Abstract
Aminoglycosides have been an essential component of the armamentarium in the treatment of life-threatening infections. Unfortunately, their efficacy has been reduced by the surge and dissemination of resistance. In some cases the levels of resistance reached the point that rendered them virtually useless. Among many known mechanisms of resistance to aminoglycosides, enzymatic modification is the most prevalent in the clinical setting. Aminoglycoside modifying enzymes catalyze the modification at different −OH or −NH2 groups of the 2-deoxystreptamine nucleus or the sugar moieties and can be nucleotidyltranferases, phosphotransferases, or acetyltransferases. The number of aminoglycoside modifying enzymes identified to date as well as the genetic environments where the coding genes are located is impressive and there is virtually no bacteria that is unable to support enzymatic resistance to aminoglycosides. Aside from the development of new aminoglycosides refractory to as many as possible modifying enzymes there are currently two main strategies being pursued to overcome the action of aminoglycoside modifying enzymes. Their successful development would extend the useful life of existing antibiotics that have proven effective in the treatment of infections. These strategies consist of the development of inhibitors of the enzymatic action or of the expression of the modifying enzymes.
Keywords: antibiotic resistance, aminoglycoside, aminoglycoside modifying enzyme, acetyltransferase, nucleotidyltransferase, phosphotransferase, kinase, antisense, RNase P, RNase H, bacterial infection
1. A brief overview of aminoglycoside antibiotics
1.1. General aspects
Aminoglycoside antibiotics are a complex family of compounds characterized for having an aminocyclitol nucleus (streptamine, 2-deoxystreptamine, or streptidine) linked to amino sugars through glycosidic bonds. In addition, other compounds such as spectinomycin, which is an aminocyclitol not linked to amino sugars, or compounds that include the aminocyclitol fortamine are also included in this family (Bryskier, 2005; Veyssier and Bryskier, 2005). Aminoglycosides are primarily used in the treatment of infections caused by gram-negative aerobic bacilli, staphylococci, and other gram-positives (Yao and Moellering, 2007). However, when used against gram-positives, aminoglycosides are recommended in combination with other antibiotics such as β-lactams or vancomycin with which they exert a synergistic effect probably due to an enhanced uptake (Eliopoulos, 1989; Scaglione et al., 1995; Yao and Moellering, 2007). Due to the nature of the mechanism of uptake of aminoglycosides, which requires respiration, anaerobic bacteria are intrinsically resistant (see below) (Bryan et al., 1979). As it would be expected from a large family of non-identical compounds, different aminoglycosides vary in their activity spectrum. Streptomycin, discovered in 1943, was the first efficient drug against tuberculosis and in 1944 a woman with this disease was cured after treatment with the antibiotic. Currently, streptomycin is still used in combination therapy to treat Mycobacterium tuberculosis (Menzies et al., 2009) and other aminoglycosides such as amikacin or kanamycin are used as second line drug in the treatment of resistant M. tuberculosis infections (Brossier et al., 2010). Besides Enterobacteriaceae and Pseudomonas aeruginosa, examples of life-threatening infections that can be treated with aminoglycosides are plague, tularemia, brucellosis, endocarditis caused by enterococci and infections caused by streptococci and enterococci (Yao and Moellering, 2007). Newer non-traditional applications of aminoglycosides include treatment of genetic disorders such as cystic fibrosis, in which about 10% of patients carry a nonsense mutation as opposed to the most common 3-bp deletion that results in the loss of a phenylalanine in the cystic fibrosis transmembrane conductance regulator (Rich et al., 1990), and Duchenne muscular dystrophy, in which 10 - 20% of patients carry a nonsense mutation in the dystrophin gene (Kellermayer, 2006). The property of aminoglycosides to decrease the fidelity of the eukaryotic elongation machinery makes them potential candidates to treat nonsense mutation related genetic disorders such as those mentioned above or others that can benefit from inducing translational readthrough (Hermann, 2007; Kellermayer, 2006; Zingman et al., 2007). Aminoglycosides, mainly gentamicin, have also been used in the treatment of Ménière’s disease by intratympanic injection (Dabertrand et al., 2010; Nakashima et al., 2000). Aminoglycoside-based drugs are also inhibitors of reproduction of the HIV virus, showing promise on the treatment of AIDS (reviewed in Houghton et al., 2010)
The most common route of administration of aminoglycosides for systemic infections is parenteral, intramuscular injection or intravenously in cases of severe infections. Oral administration is not possible for these infections due to very low levels of absorption. However, oral administration can be used for decontamination purposes such as to kill bowel flora before intestinal surgery (Vakulenko and Mobashery, 2003; Veyssier and Bryskier, 2005; Yao and Moellering, 2007). Other routes of delivery are sometimes used to increase the concentration of the drug at the site of infection or to limit nephrotoxicity or ototoxicity (Vakulenko and Mobashery, 2003; Veyssier and Bryskier, 2005; Yao and Moellering, 2007). Aminoglycosides exist in a variety of formulations, some experimental, including encapsulation in liposomes or nanoparticles, or aerosolized (Dudley et al., 2008; Kingsley et al., 2006; Pinto-Alphandary et al., 2000). A study has shown that when amikacin-encapsulated liposomes were modified they changed the organ distribution of the antibiotic (Bucke et al., 1998). Aminoglycoside antibiotics are not metabolized, they are excreted as active compounds and they show biphasic elimination with half-lives in the body of 2 – 3 hours (as long as the renal function is normal) and 37 – 100 hours (Veyssier and Bryskier, 2005; Wenk et al., 1979). They are mainly eliminated by glomerular filtration.
Binding to serum proteins, although variable among different aminoglycosides, is low. While no serum binding was demonstrable for gentamicin, tobramycin, or kanamycin, streptomycin was found to be 35% bound in a comparative study (Gordon et al., 1972). Amikacin serum protein binding in patients with spinal cord injury and in able-bodied controls was ~18% (Brunnemann and Segal, 1991). The fraction bound to serum proteins is important because it is only the unbound fraction of a drug that produces a pharmacological effect (Benet and Hoener, 2002; Heinze and Holzgrabe, 2006).
