 4 hr of incubation, as described in the in vitro polymer-mediated cytotoxicity tests.
To identify subcellular locations of pDNA, the transfected cells were seeded on coverslips, as previously described in the transfection experiments. Polyplexes (20 µL per 1 µg of pDNA) were prepared by adding YOYO-1-intercalated pDNA to the cells. After 4 hr of transfection (30 min prior to sampling), LysoTracker®Red dye and Hoechst33342 were added to stain acidic intracellular vesicles and the nucleus, respectively. The cells were rinsed with DPBS and were fixed with 4% PFA. Upon completion, the cells were evaluated with a laser scanning confocal microscope equipped with excitation lasers (408 nm for the diode, 488 nm for Ar, and 543 nm for HeNe) and variable band-pass emission filters. Confocal images were collected in 500-nm sections and used to construct images of the entire cell.
The statistical significance of the data was evaluated by conducting an unpaired Student t-test at a confidence level of p<0.05.
3. Results and discussion
3.1. Synthesis and characterization of RPC-bPEI0.8kDa polymers
Polycations (RPC) that degrade due to changes in the intracellular reduction potential were synthesized from LMW bPEI0.8kDa via thiolation and oxidation (Fig. 1). In this study, 2-iminothiolane was used to create thiol groups from primary amines. In the unique approach employed in this investigation, a primary amine reacts with 2-iminothiolane to yield a thiol group and an amidine moiety. As a result, unlike other thiolation methods, the number of positive charges at a neutral pH was preserved. Moreover, the newly generated amidines contribute to the electrostatic condensation of nucleic acids. The degree of thiolation of bPEI0.8kDa was limited to ≤ 2 thiol groups per bPEI0.8kDa because excess thiols can result in the formation of nano/micro-sized bPEI gels via crosslinking. To produce HMW degradable bPEIs equipped with disulfide bonds, DMSO (25 v/v%) was added to oxidize thiolated bPEI0.8kDa.
The structure of RPC-bPEI0.8kDa was confirmed by 1H-NMR spectroscopy. As shown in Fig. S1, the spectra of bPEI0.8kDa in D2O consisted of three major peaks centered at δ=2.6 (–CH2CH2NH3+), δ=2.7 (–NHCH2CH2-), and δ=3.0 (–NCH2CH2-). RPC-bPEI0.8kDas were characterized by the three peaks originating from bPEI0.8kDa, as well as newly generated peaks at δ=2.0 (–NHC(=NH2+)CH2CH2CH2S-), δ=3.45 (–NHC(=NH2+)CH2CH2CH2S- and –SCH2CH(COOH)NH2), δ=3.7 (–CH2CH2NHC(=NH2+)CH2CH2CH2S-), and δ=3.85 (–NHC(=NH2+)CH2CH2CH2S- and –SCH2CH(COOH)NH2). The mole ratios of bPEI0.8kDa, iminothiolane-mediated amidines (–NHC(=NH2+)CH2CH2CH2S-) and cysteine residues (-SCH2CH(COOH)NH2) in RPC-bPEI0.8kDa were calculated by integrating the corresponding peaks, as summarized in Table 2. The MWs of RPC-bPEI0.8kDas were estimated based on the viscosity of the polymer solution [9, 17], and values of 80.3 kDa, 52.7 kDa, 6.3 kDa, and 5.7 kDa were obtained for RPC-bPEI0.8kDa1, RPC-bPEI0.8kDa2, RPC-bPEI0.8kDa3, and RPC-bPEI0.8kDa4, respectively. bPEI0.8kDa-based RPCs produced via Michael addition [10] or from reaction with propylene sulfide [17] yield lower MWs than that of bPEI25kDa; thus, the results revealed that the proposed synthetic procedure can produce low or high MW RPC-bPEI0.8kDas, depending on the feed ratio.
Table 2.
Estimated mole ratios of bPEI0.8kDa, iminothiolane-mediated amidine (-NHC(=NH2+)CH2CH2CH2S-), and cysteine (-SCH2CH(COOH)NH2) moieties, the mole ratio of amidine groups per cysteine residues, and the calculated mass fraction (%) of bPEI0.8kDa in RPC-bPEI0.8kDa
| Mole ratios per bPEI0.8kDa | Mole ratio of amidine moieties per cysteine residues |
Mass fraction (%) of bPEI0.8kDa in RPC- bPEI0.8kDa |
||
|---|---|---|---|---|
| Amidine groups | Cysteine | |||
| RPC-bPEI0.8kDa1 | 2.00 | 0.31 | 6.5 | 76.8% |
| RPC-bPEI0.8kDa2 | 2.03 | 0.19 | 10.7 | 77.7% |
| RPC-bPEI0.8kDa3 | 1.66 | 0.09 | 18.4 | 81.6% |
| RPCbPEI0.8kDa4 | 1.30 | 0.11 | 11.8 | 84.6% |
The RPC-bPEIs, which can be degraded by thiol-containing compounds such as reduced glutathiones and cysteines, are anticipated to: 1) reduce polycation-induced cytotoxicity, 2) maintain polycation-mediated proton buffering capacity, 3) effectively condense nucleic acids at low complexation ratios (polycation to pDNA), and 4) release pDNA from RPC-bPEI/pDNA polyplexes under thiol/polyanion-rich environments (e.g., the cytosol and the nucleus).
