Abstract
Two yeast (Saccharomyces cerevisiae) mutants that tolerate centromere (CEN) plasmids at high copy number have been isolated. The mutations relieve the restraint normally imposed on plasmid copy number by a cloned CEN sequence. Our CEN plasmids specify resistance to G418 and are high copy plasmids only when the mutant host cells are grown on medium containing this antibiotic. The high copy number of the plasmids is independent of the specific cloned CEN sequence and recovered plasmids show no alteration in structure or function of the CEN DNA. The efficiency with which CEN plasmids go to high copy number is increased if the mutant cell is cotransformed by another CEN plasmid. The genomic mutation responsible for the high copy number (COP) is dominant and stable, and it segregates in a Mendelian manner. Homozygous COP/COP a/alpha diploids do not tolerate CEN plasmids at high copy number, suggesting that the mutation is regulated by mating type. The genomic DNA from both mutant cells contains an altered transposon (Ty) restriction fragment that cosegregates with the COP phenotype in crosses of mutant and wild-type strains. The mutations may be transposon-mediated events that identify a gene involved in centromere or mitotic spindle function.
Full text
PDF
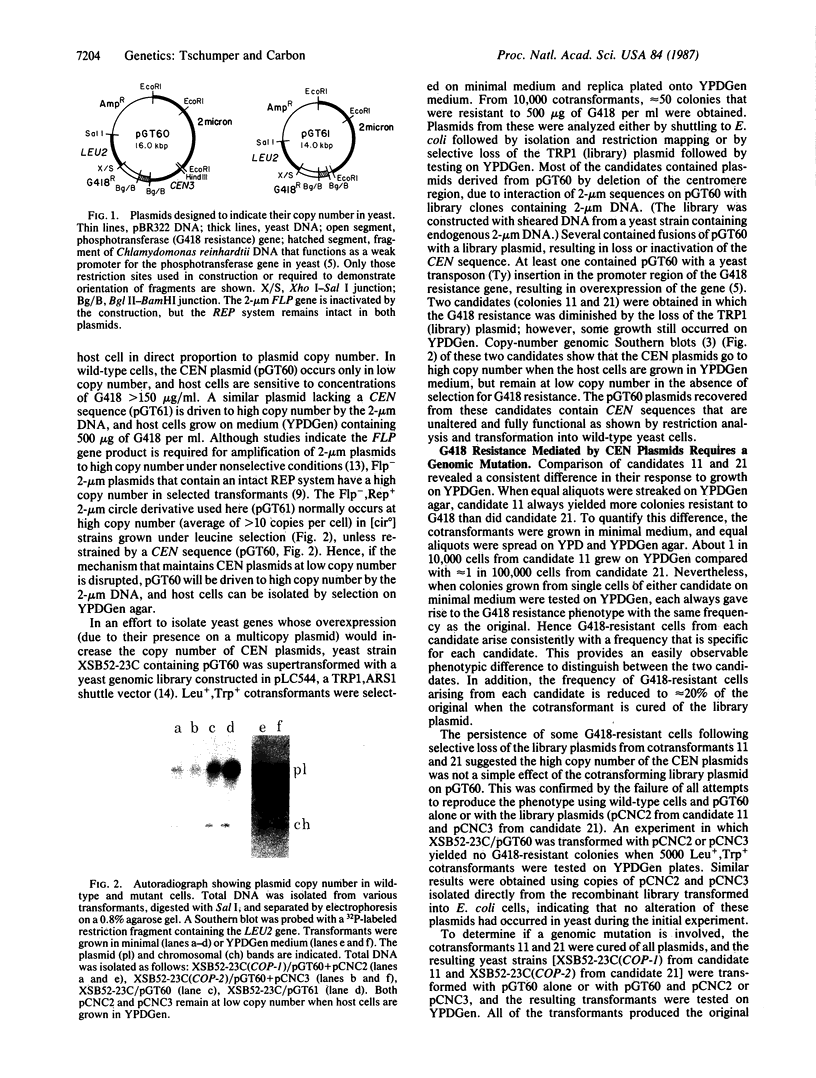



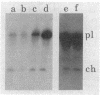
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Fitzgerald-Hayes M., Carbon J. Structural analysis and sequence organization of yeast centromeres. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1175–1185. doi: 10.1101/sqb.1983.047.01.133. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaleff D. T., Fink G. R. Genetic events associated with an insertion mutation in yeast. Cell. 1980 Aug;21(1):227–237. doi: 10.1016/0092-8674(80)90130-0. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of the centromere-linked CDC10 gene by complementation in yeast. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2173–2177. doi: 10.1073/pnas.77.4.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Futcher B., Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986 Jun;6(6):2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., Philippsen P. Functional selection and analysis of yeast centromeric DNA. Cell. 1985 Oct;42(3):913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M., Li Y. Y., Broach J. R. The yeast plasmid 2mu circle encodes components required for its high copy propagation. Cell. 1983 Aug;34(1):95–104. doi: 10.1016/0092-8674(83)90139-3. [DOI] [PubMed] [Google Scholar]
- Jimenez A., Davies J. Expression of a transposable antibiotic resistance element in Saccharomyces. Nature. 1980 Oct 30;287(5785):869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- Paquin C. E., Williamson V. M. Temperature effects on the rate of ty transposition. Science. 1984 Oct 5;226(4670):53–55. doi: 10.1126/science.226.4670.53. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. Movement of yeast transposable elements by gene conversion. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5621–5625. doi: 10.1073/pnas.79.18.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Copy number control by a yeast centromere. Gene. 1983 Aug;23(2):221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. High frequency excision of Ty elements during transformation of yeast. Nucleic Acids Res. 1986 Apr 11;14(7):2989–3001. doi: 10.1093/nar/14.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Volkert F. C., Broach J. R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986 Aug 15;46(4):541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- Webster T. D., Dickson R. C. Direct selection of Saccharomyces cerevisiae resistant to the antibiotic G418 following transformation with a DNA vector carrying the kanamycin-resistance gene of Tn903. Gene. 1983 Dec;26(2-3):243–252. doi: 10.1016/0378-1119(83)90194-4. [DOI] [PubMed] [Google Scholar]
- van Solingen P., van der Plaat J. B. Fusion of yeast spheroplasts. J Bacteriol. 1977 May;130(2):946–947. doi: 10.1128/jb.130.2.946-947.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]