The utilization of aminoglycosides is not free of adverse effects; they have been linked to drug-induced nephrotoxicity and ototoxicity, a problem that limits the doses that can be used. Nephrotoxicity is generally reversible and the most common clinical presentation is nonoliguric acute kidney injury. Other manifestations include a decrease in the glomerular ! ltration rate, enzymuria, aminoaciduria, glycosuria, hypomagnesemia, hypocalcemia, and hypokalemia (Martinez-Salgado et al., 2007; Oliveira et al., 2009). The ototoxicity effects of aminoglycosides include permanent bilaterally severe, high-frequency sensorineural hearing loss and temporary vestibular hypofunction (Guthrie, 2008). The mechanism by which aminoglycosides are ototoxic seems to be related to their ability to sequester and chelate metals forming complexes that are redox active and generate reactive oxygen species, which in turn induce cell damage (Guthrie, 2008). Free radical scavengers as well as iron chelators were shown to attenuate ototoxic effects of aminoglycosides (Nakashima et al., 2000).
1.2. Bacterial uptake
Internalization of aminoglycosides is an important process for their biological activity. Aminoglycosides penetrate the bacterial cell following a three-steps process; a first energy-independent step is followed by two energy-dependent steps (Taber et al., 1987; Tolmasky, 2007a; Vakulenko and Mobashery, 2003; Veyssier and Bryskier, 2005). Two components are needed for accumulation of aminoglycoside molecules inside the cell: ribosomes and the membrane bound respiratory chain. When aminoglycoside molecules are in contact with bacterial cells, the polycationic antibiotic molecules bind to cell’s surface anionic compounds such as lipopolysaccharide, phospholipids, and outer membrane proteins in gram-negatives, and teichoich acids and phospholipids in gram-positives. As a result of the binding to anionic sites in the outer membrane, divalent cations that cross-bridge adjacent lipopolysaccharide molecules are displaced resulting in an increase in permeability that leads to the so called “self-promoted uptake” penetration of aminoglycoside molecules to the periplasmic space (Vanhoof et al., 1995). The following process is blocked by inhibitors of electron transport and oxidative phosphorylation (Muir et al., 1984) and is known as “energy-dependent phase I”. Although alternatives have been proposed (Nichols and Young, 1985) it is generally accepted that this phase is characterized by the uptake into the cytoplasm of a small number of aminoglycoside molecules in an energy-dependent fashion, and since it needs a functional electron transport system anaerobes tend not to be susceptible to these antibiotics (Bryan and van der Elzen, 1977). The small number of molecules that reach the cytoplasm during energy-dependent phase I induce errors in protein synthesis and the mistranslated membrane proteins cause damage to the integrity of the cytoplasmic membrane when they are inserted triggering the following step known as “energy-dependent phase II”. This mechanism is supported by the need for protein synthesis for triggering the energy-dependent phase II (Hurwitz et al., 1981). The aberrant proteins in the damaged membrane facilitate transport of more molecules of antibiotic that increase the level of interference with normal protein synthesis leading to yet more damage in the membrane resulting in an autocatalytic accelerated rate of uptake that ultimately results in death of the cell (Davis, 1988; Nichols, 1989; Taber et al., 1987). The presence of capsule or exopolysaccharide layers seems not to affect diffusion of the aminoglycosides (Nichols et al., 1988).
1.3. Molecular mechanisms of action
Studies on the effect of aminoglycosides on protein synthesis resulted not only in an understanding of the mode of action of these antibiotics but also in contributions to the understanding of the molecular mechanisms of translation fidelity (Davies, 2006; Davis, 1987; Houghton et al.; Magnet and Blanchard, 2005; Majumder et al., 2007; Vakulenko and Mobashery, 2003). It is clear now that pairing of the codon/anticodon nucleotides cannot account for the levels of fidelity observed in selection of the correct aminoacyl-tRNA (Ogle et al., 2001; Ogle et al., 2003). The ribosome plays an active role in stabilization of the cognate tRNA/mRNA association and rejection of near-cognate tRNAs. One of the earliest observations that led to the idea that the ribosome is an active player in the faithful decoding mechanism by modulating tRNA/mRNA interactions was the production of an enzyme that was otherwise absent due to a premature stop codon in specific Escherichia coli auxotrophic mutants upon addition of streptomycin (Spotts and Stanier, 1961). These experiments no only helped understanding mechanisms of translation fidelity during protein synthesis but also contributed to the clarification of the misreading-inducing properties of aminoglycosides. It is now well established that the A site is the decoding center of the ribosome, located on the 16S RNA (which together with about 21 proteins composes the 30S subunit of the ribosome). Regions of the 16S RNA establish contact with the cognate codon/anticodon pair and modify their structure resulting in what is known as the closed conformation of the 30S RNA subunit as opposed to the open structure of the empty A site (reviewed in Ogle et al., 2003; Ogle and Ramakrishnan, 2005; Zaher and Green, 2009). Conversely, binding of a near cognate tRNA does not induce the closed state.
The intimate mechanisms by which aminoglycosides interfere with translational fidelity are becoming ever more clear with the dilucidation of crystal structures of complexes between different aminoglycosides and the A site as well as the effects caused by these interactions. Structures of a number of aminoglycosides bound to oligonucleotides containing the decoding A site or the entire subunit have recently been determined by NMR or X-ray crystallography (reviewed in Jana and Deb, 2006; Ogle et al., 2003; Ogle and Ramakrishnan, 2005; Vicens and Westhof, 2003; Zaher and Green, 2009). These studies showed that not all classes of aminoglycosides bind to identical sites of the 16S rRNA but the common effect of their binding is a change of conformation of the A site to one that mimics the closed state induced by interaction between cognate tRNA and mRNA eliminating the proofreading capabilities of the ribosome and thereby promoting mistranslation. With the exception of spectinomycin and kasugamycin, aminoglycosides are bactericidal and their lethality is thought to be due to the secondary effects of inducing mistranslation (Bakker, 1992; Busse et al., 1992; Davis, 1987, 1989; Magnet and Blanchard, 2005; Vakulenko and Mobashery, 2003).
Aminoglycosides such as neomycin and paromomycin have also been shown to inhibit 30S ribosomal subunit assembly although this also could be a secondary effect to protein mistranslation (Mehta and Champney, 2003). Other effects of aminoglycosides include their ability to induce RNA cleavage (Belousoff et al., 2009) or interfere with essential functions such as RNase P, which has been shown to be inhibited by neomycin B due to interference of the antibiotic molecule with the binding of divalent metal ions to the RNA moiety of RNase P (Mikkelsen et al., 1999). These properties could be exploited to develop new aminoglycosides directed to targets other than the ribosome.