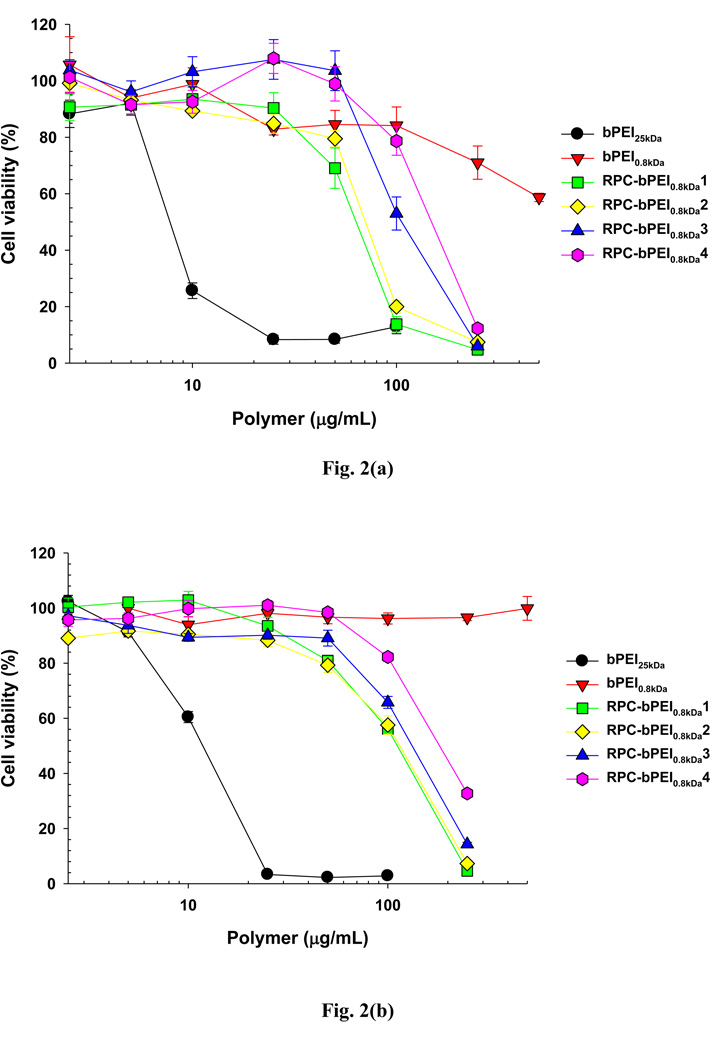
After 24 hr of treatment, the cytotoxicities of the RPC-bPEI0.8kDas to HEK293 and MCF7 cells (Fig. 2) were determined. The results indicated that the cytotoxicities of RPC-bPEI0.8kDas to HEK293 cells were greater than that of bPEI0.8kDa due to the relatively high MW of the RPC-bPEI0.8kDas. However, although the MW of RPC-bPEI0.8kDa1 and RPC-bPEI0.8kDa2 was significantly higher than that of bPEI25kDa, the IC50s of RPC-bPEI0.8kDa1 (62 µg/mL) and RPC-bPEI0.8kDa2 (70 µg/mL) were approximately 8~9-fold higher than that of bPEI25kDa (8 µg/mL). Moreover, the cytotoxicities of RPC-bPEI0.8kDa3 and RPC-bPEI0.8kDa4, which possess a MW of 5~6 kDa, were 14~19 times lower (IC50: 110 µg/mL and 150 µg/mL, respectively) than that of bPEI25kDa. Similarly, with MCF7 cells, the cytotoxicities of the RPC-bPEI0.8kDas (IC50: 110 µg/mL for RPC-bPEI0.8kDa1 and RPC-bPEI0.8kDa2, 135 µg/mL for RPC-bPEI0.8kDa3, and 185 µg/mL for RPC-bPEI0.8kDa4) were 9~15-fold less than that of bPEI25kDa (IC50: 12 µg/mL). These results are consistent with the reported cytotoxicities (4~18 folds lower) of other oligoethylenimine-based RPCs with MWs lower than that of bPEI25kDa (the toxicities of the materials were tested on different cell lines) [9, 17]. In general, the toxicity of HMW PEI is greater than that of LMW PEI because HMW PEI interacts more effectively with components that are essential for cell survival such as intracellular membranes, vital proteins, and nucleic acids than its LMW counterpart [3, 24]. Thus, the reduced cytotoxicity of HMW RPC-bPEI0.8kDa may be due to the degradation of the polymer in the intracellular environment after cellular uptake.
Fig. 2.
Cytotoxicity of RPC-bPEI0.8kDa and bPEI (bPEI0.8kDa and bPEI25kDa) in (a) HEK293 cells and (b) MCF7 cells (n=6, Mean±SEM).
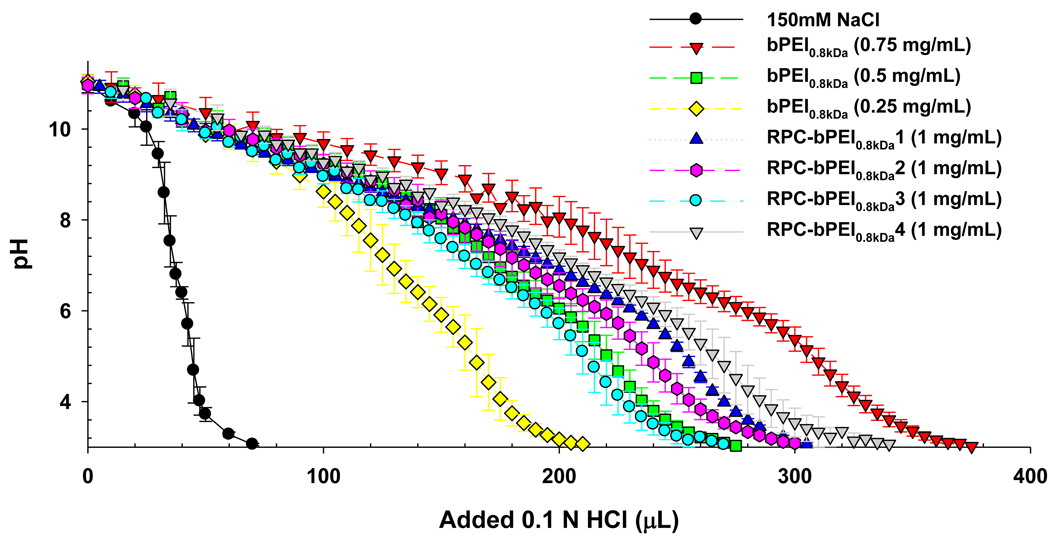
The RPC-bPEI0.8kDas were titrated [18] to evaluate the buffering capacity of RPC-bPEI0.8kDa between pH 7.4 and pH 5.1. The proton buffering capacities of RPC-bPEI0.8kDa were compared to that of bPEI0.8kDa at different concentrations (0.25 mg/mL to 0.75 mg/mL). As shown in Fig. 3, the buffering capacity of RPC-bPEI0.8kDa1, RPC-bPEI0.8kDa2, RPC-bPEI0.8kDa3, and RPC-bPEI0.8kDa4 was approximately 70%, 61%, 52%, and 69% of the buffering capacity of bPEI0.8kDa. Thus, the reduced buffering capacity of RPC-bPEI0.8kDas may be attributed to the relative decrease in the mass fraction of bPEI0.8kDa. As shown in Table 2, the mass fraction of bPEI0.8kDa in the RPCs decreased to approximately 75%~85% due to the introduction of amidine moieties (-NHC(=NH2+)CH2CH2CH2S-) and cysteine residues (–SCH2CH(COOH)NH2).
Fig. 3.
Acid-base titration curves of RPC-bPEI0.8kDa and bPEI0.8kDa (n=3, Mean±SD).
Another factor affecting the proton buffering capacity of the polymers is the hydrophobicity/hydrophilicity of the local environment [25]. Replacing a primary amine (-NH3+) with an amidine moiety (-NHC(=NH2+)CH2CH2CH2S-) may locally increase the ‘-CH2CH2CH2-’-induced hydrophobicity of neighboring amine groups, which can suppress the protonation of amines and further reduce the buffering capacity of RPC-bPEI0.8kDas. However, the cysteine residues of RPC-bPEI0.8kDa may compensate for the hydrophobicity of amidine groups by introducing hydrophilic amines and carboxylic acids into the polymer. Namely, as the mole ratio of amidine groups to cysteine residues in RPC-bPEI0.8kDa increased (i.e., the hydrophobicity increases), the buffering capacity of the polymers decreased below the theoretical value (based on the mass fraction of bPEI0.8kDa in RPC-bPEI0.8kDa), as shown in Table 2.