Experiments exposing E. coli cells to sublethal concentrations of amikacin showed that one of the most susceptible cellular mechanisms is formation of the Z ring, which leads to anomalies in cell division. At these low concentrations of the antibiotic the chromosomes continued replication and were properly located (Possoz et al., 2007).
2. Bacterial resistance to aminoglycoside antibiotics
2.1. Mechanisms of resistance
Aminoglycoside resistance occurs through several mechanisms that can coexist simultaneously in the same cell (Alekshun and Levy, 2007; Houghton et al.; Magnet and Blanchard, 2005; Taber et al., 1987; Tolmasky, 2007a). Described mechanisms include modification of the target by mutation of the 16S rRNA or ribosomal proteins (Galimand et al., 2005; O’Connor et al., 1991); methylation of 16S rRNA, a mechanism found in most aminoglycoside-producing organisms and in clinical strains (Doi and Arakawa, 2007; Galimand et al., 2005); reduced permeability by modification of outer membrane’s permeability or diminished inner membrane transport (Hancock, 1981; MacLeod et al., 2000; Over et al., 2001); export outside the cell by active efflux pumps (Aires et al., 1999; Magnet et al., 2001; Rosenberg et al., 2000), one of which has recently been shown to be involved in adaptive resistance (Hocquet et al., 2003); active swarming, a probably non-specific mechanism recently shown in P. aeruginosa cells, which exhibited adaptive antibiotic resistance against several antibiotics (Overhage et al., 2008); sequestration of the drug by tight binding to an acetyltransferase of very low activity (Magnet et al., 2003); and enzymatic inactivation of the antibiotic molecule, the most prevalent in the clinical setting and the subject of this review.
3. Aminoglycoside modifying enzymes
Aminoglycoside modifying enzymes catalyze the modification at −OH or −NH2 groups of the 2-deoxystreptamine nucleus or the sugar moieties and can be acetyltransferases (AACs), nucleotidyltranferases (ANTs), or phosphotransferases (APHs) (Fig. 1). The combination of mutagenesis, which leads to continuous generation of new enzyme variants that can utilize an ever growing number of antibiotics as substrates, with the coding genes’ ability to transfer at the molecular level as part of integrons, gene cassettes, transposons, or integrative conjugative elements and at the cellular level through conjugation, as part of mobilizable or conjugative plasmids, natural transformation or transduction results in the ability of this resistance mechanism to reach virtually all bacterial types (Tolmasky, 2007b).
Fig. 1.
Representative aminoglycosides and modification sites by AAC, ANT, and APH enzymes. An example of each kind of modification is shown on one of the substrates. The square and oval on positions 2′ and 6″ in paromomycin I indicate that although this molecule is preferentially acetylated at the position 1, 1,2′-di-N-acetylparomomycin and 1,6″-di-N-acetylparomomycin are also found as products of the enzymatic reaction (Sunada et al., 1999). AAC(3)-X can catalyze acetylation at the 3″-amino group in arbekacin and amikacin (Hotta et al. 1998).
The number of aminoglycoside modifying enzymes identified to date as well as the hosts and genetic environments is impressive, therefore the citations and examples described here should be considered representative rather than comprehensive. Furthermore, putative genes coding for aminoglycoside modifying enzymes are being found in complete genome sequences. These genes, for which there is no further information other than the annotation, are not discussed in this review. Summaries including relevant data on aminoglycoside modifying enzymes are shown in Fig. 1 and Tables 1 - 3.
Table 1.
Aminoglycoside N-acetyltransferases
| AACs | Gene names | Genetic location |
Accession number |
Host | References |
|---|---|---|---|---|---|
| AAC(1) |
E. coli, Actinomycete, Campylobacter spp. |
(Gomez-Luis et al., 1999; Lovering et al., 1987; Sunada et al., 1999) |
|||
| AAC(3)-Ia C |
aac(3)-Ia,
aacC1 |
Plasmid, transposon, integron |
X15852, AF550679 |
S. marcescens, E. coli,
Acinetobacter baumannii, Klebsiella pneumoniae, Klebsiella oxytoca, P. aeruginosa, Salmonella typhimurium, Proteus mirabilis |
(Javier Teran et al., 1991; Wohlleben et al., 1989) |
| AAC(3)-Ib | aac(3)-Ib | Integron | L06157 | P. aeruginosa | (Schwocho et al., 1995) |
| AAC(3)-Ic | aac(3)-Ic | Integron | AJ511268 | P. aeruginosa | (Riccio et al., 2003) |
| AAC(3)-Id | aac(3)-Id | Genomic island, integron |
AY458224 |
S. enterica, P. mirabilis,
Vibrio fluvialis |
(Doublet et al., 2004) |
| AAC(3)-Ie |
aac(3)-Ie,
aacCA5 |
Integron |
AY463797, DQ520937, AY463797 |
S. enterica, P. mirabilis,
P. aeruginosa |
(Gionechetti et al., 2008; Levings et al., 2005) |
| AAC(3)-IIa |
aac(3)-IIa,
aaC3, aacC5, aacC2, aac(3)-Va |
Plasmid | X13543 |
K. pneumoniae, E.
cloacae, Actinobacillus pleuropneumoniae, S. typhimurium, Citrobacter freundii |
(Allmansberger et al., 1985) |
| AAC(3)-IIb |
aac(3)-IIb,
aac(3)-Vb |
M97172 |
E. coli, A. faecalis, S.
marcescens |
(Rather et al., 1992) (Dahmen et al., 2010) |
|
| AAC(3)-IIc |
aac(3)-IIc,
aacC2 |
Plasmid | X54723 | E. coli, P. aeruginosa | (Dubois et al., 2008) |
| AAC(3)-IIIa |
aac(3)-IIIa,
aacC3 |
Chromosome | X55652 | P. aeruginosa | (Vliegenthart et al., 1991a) |
| AAC(3)-IIIb | aac(3)-IIIb | L06160 | P. aeruginosa | ||
| AAC(3)-IIIc |
aac(3)-IIIc,
ant(2″)-Ib |
L06161 | P. aeruginosa | ||
| AAC(3)-IVa | aac(3)-IVa | Plasmid |
X01385, AY216678, AJ493432 |
E. coli, C. jejuni, P.
stutzeri |
(Brau et al., 1984; Heuer et al., 2002) |
| AAC(3)-VIa | aac(3)-VIa | Plasmid |
M88012, NC_009140 NC_009838 |
E. cloacae, S. enterica,
E. coli |
(Rather et al., 1993a) (Call et al., 2010) |
| AAC(3)-VIIa |
aac(3)-VIIa,
aacC7 |
Chromosome | M22999 | Streptomyces rimosus | (Lopez-Cabrera et al., 1989) |
| AAC(3)-VIIIa |
aac(3)-VIIIa,
aacC8 |
Chromosome | M55426 | Streptomyces fradiae | (Salauze et al., 1991) |
| AAC(3)-IXa |
aac(3)-IXa,
aacC9 |
Chromosome | M55427 | Micromonospora chalcea | (Salauze et al., 1991) |
| AAC(3)-X | aac(3)-Xa | Chromosome | AB028210 | Streptomyces griseus | (Ishikawa et al., 2000) |
| AAC(2′)-Ia | aac(2′)-Ia | Chromosome | L06156 | P. stuartii | (Rather et al., 1993b) |
| AAC(2′)-Ib | aac(2′)-Ib | Chromosome | CP001172 |
M. fortuitum, A.
baumannii |
(Adams et al., 2008; Ainsa et al., 1997) |
| AAC(2′)-Ic C | aac(2′)-Ic | Chromosome |
CP001658, NC_002945 |
M. tuberculosis, M. bovis | (Ainsa et al., 1997) |
| AAC(2′)-Id | aac(2′)-Id | Chromosome | NC_008596 | M. smegmatis | (Ainsa et al., 1997) |
| AAC(2′)-Ie | aac(2′)-Ie | Chromosome | M. leprae | (Ainsa et al., 1997) | |
| Putative AAC(2′) | Chromosome | AM743169 | S. maltophilia | (Crossman et al., 2008) | |
| AAC(6′)-Ia |
aac(6′)-Ia,
aacA1 |
Plasmid, transposon, integron |
M18967, AF047479, M86913 |
Citrobacter diversus, E.
coli, K. pneumoniae, Shigella sonnei |
(Tenover et al., 1988), (Parent and Roy, 1992) |
| AAC(6′)-Ib C |
aac(6′)-Ib, aac(6′)-4
aacA4 |
Plasmid, transposon, integron |
M21682, M23634, AF479774 |
K. pneumoniae, P.
mirabilis, P. aeruginosa, S. enterica, K. oxytoca, S. maltophilia, E. cloacae |
(Nobuta et al., 1988; Tran van Nhieu and Collatz, 1987) |
| AAC(6′)-Ib’ | aac(6′)-Ib’, aac(6′)-Ib6 | Integron |
L25617, AJ584652, L25666 |
P. fluorescens , P.
aeruginosa |
(Lambert et al., 1994b; Mendes et al., 2004); (Casin et al., 1998) |
| AAC(6′)-Ic | aac(6′)-Ic | Chromosome | M94066 | S. marcescens | (Shaw et al., 1992) |
| AAC(6′)-Ie |
aac(6′)-Ie, aac(6′)-bifuncional |
Transposon | M18086 |
S. aureus, Macrococcus
caseolyticus, E. faecalis, Enterococcus faecium |
(Rouch et al., 1987) |
| AAC(6′)-If | aac(6′)-If | Plasmid | X55353 | E. cloacae | (Teran et al., 1991) |
| AAC(6′)-Ig | aac(6′)-Ig | Chromosome | L09246 |
Acinetobacter
haemolyticus |
(Lambert et al., 1993) |
| AAC(6′)-Ih | aac(6′)-Ih | Plasmid | L29044 | A. baumannii | (Lambert et al., 1994a) |
| AAC(6′)-Ii C | aac(6′)-Ii | Chromosome | L12710 | Enterococcus spp. | (Costa et al., 1993; Draker et al., 2003; Wybenga-Groot et al., 1999) |
| AAC(6′)-Ij | aac(6′)-Ij | Chromosome | L29045 |
Acinetobacter
genomosp. 13 |
(Lambert et al., 1994a) |
| AAC(6′)-Ik | aac(6′)-Ik | Chromosome | L29510 | Acinetobacter sp. | (Rudant et al., 1994) |
| AAC(6′)-Ip |
aac(6′)-Il, aac(6′)-Im,
aac(6′)-Ip |
Integron | Z54241 | C. freundii | (Hannecart-Pokorni et al., 1997) |
| AAC(6′)-Iq | aac(6′)-Iq | Plasmid, integron |
AF047556 | K. pneumoniae | (Centron and Roy, 1998) |
| AAC(6′)-Im | aac(6′)-Im | Plasmid | AF337947 | E. coli, E. faecium | (Chow et al., 2001) |
| AAC(6′)-Il |
aac(6′)-Il,
aacA7 |
Plasmid, integron |
U13880 | Enterobacter aerogenes | (Bunny et al., 1995) |
| AAC(6′)-Ir | aac(6′)-Ir | Chromosome | AF031326 |
Acinetobacter
genomosp. 14 |
(Rudant et al., 1999) |
| AAC(6′)-Is | aac(6′)-Is | Chromosome | AF031327 |
Acinetobacter
genomosp. 15 |
(Rudant et al., 1999) |
| AAC(6′)-Isa | aac(6′)-Isa | Plasmid | AB116646 | Streptomyces albulus | (Hamano et al., 2004) |
| AAC(6′)-It | aac(6′)-It | Chromosome | AF031328 | A. genomosp. 16 | (Rudant et al., 1999) |
| AAC(6′)-Iu | aac(6′)-Iu | Chromosome | AF031329 | A. genomosp. 17 | (Rudant et al., 1999) |
| AAC(6′)-Iv | aac(6′)-Iv | Chromosome | AF031330 | Acinetobacter sp. | (Rudant et al., 1999) |
| AAC(6′)-Iw | aac(6′)-Iw | Chromosome | AF031331 | Acinetobacter sp. | (Rudant et al., 1999) |
| AAC(6′)-Ix | aac(6′)-Ix | Chromosome | AF031332 | Acinetobacter sp. | (Rudant et al., 1999) |
| AAC(6′)-Iy C | aac(6′)-Iy | Chromosome | AF144881 | S. enteritidis, S. enterica | (Magnet et al., 1999) |
| AAC(6′)-Iz | aac(6′)-Iz | Chromosome | AF140221 | S. maltophilia | (Lambert et al., 1999) |
| AAC(6′)-Iaa | aac(6′)-Iaa | Chromosome | NC_003197 | S. typhimurium | (Salipante and Hall, 2003) |