3.2. Physicochemical characteristics of RPC-bPEI0.8kDa/pDNA complexes
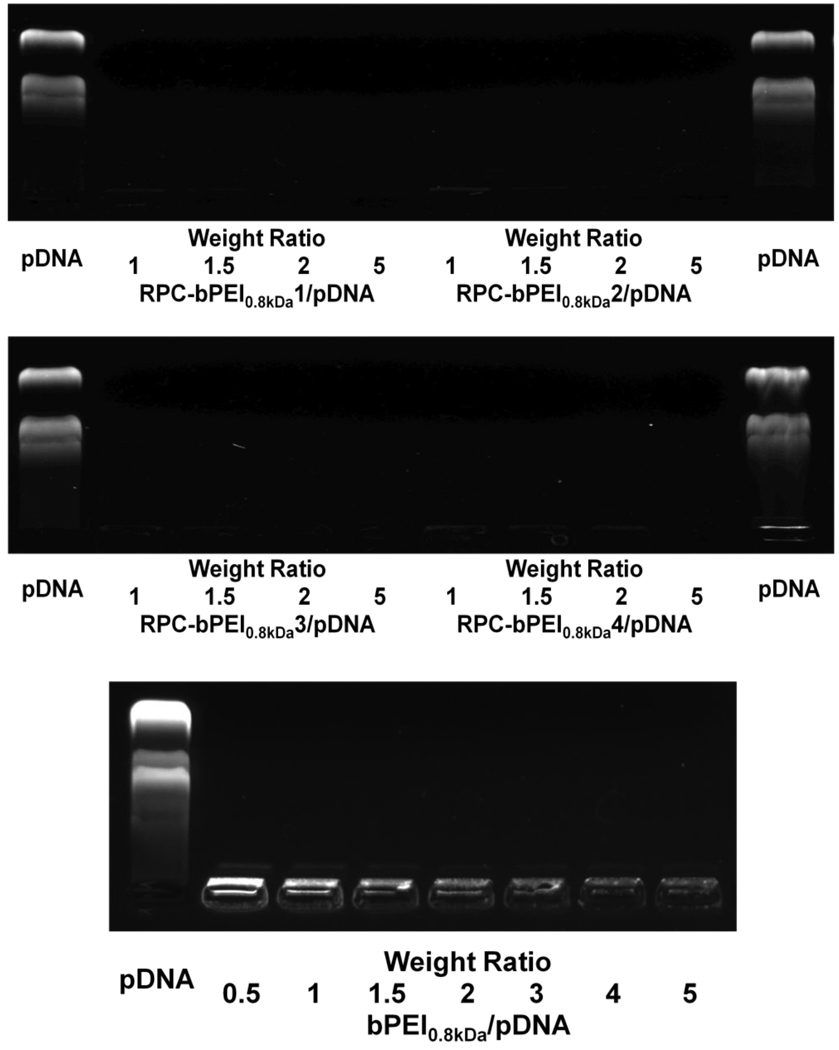
The gene condensing capabilities of RPC-bPEI0.8kDas were evaluated by conducting a gel retardation assay to determine whether the gene was shielded by polycations or exposed on the surface of the polyplex. Although bPEI0.8kDa/pDNA polyplexes prepared at a weight ratio (WR) of 1 (N/P~7.7) were completely retarded at the loading well, pDNA on the polyplexes were partially exposed, as evidenced by the moderate fluorescent signal of EtBr-intercalated pDNA (Fig. 4). As the ratio of bPEI0.8kDa to pDNA increased (i.e., increasing WR), the EtBr signals of the polyplexes decreased. RPC-bPEI0.8kDa/pDNA polyplexes formed at a WR ≤ 0.5 contained uncomplexed or exposed pDNA (Fig. S2). However, RPC-bPEI0.8kDa/pDNA polyplexes formed at a WR ≥ 1 were fully retarded at the loading well, and pDNA was completely shielded (Fig. 4). In general, higher MW polycations can complex pDNA more effectively [24].
Fig. 4.
Condensation of pDNA with RPC-bPEI0.8kDa in agarose gel.
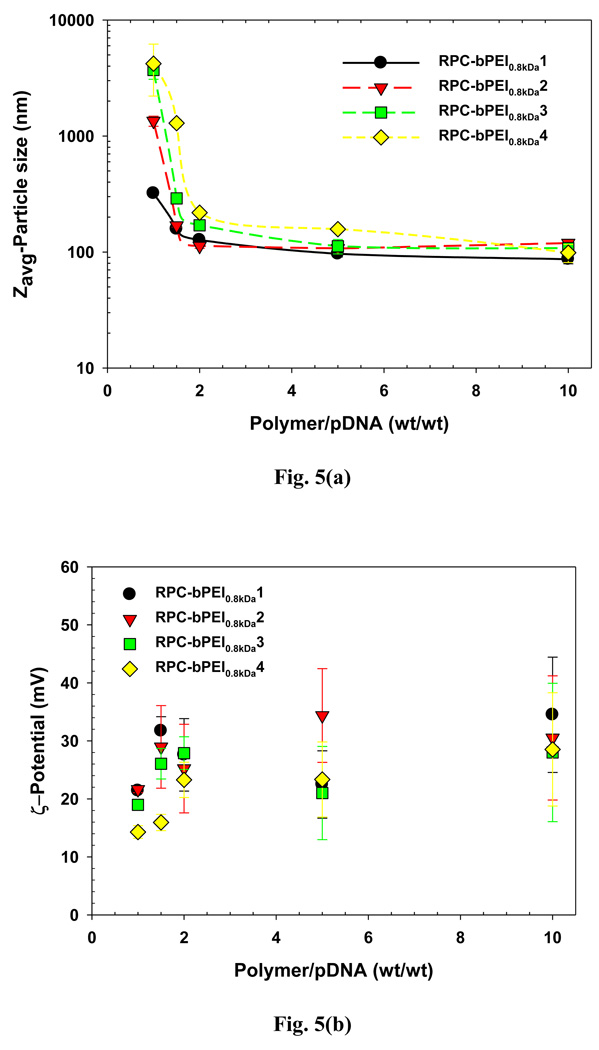
To further confirm pDNA complexation with RPC-bPEI0.8kDa, the size and surface charge of the polyplexes were determined. bPEI0.8kDa produced at a WR of 40 formed gene complexes that were approximately 2 µm in size. However, as shown in Fig. 5(a), at a fixed WR, the particle size of RPC-bPEI0.8kDa/pDNA polyplexes decreased with an increase in the MW of RPC-bPEI0.8kDa. At a WR of 1.5, the particle size of gene complexes derived from RPC-bPEI0.8kDa1 and RPC-bPEI0.8kDa2 were less than 200 nm. However, at a WR ≥ 2, the diameter of RPC-bPEI0.8kDa1 and RPC-bPEI0.8kDa2 polyplexes was approximately 100 nm. Regardless of the MW of RPC-bPEI0.8kDa, all RPC-bPEI0.8kDa/pDNA polyplexes formed at a WR ≥ 2 exhibited a diameter of 100~200 nm, which is the optimal size range for in vivo studies. In addition, as shown in Fig. 5 (b), all RPC-bPEI0.8kDa/pDNA polyplexes exhibited positive surface charges. In particular, complexes with a WR ≥ 2 displayed a surface zeta potential of 20~35 mV.
Fig. 5.
(a) Particle size and (b) surface charge of RPC-bPEI0.8kDa/pDNA complexes in HBG.
The thiol-rich environment (about 20 mM) of the cytosol was mimicked with DTT (a reducing reagent) to determine whether RPC-bPEI0.8kDa/pDNA polyplexes dissociate and release pDNA via the thiol-triggered degradation of RPC-bPEI0.8kDa. Prior to studying the RPC-bPEI0.8kDa/pDNA polyplexes, the gene condensing capabilities of the reduced polymer fragments were examined. RPC-bPEI0.8kDa was treated with 20 mM DTT for 48 hr to achieve the complete degradation of RPC-bPEI0.8kDa into bPEI0.8kDa derivatives. With DTT-pretreated RPC-bPEI0.8kDa, the formed pDNA complex was subjected to electrophoresis. pDNA was not released from the complexes, but pDNA was partially exposed in the loading well (Fig. S3). These results are similar to those of bPEI0.8kDa complexes prepared at a low WR (Fig. 4). However, as shown in Fig. S4, DTT-pretreated RPC-bPEI0.8kDa/pDNA complexes prepared at a WR of 4 were 1~2 µm in diameter and were similar to bPEI0.8kDa/pDNA produced at a WR of 4 (about 2 µm). Moreover, the particle sizes of DTT-pretreated RPC-bPEI0.8kDa/pDNA complexes were significantly different from that of RPC-bPEI0.8kDa/pDNA complexes (100~200 nm, Fig. 5(a)). These observations indicate that RPC-bPEI0.8kDa degrades in a thiol-rich environment; however, fragmentation did not induce the release of pDNA.
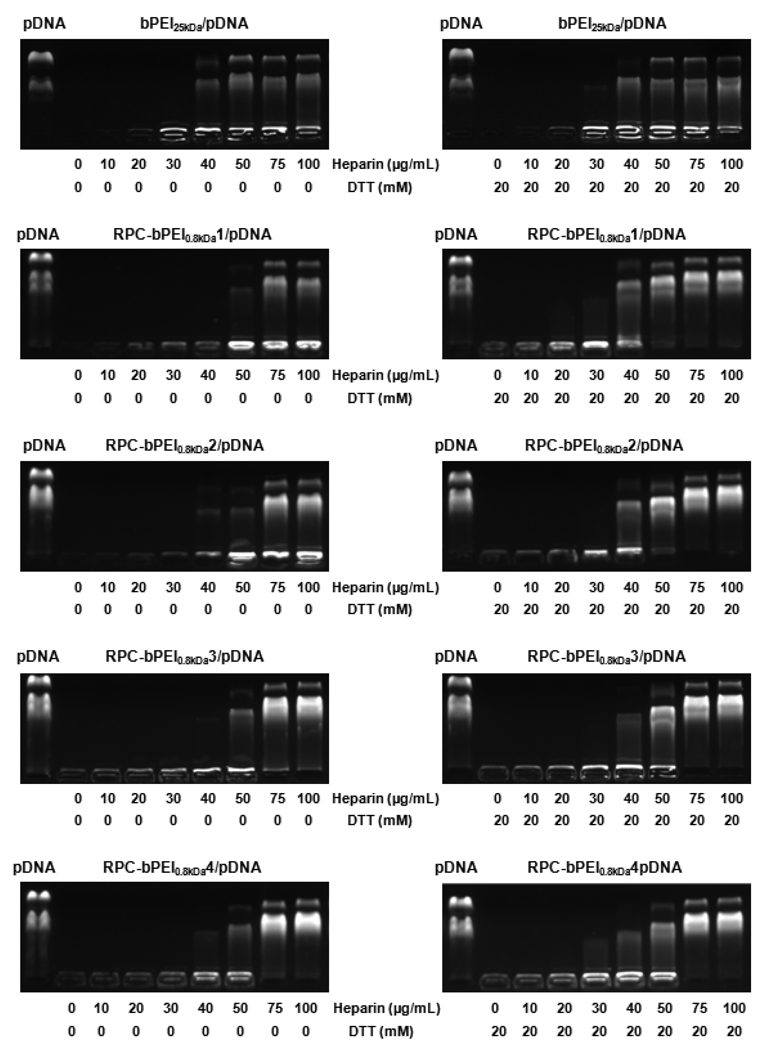
Unpackaging of pDNA from the polyplexes was observed due to the presence of glycosaminoglycans in plasma [26–28], cytosolic RNA in the cytoplasm [29], and chromosomal DNA in the nucleus [30]. Thus, a model polyanion (heparin) was added to the DTT solution (20 mM) to mimic extracellular (lower polyanion concentrations; about 20 µg/mL) or intracellular environments (higher polyanion concentrations).
The polyplexes were exposed to heparin (0~100 µg/mL), and the results indicated that pDNA from the bPEI25kDa/pDNA (N/P 5) complex was readily released or exposed at lower heparin concentrations than that of the RPC-bPEI0.8kDa/pDNA (WR 2) complex (especially, HMW RPC-bPEI0.8kDa) (Fig. 6), which suggests that RPC-bPEI0.8kDa/pDNA complexes are more stable than bPEI25kDa/pDNA complexes. Compared to pure heparin, DTT (20 mM) induced the release or exposure of pDNA from RPC-bPEI0.8kDa/pDNA (WR 2) at lower heparin concentrations. Alternatively, the release of pDNA from non-reducible bPEI25kDa/pDNA complexes was not affected by thiols (Fig. 6).
Fig. 6.
DTT-triggering pDNA release from RPC-bPEI0.8kDa/pDNA complexes (WR 2) in the presence of a solution of heparin in 150 mM NaCl.