| AAC(6′)-Iad | aac(6′)-Iad | Plasmid | AB119105 |
Acinetobacter
genomosp. 3 |
(Doi et al., 2004) |
| AAC(6′)-Iae | aac(6′)-Iae | Integron | AB104852 |
P. aeruginosa, S.
enterica |
(Sekiguchi et al., 2005) |
| AAC(6′)-Iaf | aac(6′)-Iaf | Plasmid, integron |
AB462903 | P. aeruginosa | (Kitao et al., 2009) |
| AAC(6′)-Iai | aac(6′)-Iai | Plasmid, integron |
EU886977 | P. aeruginosa | |
| AAC(6′)-Ib3 | aac(6′)-Ib3, aac(6′)-Ib5 | integron | X60321 | P. aeruginosa | (Mabilat et al., 1992); (Casin et al., 1998) |
| AAC(6′)-Ib4 | aac(6′)-Ib4 | S49888 | Serratia spp. | (Toriya et al., 1992) | |
| AAC(6′)-Ib7 | aac(6′)-Ib7 | Plasmid | Y11946 | E. cloacae, C. freundii | (Casin et al., 1998), |
| AAC(6′)-Ib8 | aac(6′)-Ib8 | Plasmid | Y11947 | E. cloacae | (Casin et al., 1998) |
| AAC(6′)-Ib9 | aac(6′)-Ib9 | Integron | AF043381 | P. aeruginosa | (Mugnier et al., 1998a) |
| AAC(6′)-Ib10 | aac(6′)-Ib10 | Integron | P. aeruginosa | (Mugnier et al., 1998b) | |
| AAC(6′)-Ib11 C | aac(6′)-Ib11 | Integron | AY136758 | S. enterica | (Casin et al., 2003) |
| AAC(6′)-29a | aac(6′)-29a | Integron | AF263519 | P. aeruginosa | (Poirel et al., 2001) |
| AAC(6′)-29b | aac(6′)-29b | Integron | AF263520 | P. aeruginosa | (Poirel et al., 2001) |
| AAC(6′)-31 | aac(6′)-31 | Integron | AM28348, AM283490 |
Pseudomonas putida, A.
baumannii, K. pneumoniae |
(Mendes et al., 2007) |
| AAC(6′)-32 | aac(6′)-32 | Plasmid, integron |
EF614235 | P. aeruginosa | (Gutierrez et al., 2007) |
| AAC(6′)-33 | aac(6′)-33 | Integron | GQ337064 | P. aeruginosa | (Viedma et al., 2009) |
| AAC(6′)-I30 | aac(6′)-I30 | Integron | AY289608 | S. enterica | (Mulvey et al., 2004) |
| AAC(6′)-Iid | aac(6′)-Iid | Chrosmome | AJ584700 | Enterococcus durans | (Del Campo et al., 2005) |
| AAC(6′)-Iih | aac(6′)-Iih | Chromosome | AJ584701 | Enterococcus hirae | (Del Campo et al., 2005) |
| AAC(6′)-Ib-Suzhou | aac(6′)-Ib-Suzhou | EF37562, EU085533 |
E. cloacae, K.
pneumoniae |
(Huang et al., 2008) | |
| AAC(6′)-Ib-Hangzhou | aac(6′)-Ib-Hangzhou | FJ503047 | A. baumannii | ||
| AAC(6′)-SK | aac(6′)-sk | Chromosome | AB164230 |
Streptomyces
kanamyceticus |
(Matsuhashi et al., 1985) |
| AAC(6′)-IIa | aac(6′)-IIa | Plasmid, integron |
M29695 |
P. aeruginosa, S.
enterica |
(Shaw et al., 1989) |
| AAC(6′)-IIb | aac(6′)-Iib | Integron | L06163 | P. fluorescens | |
| AAC(6′)-IIc | aac(6′)-IIc | Plasmid, integron |
NC_012555 | E. cloacae | (Chen et al., 2009) |
| AAC(6′)-Ib-cr | aac(6′)-Ib-cr | Plasmid, transposon, integron |
DQ303918 | Enterobacteriaceae | (Robicsek et al., 2006) |
| AAC(6′)-Ie-APH(2″)-Ia | aac(6′)-aph (2″) | Plasmid, transposon |
M18086, M13771 |
S.aureus, E. faecalis, E.
faecium, Staphylococcus. warneri |
(Rouch et al., 1987) |
| ANT(3″)-Ii-AAC(6′)-IId |
ant(3″)-Ii-aac(6′)-IId,
ant(3″)-Ih-aac(6′)-IId |
Integron | AF453998 | S. marcescens | (Centron and Roy, 2002) |
| AAC(6′)-30/AAC(6′)-Ib’ | aac(6′)-30/aac(6′)-Ib’ | Integron | AJ584652 | P. aeruginosa | (Mendes et al., 2004) |
| AAC(3)-Ib/AAC(6′)-Ib” | aac(3)-Ib/aac(6′)-Ib” | Integron | AF355189 | P. aeruginosa | (Dubois et al., 2002) |
Only representative hosts, references and accession numbers are shown.
C, three dimensional structure has been resolved. AAC(3)-Ia pdb id: 1BO4 (Wolf et al., 1998). AAC(2′)-Ic pdb id: 1M44, 1M4D (in complex with CoA and tobramycin), 1M4G (in complex with CoA and ribostamycin), 1M4I (in complex with CoA and kanamycin A) (Vetting et al., 2002). AAC(6′)-Ib pdb id: 1V0C (in complex with kanamycin C and AcetylCoA), 2BUE (in complex with ribostamycin and CoA), 2VQY (in complex with parmomycin and AcetylCoA (Vetting et al., 2008); 2PRB (in complex with CoA), 2QIR (in complex with CoA and kanamycin) (Maurice et al., 2008). AAC(6′)-Ib11 pdb id: 2PR8 (Maurice et al., 2008). AAC(6′)-Ii pdb id: 2A4N (in complex with CoA) (Burk et al., 2005), 1N71 (in complex with CoA) (Burk et al., 2003), 1B87 (in complex with AcetylCoA) (Wybenga-Groot et al., 1999). AAC(6′)-Iy pdb id: 2VBQ (in complex with bisubstrate analog CoA-S-monomethy-acetylneamine) (Magalhaes et al., 2008), 1S3Z (in complex with CoA and ribostamycin), 1S5K (in complex with CoA and N-terminal His(6)-tag, crystal form 1), 1S60 (in complex with CoA and N-terminal His(6)-tag, crystal form 2) (Vetting et al., 2004).