Various authors report that reducible polyplexes made of linear PEI or chemicals other than bPEI display thiol-triggered gene release in the absence of polyanions [6, 7]. However, to release pDNA under non-cellular conditions, the proposed RPC-bPEI0.8kDa/pDNA complexes required thiol and heparin. The release conditions of the proposed RPC-bPEI0.8kDa/pDNA complexes may be attributed to the physicochemical and gene condensing characteristics of the starting materials. For instance, bPEI0.8kDa has a high charge density and its pDNA complexes were completely retarded. Additionally, the amidine groups of DAPI can bind to pDNA; thus, the newly generated amidine groups in RPC-bPEI0.8kDa may enhance the complexation of pDNA [31]. Unlike intracellular conditions, the continued reduction of disulfide bonds in non-cellular conditions is limited because the reaction reaches equilibrium over time. However, in an intracellular environment, thiols can be continuously supplied by glutathione reductase, and a GSH/GSSG ratio of 30 and 100 is maintained in the cell [3, 32]. Thus, although RPC-bPEI0.8kDa/pDNA may be stable in extracellular environments, the release of pDNA may occur under intracellular conditions.
3.3. Transfection-mediated biological characteristics of RPC-bPEI0.8kDa/pDNA complexes
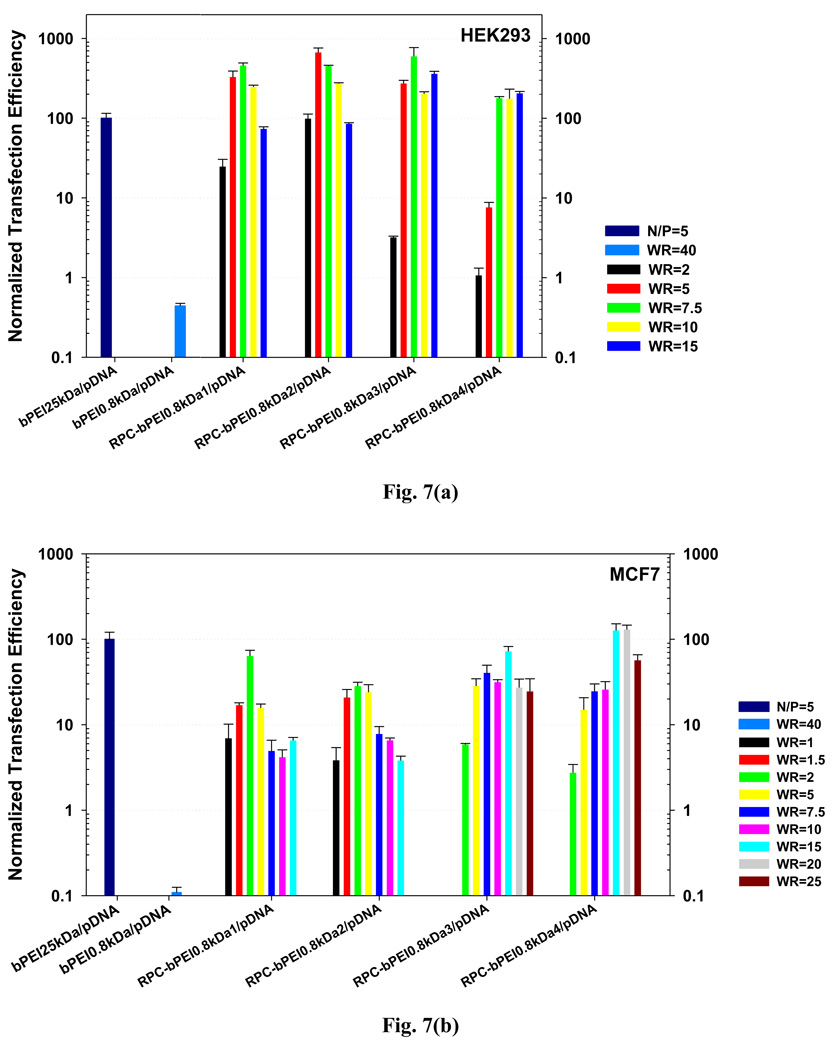
Complexes of RPC-bPEI0.8kDa and a luciferase reporter gene were prepared at various WRs. The complexes were applied to HEK293 and MCF7 cells, and their transfection efficiencies were compared to that of bPEI25kDa/pDNA (N/P 5) and bPEI0.8kDa/pDNA (WR 40). Complexation conditions resulting in low polyplex toxicity were employed to exclude the effects of polyplex toxicity-mediated damage on the polymeric transfection efficiency (Fig. S5). When the transfection efficiency of bPEI25kDa/pDNA (N/P 5)-transfected HEK293 cells was set to 100, the expression (p<0.001) of luciferase from RPC-bPEI0.8kDa/pDNA complexes was 2–7 times higher, depending on the RPC-bPEI0.8kDa and the gene complexation ratio. Moreover, the transfection efficiencies of RPC-bPEI0.8kDa/pDNA complexes were 1500-fold greater (p<0.001) than that of bPEI0.8kDa/pDNA (WR 40). Similarly, the results of in vitro transfection studies of RPC-bPEI0.8kDa/pDNA complexes with MCF7 cells indicated that the transfection efficiency was equal to that of bPEI25kDa/pDNA (N/P 5) and was 1200-fold greater (p<0.001) than that of bPEI0.8kDa/pDNA (WR 40).
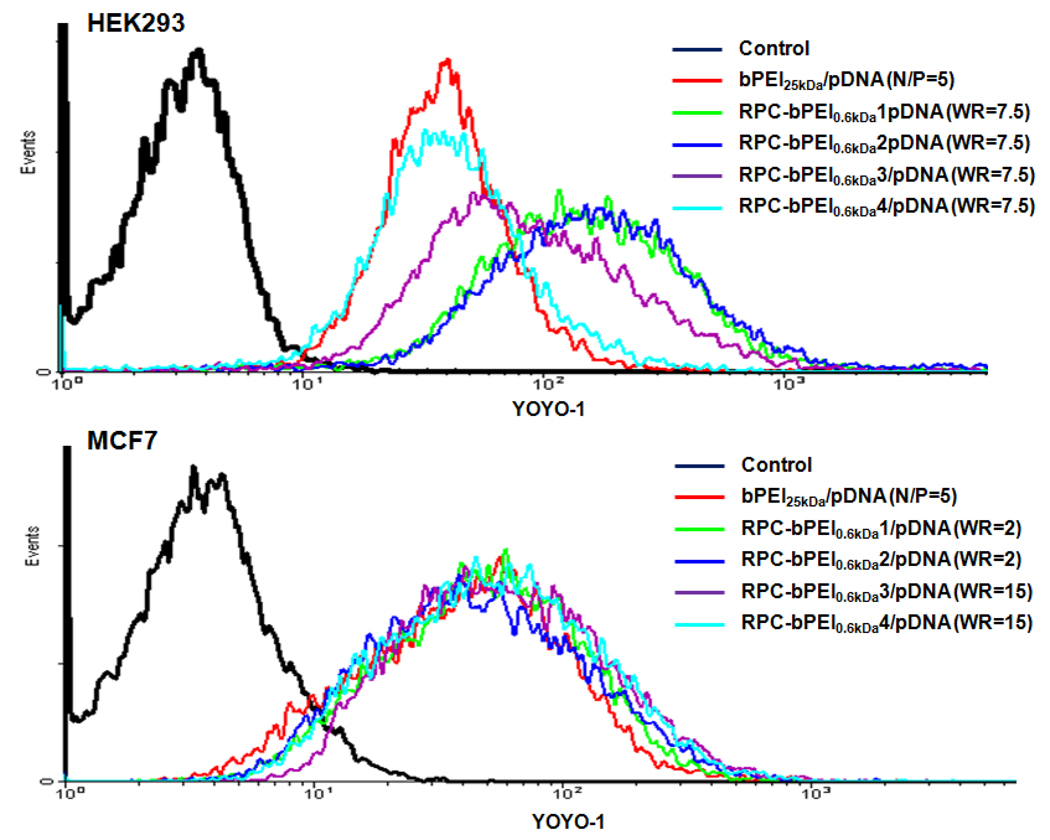
The aforementioned results were supported by those obtained from the RPC-bPEI0.8kDa/pDNA uptake studies, which were conducted 4 hr after transfection. At a WR of 7.5, the highest RPC-bPEI0.8kDa/pDNA-mediated HEK293 transfection efficiency was observed, and the polyplexes of RPC-bPEI0.8kDa1, RPC-bPEI0.8kDa2, and RPC-bPEI0.8kDa3 displayed higher YOYO-1 intensities (i.e., more polyplex uptake) inside the cells than RPC-bPEI0.8kDa4/pDNA and bPEI25kDa/pDNA (N/P 5) (Fig. 8). Alternatively, the cellular uptakes of RPC-bPEI0.8kDa4/pDNA and bPEI25kDa/pDNA (N/P 5) were not significantly different. In the transfection of MCF7, cellular internalization levels of RPC-bPEI0.8kDa1/pDNA (WR 2), RPC-bPEI0.8kDa2/pDNA (WR 2), RPC-bPEI0.8kDa3/pDNA (WR 15), and RPC-bPEI0.8kDa4/pDNA (WR 15) were nearly identical to that of bPEI25kDa/pDNA (N/P 5), as shown in Fig. 8.
Fig. 8.
RPC-bPEI0.8kDa/pDNA- and bPEI25kDa/pDNA-uptake of (a) HEK293 cells and (b) MCF7 cells.
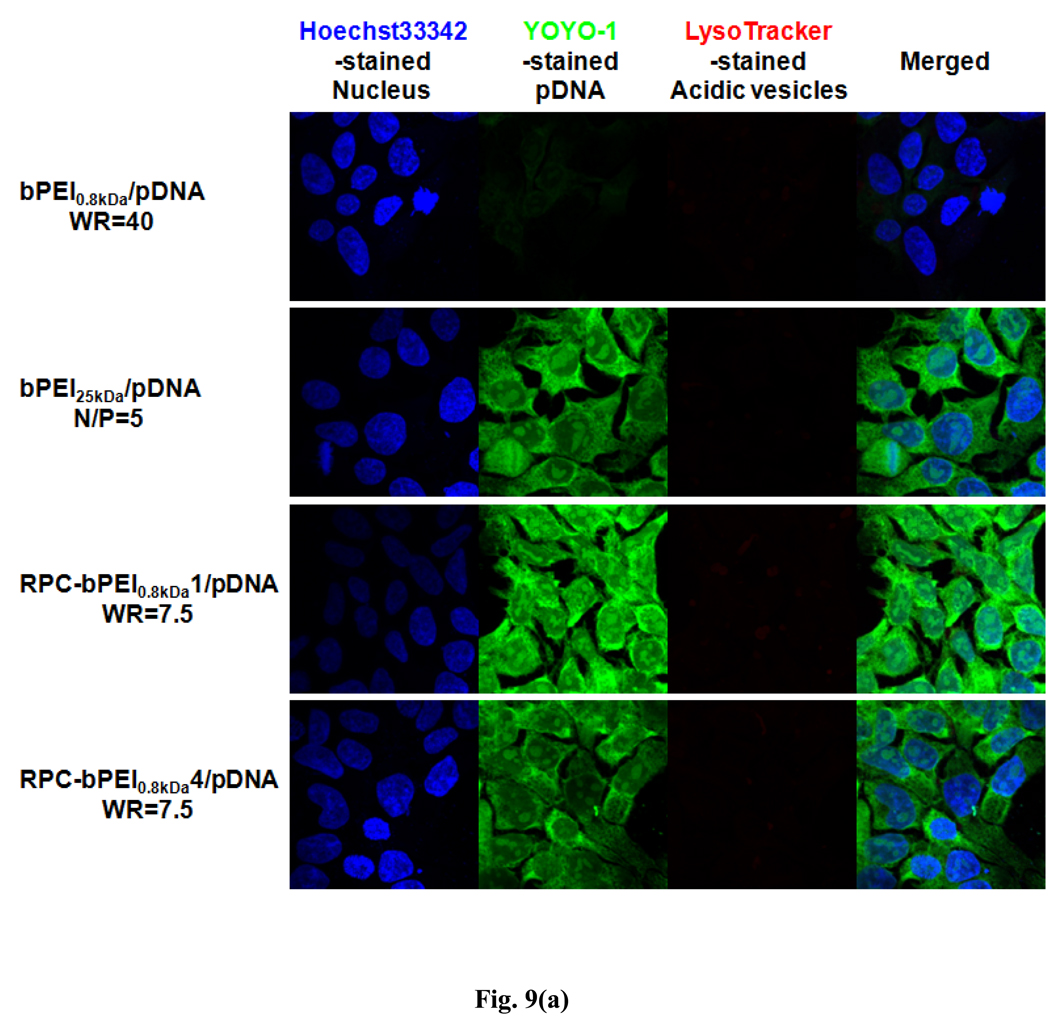
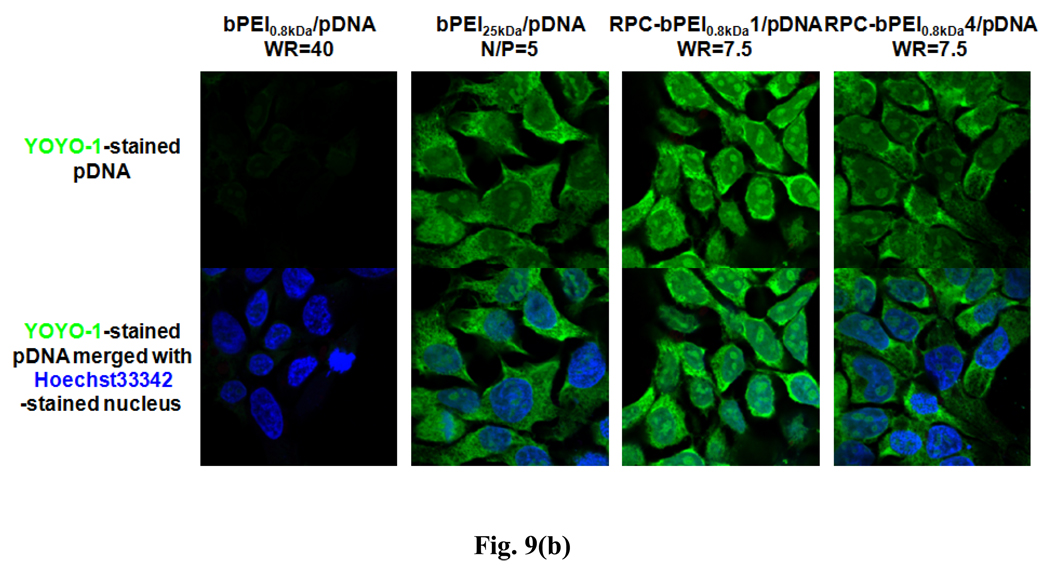
To verify the transfection results of RPC-bPEI0.8kDa/pDNA polyplexes in HEK293 cells, the intracellular localization of YOYO-1-intercalated pDNA was identified by subcellular compartment staining with LysoTracker®Red dye (for the acidic late endosomal and lysosomal compartments) and Hoechst 33342 (for the nucleus). As shown in Fig. 9(a), the accumulated images indicated that less bPEI0.8kDa/pDNA (WR 40) was incorporated into the cell than RPC-bPEI0.8kDa/pDNA (WR 7.5) and bPEI25kDa/pDNA (N/P 5). Moreover, similar to the polyplex uptake results, the YOYO-1 intensity of RPC-bPEI0.8kDa1/pDNA (WR 7.5)-transfected HEK293 cells was stronger than that of RPC-bPEI0.8kDa4/pDNA (WR 7.5)- and bPEI25kDa/pDNA (N/P 5)-transfected cells. In addition, bPEI25kDa/pDNA (N/P 5) polyplexes were not detected in the acidic vesicles of the cell due to their strong proton buffering capacity and propensity for endosomal escape. Interestingly, similar to the results of bPEI25kDa/pDNA (N/P 5) and bPEI0.8kDa/pDNA (WR 40), only minor amounts of RPC-bPEI0.8kDa1/pDNA and RPC-bPEI0.8kDa4/pDNA were trapped inside the acidic vesicles of the cell. Although approximately 70% of the proton buffering capacity of bPEI0.8kDa was preserved in the RPC-bPEI0.8kDas, the proton buffering capacity of RPC-bPEI0.8kDa/pDNA complexes may be strong enough to favor endosomal escape.
Fig. 9.
Intracellular localization of pDNA delivered by RPC-bPEI0.8kDa, bPEI25kDa, and bPEI0.8kDa in polyplex-transfected HEK293 cells 4 hr after transfection: (a) accumulated images and (b) center-sectional images. Nuclei, pDNA, and acidic vesicles were distinguished using Hoechst 33342 (blue), YOYO-1 (green), and LysoTracker® dye (red).