Table 3.
Aminoglycoside O-phosphotransferases
| APHs | gene names | Genetic location |
Accesion number |
Host | References |
|---|---|---|---|---|---|
| APH(4)-Ia | aph(4)-Ia, hph | Plasmid | V01499 | E. coli | (Kaster et al., 1983) |
| APH(4)-Ib | aph(4)-Ib, hyg | Chromosome | X03615 |
Streptomyces
hygroscopicus |
(Zalacain et al., 1986) |
| APH(6)-Ia | aph(6)-Ia, aphD, strA | Chromosome | Y00459 | S. griseus | (Distler et al., 1987) |
| APH(6)-Ib | aph(6)-Ib, sph | Chromosome | X05648 | S. glaucescens | (Vogtli and Hutter, 1987) |
| APH(6)-Ic | aph(6)-Ic, str | Transposon | X01702 |
S. enterica, P.
aeruginosa, E. coli |
(Mazodier et al., 1985; Steiniger-White et al., 2004) |
| APH(6)-Id | aph(6)-Id, strB, orfI | Plasmid, integrative conjugative element, chromosomal genomic islands |
M28829 |
K. pneumoniae, Salmonella spp., E. coli, Shigella flexneri, Providencia alcalifaciens, Pseudomonas spp., V. cholerae, Edwardsiella tarda, Pasteurella multocida, Aeromonas bestiarum |
(Daly et al., 2005; Gordon et al., 2008; Meyer, 2009; Scholz et al., 1989) |
| APH(9)-Ia C | aph(9)-Ia | Chromosome |
U94857, CR628337 |
L. pneumophila | (Suter et al., 1997) |
| APH(9)-Ib | aph(9)-Ib, spcN | Chromosome | U70376 | S. flavopersicus | (Lyutzkanova et al., 1997) |
| APH(3′)-Ia | aph(3′)-Ia, aphA-1 | Transposon | V00359 | E. coli, S. enterica | (Oka et al., 1981) |
| APH(3′)-Ib | aph(3′)-Ib, aphA-like | Plasmid | M20305 | E. coli | (Pansegrau et al., 1987) |
| APH(3′)-Ic |
aph(3′)-Ic, apha1-1AB,
apha7 |
Plasmid, transposon, genomic island |
M37910 |
K. pneumoniae, A. baumannii, S. marcescens, Corynebacterium spp., Photobacterium spp., Citrobacter spp. |
(Lee et al., 1990; Tauch et al., 2000) |
| APH(3′)-IIa C | aph(3′)-Iia, aphA-2 | Transposon | V00618 | E. coli | (Beck et al., 1982) |
| APH(3′)-IIb | aph(3′)-IIb | Chromosome | NC_002516 | P. aeruginosa | (Stover et al., 2000) |
| APH(3′)-IIc | aph(3′)-IIc | Chromosome | S. maltophilia | (Okazaki and Avison, 2007) | |
| APH(3′)-IIIa C | aph(3′)-IIIa | Plasmid | V01547 |
S. aureus, Enterococcus spp. |
(Trieu-Cuot and Courvalin, 1983) |
| APH(3′)-IVa | aph(3′)-Iva, aphA4 | Chromosome | X01986 | B. circulans | (Herbert et al., 1983) |
| APH(3′)-Va | aph(3′)-Va, aphA-5a | Chromosome | K00432 | Streptomyces fradiae | (Thompson and Gray, 1983) |
| APH(3′)-Vb | aph(3′)-Vb, aphA-5b, rph | Chromosome | M22126 |
Streptomyces
ribosidificus |
(Hoshiko et al., 1988) |
| APH(3′)-Vc | aph(3′)-Vc, aphA-5c | Chromosome | S81599 | M. chalcea | (Salauze et al., 1991) |
| APH(3′)-VIa | aph(3′)-Via, aphA-6 | Plasmid | X07753 | A. baumannii | (Martin et al., 1988) |
| APH(3′)-VIb | aph(3′)-VIb | Plasmid |
K. pneumoniae, S.
marcescens |
(Gaynes et al., 1988) | |
| APH(3′)-VIIa | aph(3′)-VIIa, aphA-7 | Plasmid | M29953 | C. jejuni | (Tenover et al., 1989) |
| APH(2″)-Ia |
aph(2″)-Ia, aph(2″)-
bifunctional |
Plasmid | AP003367 |
S. aureus, Clostridium
difficile, Streptococcus mitis, E. faecium |
(Ferretti et al., 1986) |
| APH(2″)-IIa C | aph(2″)-Iia, aph(2′)-Ib | Chromosome |
AF207840, AF337947 |
E. faecium, E. coli | (Kao et al., 2000) |
| APH(2″)-IIIa C | aph(2″)-IIIa, aph(2′)-Ic | Plasmid | U51479 |
Enterococcus
gallinarum |
(Chow et al., 1997) |
| APH(2″)-IVa C | aph(2″)-Iva, aph(2″)-Id | Chromosome | AF016483 | E. casseliflavus | (Tsai et al., 1998) |
| APH(2″)-Ie | aph(2″)-Ie | Plasmid, transposon |
AY939911 |
E. faecium , E.
casseliflavus |
(Chen et al., 2006) |
| APH(3″)-Ia | aph(3″)-Ia, aphE, aphD2 | Chromosome | X53527 | S. griseus | (Trower and Clark, 1990) |
| APH(3″)-Ib | aph(3″)-Ib, strA, orfH | Plasmid, transposon, integrative conjugative elements, chromosome |
M28829 |
Enterobacteriaceae, Pseudomonas spp. |
(Scholz et al., 1989) |
| APH(3″)-Ic | aph(3″)-Ic | Chromosome | DQ336355 | M. fortuitum | (Ramon-Garcia et al., 2006) |
| APH(7″)-Ia | aph(7″)-Ia, aph7! | Chromosome | S. hygroscopicus | (Berthold et al., 2002) |
Only representative hosts, references and accession numbers are shown.
C, three dimensional structure has been resolved. APH(9)-Ia pdb id: 3I0O (in complex with ADP and Spectinomcyin), 3I0Q (in complex with AMP), 3I1A (Fong et al., 2010). APH(3′)-IIa pdb id: 1ND4 (Nurizzo et al., 2003). APH(3′)-IIIa pdb id: 1J7I, 1J7L (in complex with ADP), 1J7U (in comlex with APPNP) (Burk et al., 2001), 1L8T (in complex with ADP and kanamycin A) (Fong and Berghuis, 2002) 3H8P (in complex with AMPPNP and butirosin A) (Fong and Berghuis, 2009), 2BKK (in complex with the inhibitor AR_3A) (Kohl et al., 2005). APH(2″)-IIa pdb id: 3HAV (in complex with ATP and streptomycin), 3HAM (in complex with gentamicin) (Young et al., 2009).
3.1. Aminoglycoside modifying enzymes: nomenclature
There are two main nomenclatures currently in use to identify aminoglycoside modifying enzymes. One of them consists of a three-letter identifier of the activity followed by the site of modification between parenthesis (class), a roman number particular to the resistant profile they confer to the host cells (subclass), and a low case letter that is an individual identifier (Shaw et al., 1993). The parenthesis and the subclass are usually separated by a hyphen but lately some authors have removed it (Oteo et al., 2006). For example, AAC(6′)-Ia represents an N-acetyltransferase that catalyzes acetylation at the 6′ position conferring a resistance profile identical to the other AAC(6′)-I enzymes (AAC(6′)-Ib – AAC(6′)-Iaf). In the other nomenclature system the genes are designated aac, aad and aph followed by a capital letter that identifies the site of modification (Novick et al., 1976). Thus, aacA, aacB, and aacC identify aminoglycoside 6′-N- acetyltransferase, aminoglycoside 2′-N- acetyltransferase, and aminoglycoside 3-N-acetyltransferase respectively. A number is then added to provide a unique identifier to different genes. Each of the nomenclatures has its own advantages and disadvantages and different authors prefer one to the other but, as it has been suggested before (Tolmasky, 2007a; Vanhoof et al., 1998), it would be convenient to reach consensus and use only one of them to avoid confusion and facilitate following the advances in the field. The confusion is sometimes compounded by different additions or modifications in naming new genes or variants (see below). We suggest that returning to a simpler nomenclature with the support of an internet repository site could facilitate the naming of the genes, avoid duplications, and facilitate further changes when new enzymes with new, and may be unexpected, characteristics are discovered.
3.2. Aminoglycoside modifying enzymes: aminoglycoside N-acetyltransferases (AACs)
AACs belong to the ubiquitous GCN5-related N-acetyltransferase (GNAT) superfamily of proteins, which include about 10,000 proteins (Vetting et al., 2005). GNAT enzymes catalyze the acetylation of −NH2 groups in the acceptor molecule using acetyl coenzyme A as donor substrate, in the case of AACs the acceptor is an aminoglycoside antibiotic. The AACs catalyze acetylation at the 1 [AAC(1)], 3 [AAC(3)], 2′ [AAC(2′)], or 6′ [AAC(6′)] positions (Fig. 1 and Table 1). The three dimensional structures of several acetyltransefrases have been resolved (see Table 1), mechanistic and structural aspects of these and other representatives of these enzymes have been thoroughly studied and reviewed (Azucena and Mobashery, 2001; Houghton et al., 2010; Tolmasky, 2007a; Vetting et al., 2005; Wright and Berghuis, 2007).
3.2.1. AAC(1)
To date AAC(1) enzymes have been found in E. coli , Campylobacter spp., and an actinomycete (Gomez-Luis et al., 1999; Lovering et al., 1987; Sunada et al., 1999). The AAC(1) isolated from E. coli catalyzes acetylation of apramycin, butirosin, lividomycin and paromomycin at the 1 position, and catalyzes di-acetylation of ribostamycin and neomycin. The AAC(1) isolated from an actinomycete (strain #8) differed in substrate profile from that one from E. coli as apramycin was not acetylated by this enzyme. Furthermore paromomycin was preferentially acetylated at position 1, but 1,2′-di-N-acetylparomomycin and 1,6″′-di-N-acetylparomomycin were also found as products of the enzymatic reaction (Sunada et al., 1999). These studies also determined that these modifications were not accompanied by a significant reduction of the antibiotic activity. The substrate profile of the AAC(1) isolated from Campylobacter spp. was similar to that of the E. coli enzyme. This was the only instance in which an AAC(1) was found in clinical isolates. The authors suggested that the gene is located in the chromosome, but these results await confirmation. Although all three enzymes have been named AAC(1), the difference in substrate profile of at least one of them would justify to named them with a subclass number.
3.2.2. AAC(3)
There are nine recognized subclasses of AAC(3) enzymes described to date, all of them in gram-negatives. The subclass AAC(3)-V has been eliminated after confirmation that the only enzyme in this group is identical to AAC(3)-II (Shaw et al., 1993). The subclass AAC(3)-I includes five enzymes that confer resistance to gentamicin, sisomicin, and fortimicin (astromicin) and are present in a large number of Enterobacteriaceae and other gram-negative clinical isolates. The X-ray structure of AAC(3)-Ia from Serratia marcescens (Javier Teran et al., 1991) complexed to CoA has been determined at 2.3 Å resolution (Wolf et al., 1998), as it is the case with several acetyltransferases this enzyme seems to exist as a dimer under physiological conditions.
All five genes have been found as part of gene cassettes in integrons. The latest gene in this subclass to be reported is aac(3)-Ie, which was found in integrons in Proteus vulgaris, P. aeruginosa, and within a Salmonella enterica subsp. enterica genomic island (Gionechetti et al., 2008; Wilson and Hall, 2010).