To analyze the intracellular distribution of the polyplexes, the center-sectioned images of the cells were combined, as shown in Fig. 9(b). The results indicated that the internalized complexes of bPEI0.8kDa/pDNA and bPEI25kDa/pDNA were more localized in the cytoplasm than in the nucleus. Moreover, LMW RPC-bPEI0.8kDa4/pDNA complexes displayed a greater propensity for cytoplasmic distribution. Alternatively, the distribution of HMW RPC-bPEI0.8kDa1/pDNA in the nucleus was greater than that of the cytosol. In particular, the majority of RPC-bPEI0.8kDa1/pDNA complexes in the cytosplasm were accumulated in the perinuclear area. LMW RPC-bPEI0.8kDa4/pDNA and HMW RPC-bPEI0.8kDa1/pDNA have similar proton buffering capacities at the endosomal pH; nevertheless, differences in the intracellular localization of RPC-bPEI0.8kDa/pDNA polyplexes could be caused by differences in pDNA unpackaging rates, which may be attributed to differences in the MWs of the complexes. In the cytoplasm, LMW RPC-bPEI0.8kDa4/pDNA complexes dissociate more rapidly than HMW RPC-bPEI0.8kDa1/pDNA complexes. However, compared to complexed pDNA, free pDNA within the cytoplasm may move slowly [33–36] and become degraded by cytosolic nucleases [37].
The results of the present study suggest that iminothiolane-mediated RPC synthesis can be used to produce HMW RPCs or LMW RPCs, depending on the feed composition. Further, it is expected that the synthetic protocol will aid in the design of novel and effective reducible polymeric gene carriers from small natural molecules such as amino acids and spermine.
The cytotoxicity of high MW RPC-bPEI0.8kDa was lower than that of bPEI25kDa, indicating that intracellular interactions between polycations and intracellular compartments and proteins may be more significant than polycation-mediated damage of the plasma membrane [3]. Moreover, compared to the MW of bPEI25kDa, the MWs of RPC-bPEI0.8kDa1 and RPC-bPEI0.8kDa2 may be higher in extracellular environments and lower in the cytosol/nucleus.
Additionally, the selective intracellular localization of RPC-bPEI0.8kDa/pDNA complexes suggests that HMW RPC-bPEI0.8kDa and LMW RPC-bPEI0.8kDa are effective for the nuclear delivery of pDNA and the cytoplasmic delivery of siRNA and antisense oligonucleotide, respectively.
4. Conclusion
Gene-condensable and reducible polycations with various MWs and buffering capacities were synthesized from LMW bPEI. RPC-bPEI0.8kDa was less cytotoxic than bPEI25kDa, and the transfection efficiency of RPC-bPEI0.8kDa-mediated polyplexes was equivalent or superior to that of bPEI25kDa/pDNA complexes. RPC-bPEI0.8kDa/pDNA complexes exhibited different intracellular localization preferences, and the distribution of the complexes within the cell was dependent on the MW of the polyplex. Thus, the results of this study suggest that RPC-bPEI0.8kDa polymers have the potential for effective polymeric gene delivery.
Supplementary Material
Fig. 7.
Normalized transfection efficiencies of RPC-bPEI0.8kDa/pDNA-, bPEI0.8kDa/pDNA- and bPEI25kDa/pDNA in (a) HEK293 cells and (b) MCF7 cells (the transfection efficiency of bPEI25kDa/pDNA (N/P=5)-transfected cells was set to 100.) (n≥4, Mean±SEM).
ACKNOWLEDGMENTS
This work was supported by NIH GM82866. HJK appreciates Kyung Hee University Program (2009) for his sabbatical leave. The authors appreciate Soyoung Kim (West High School, Salt Lake City, UT, 84103, USA) for her volunteer technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kang HC, Lee ES, Na K, Bae YH. Stimuli-sensitive nanosystems: For drug and gene delivery. In: Torchilin VP, editor. Multifunctional Pharmaceutical Nanocarriers. New York: Springer; 2008. pp. 161–199. [Google Scholar]
- 2.Kim YH, Park JH, Lee M, Kim YH, Park TG, Kim SW. Polyethylenimine with acid-labile linkages as a biodegradable gene carrier. J Control Release. 2005;103:209–219. doi: 10.1016/j.jconrel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Manickam DS, Oupicky D. Polyplex gene delivery modulated by redox potential gradients. J Drug Target. 2006;14:519–526. doi: 10.1080/10611860600834409. [DOI] [PubMed] [Google Scholar]
- 4.Meng F, Hennink WE, Zhong Z. Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials. 2009;30:2180–2198. doi: 10.1016/j.biomaterials.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Oupicky D, Bisht HS, Manickam DS, Zhou QH. Stimulus-controlled delivery of drugs and genes. Expert Opin Drug Deliv. 2005;2:653–665. doi: 10.1517/17425247.2.4.653. [DOI] [PubMed] [Google Scholar]
- 6.Ou M, Xu R, Kim SH, Bull DA, Kim SW. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30:5804–5814. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, et al. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjug Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 9.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Russ V, Wagner E. Influence of the molecular weight of bioreducible oligoethylenimine conjugates on the polyplex transfection properties. AAPS J. 2009;11:445–455. doi: 10.1208/s12248-009-9122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma N, Li Y, Xu H, Wang Z, Zhang X. Dual redox responsive assemblies formed from diselenide block copolymers. J Am Chem Soc. 2010;132:442–443. doi: 10.1021/ja908124g. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Mo H, Koo H, Park JY, Cho MY, Jin GW, et al. Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery. Bioconjug Chem. 2007;18:13–18. doi: 10.1021/bc060113t. [DOI] [PubMed] [Google Scholar]
- 13.Koo H, Jin GW, Kang H, Lee Y, Nam K, Bai CZ, et al. Biodegradable branched poly(ethylenimine sulfide) for gene delivery. Biomaterials. 2010;31:988–997. doi: 10.1016/j.biomaterials.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Blacklock J, Handa H, Manickam DS, Mao G, Mukhopadhyay A, Oupicky D. Disassembly of layer-by-layer films of plasmid DNA and reducible TAT polypeptide. Biomaterials. 2007;28:117–124. doi: 10.1016/j.biomaterials.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 15.Manickam DS, Bisht HS, Wan L, Mao G, Oupicky D. Influence of TAT-peptide polymerization on properties and transfection activity of TAT/DNA polyplexes. J Control Release. 2005;102:293–306. doi: 10.1016/j.jconrel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2002;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 17.Peng Q, Zhong Z, Zhuo R. Disulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectors. Bioconjug Chem. 2008;19:499–506. doi: 10.1021/bc7003236. [DOI] [PubMed] [Google Scholar]
- 18.Kang HC, Bae YH. pH-Tunable endosomolytic oligomers for enhanced nucleic acid delivery. Adv Funct Mater. 2007;17:1263–1272. [Google Scholar]
- 19.Kang HC, Bae YH. Transfection of insulin-secreting cell line and rat islets by functional polymeric gene vector. Biomaterials. 2009;30:2837–2845. doi: 10.1016/j.biomaterials.2009.01.035. [DOI] [PubMed] [Google Scholar]
- 20.Kang HC, Samsonova O, Bae YH. Trafficking microenvironmental pH of gene vector polycation in drug-sensitive and multidrug-resistant MCF7 breast cancer cell. Biomaterials. 2010;31:3071–3078. doi: 10.1016/j.biomaterials.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HC, Kim S, Lee M, Bae YH. Polymeric gene carrier for insulin secreting cells: Poly(l-lysine)-g-sulfonylurea for receptor mediated transfection. J Control Release. 2005;105:164–176. doi: 10.1016/j.jconrel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Park KM, Kang HC, Cho JK, Chung IJ, Cho SH, Bae YH, et al. All-trans-retinoic acid (ATRA)-grafted polymeric gene carriers for nuclear translocation and cell growth control. Biomaterials. 2009;30:2642–2652. doi: 10.1016/j.biomaterials.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 23.Kang HC, Bae YH. Polymeric gene transfection on insulin-secreting cells: sulfonylurea receptor-mediation and transfection medium effect. Pharm Res. 2006;23:1797–1808. doi: 10.1007/s11095-006-9027-0. [DOI] [PubMed] [Google Scholar]
- 24.Kunath K, von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 25.Suh J, Paik HJ, Hwang BK. Ionization of poly(ethyleneimine) and poly(allylamine) at various pH's. Bioorg Chem. 1994;22:318–327. [Google Scholar]
- 26.Ruponen M, Yla-Herttuala S, Urtti A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. Biochim Biophys Acta. 1999;1415:331–341. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]
- 27.Moret I, Peris JE, Guillem VM, Benet M, Revert F, Dasi F, et al. Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline pH, heparin and serum. J Control Release. 2001;76:169–181. doi: 10.1016/s0168-3659(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 28.Lu H, McDowell LM, Studelska DR, Zhang L. Glycosaminoglycans in human and bovine serum: detection of twenty-four heparan sulfate and chondroitin sulfate motifs including a novel sialic acid-modified chondroitin sulfate linkage hexasaccharide. Glycobiol Insights. 2010;2:13–28. [PMC free article] [PubMed] [Google Scholar]
- 29.Huth S, Hoffmann F, von Gersdorff K, Laner A, Reinhardt D, Rosenecker J, et al. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J Gene Med. 2006;8:1416–1424. doi: 10.1002/jgm.975. [DOI] [PubMed] [Google Scholar]
- 30.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 31.Baraldi PG, Bovero A, Fruttarolo F, Preti D, Tabrizi MA, Pavani MG, et al. DNA minor groove binders as potential antitumor and antimicrobial agents. Med Res Rev. 2004;24:475–528. doi: 10.1002/med.20000. [DOI] [PubMed] [Google Scholar]
- 32.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 33.Suh J, Dawson M, Hanes J. Real-time multiple-particle tracking: applications to drug and gene delivery. Adv Drug Deliv Rev. 2005;57:63–78. doi: 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci USA. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 36.Kang HC, Lee M, Bae YH. Polymeric gene carriers. Crit Rev Eukaryot Gene Expr. 2005;15:317–342. doi: 10.1615/critreveukargeneexpr.v15.i4.30. [DOI] [PubMed] [Google Scholar]
- 37.Pollard H, Toumaniantz G, Amos JL, Avet-Loiseau H, Guihard G, Behr JP, et al. Ca2+-sensitive cytosolic nucleases prevent efficient delivery to the nucleus of injected plasmids. J Gene Med. 2001;3:153–164. doi: 10.1002/jgm.160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.