The subclass AAC(3)-II, which is characterized by resistance to gentamicin, netilmicin, tobramycin, sisomicin, 2′-N-ethylnetilmicin, 6′-N-ethylnetilmicin and dibekacin (Shaw et al., 1993), includes three enzymes: AAC(3)-IIa and AAC(3)-IIb, which were previously published as AAC(3)-Va and AAC(3)-Vb (see letter and reply van de Klundert and Vliegenthart, 1993), and AAC(3)-IIc. While AAC(3)-IIa has been found in a large variety of genera, AAC(3)-IIb and AAC(3)-IIc have been found in E. coli, Alcaligenes faecalis and S. marcescens or E. coli and P. aeruginosa respectively (Dubois et al., 2006; Dubois et al., 2008; Oteo et al., 2006; Shaw et al., 1993). A recent survey of Enterobacteriaceae clinical isolates from a Tunisian Hospital showed the presence of undetermined AAC(3)-II enzymes, although the authors suggest the possibility of AAC(3)-IIb, in all genera tested (Dahmen et al., 2010).
There are three enzymes belonging to the subclass AAC(3)-III, all isolated from P. aeruginosa isolates. When cloned, the aac(3)-IIIa gene was expressed in P. aeruginosa but not in E. coli (Vliegenthart et al., 1991b). This does not seem to be due to an inactive promoter in E. coli. The authors proposed that most probably the mRNA is not completely synthesized or the initiation of translation of the gene is obstructed (Vliegenthart et al., 1991b). There were other early reports of AAC(3)-III enzymes in other genera, e.g., Klebsiella pneumoniae, but they seem to be misnamed (for clarification see Vliegenthart et al., 1991b).
The only representative of AAC(3)-IV has been identified in clinical strains of E. coli (originally thought to be Salmonella) (Brau et al., 1984), Campylobacter jejuni, and in environmental Pseudomonas stutzeri (Heuer et al., 2002).
Although only AAC(3)-VIa is recognized in the literature within subclass AAC(3)-VI, comparison of the original sequence from Enterobacter cloacae, with the more recently isolated genes from E. coli, and S. enterica show a one amino acid difference (Call et al., 2010; Rather et al., 1993a).
Subclasses AAC(3)-VII, AAC(3)-VIII, AAC(3)-IX, and AAC(3)-X are represented in strains of actinomycetes (Ishikawa et al., 2000; Lopez-Cabrera et al., 1989; Salauze et al., 1991). This latter enzyme was of interest because besides catalyzing acetylation of kanamycin and dibekacin at the 3-amino group it also mediates acetylation the 3″-amino group in arbekacin and amikacin, making this the first AAC detected to have also AAC(3″) activity. Interestingly, while 3″-N-acetylamikacin lost most or all antibiotic activity, 3″-N-acetylarbekacin was still active (Hotta et al., 1998).
3.2.3. AAC(2′)
These enzymes have been found in gram-negatives and Mycobacterium, they mediate modification of several aminoglycosides including gentamicin, tobramycin, dibekacin, kanamycin and netimicin. Only one subclass exists, which includes AAC(2′)-Ia (Providencia stuartii), AAC(2′)-Ib (Mycobacterium fortuitum and Acinetobacter baumannii), AAC(2′)-Ic (M. tuberculosis and Mycobacterium bovis), AAC(2′)-Id (Mycobacterium smegmatis), and a putative AAC(2′)-Ie identified in the Mycobacterium leprae genome (Adams et al., 2008; Ainsa et al., 1997; Hegde et al. 2001; Rather et al., 1993b). A putative AAC(2′) enzyme has been proposed to be part of multidrug resistance in Stenotrophomonas maltophilia but it has not been named further (Crossman et al., 2008). Our Blast analysis of the amino acid sequence of this protein against those in GenBank did not show 100% homology with any of the AAC(2′) know enzymes.
3.2.4. AAC(6′)
AAC(6′) enzymes are by far the most common, they are present in gram-negatives as well as gram-positives, the genes have been found in plasmids and chromosomes, and are often part of mobile genetic elements, some of them with unusual structures (Centron and Roy, 2002; Soler Bistue et al., 2008; Tolmasky, 2007a; Tolmasky, 2000). Accordingly, there is a very large volume of information available about them. There are two main subclasses of AAC(6′) enzymes that specify resistance to several aminoglycosides and differ in their activity against amikacin and gentamicin C1. While AAC(6′)-I shows high activity against amikacin and gentamicin C1a and C2 but very low towards gentamicin C1, AAC(6′)-II enzymes actively mediate acetylation of all three forms of gentamicin but not amikacin (Rather et al., 1992; Shaw et al., 1993; Tolmasky, 2007a; Tolmasky et al., 1986; Woloj et al., 1986). A novel enzyme that includes fluoroquinolones as substrates, could be considered a third class because of the change in pattern of substrates but it has been named AAC(6′)-Ib-cr, most probably because it is an evolutionary product of AAC(6′)-Ib by modification of two amino acids, Trp102Arg and Asp179Tyr (Robicsek et al., 2006). Unfortunately, due to the high variability and number of enzymes belonging to this class, the fast pace of research on these enzymes, and the fact that a large number of enzymes have different degrees of similarity in sequence and phenotype, there is a good deal of confusion and lack of consistency in nomenclature and classification of many members. In at least one instance two simultaneously discovered enzymes were named identically (Vanhoof et al., 1998). Enzymes with AAC(6′)-II resistance profiles but with higher identity to AAC(6′)-I enzymes at the amino acid level were named AAC(6′)-I (Casin et al., 2003; Lambert et al., 1994b). Different enzymes have been named identically, for example an acetyltransferase encoded by plasmid pBWH301 was named AAC(6′)-Il (accession number U13880) (Bunny et al., 1995), and the same name was used to name an acetyltransferase from C. freundii Cf155 (accession number Z54241) (Hannecart-Pokorni et al., 1997). This latter enzyme was subsequently renamed AAC(6′)-Im (Vanhoof et al., 1998). A search in PubMed shows the title of this paper as “AAC(6′)-Im [corrected]”. However, this enzyme was also called AAC(6′)-Ip by Centrón et al (Centron and Roy, 1998). Another enzyme identified later in E. coli and Entererococcus faecium was named AAC(6′)-Im (Chow et al., 2001).
The AAC(6′)-I subclass is so highly populated that a double low case letter was necessary to identified them, at the moment the latest published enzyme named as such is the AAC(6′)-Iaf (Kitao et al., 2009). An AAC(6′)-Iai can be found in GenBank but not AAC(6′)-Iag or AAC(6′)-Iah. Variants of AAC(6′)-Ib have been identified with subscripts e.g., AAC(6′)-Ib3, AAC(6′)-Ib4, AAC(6′)-Ib6, and AAC(6′)-Ib7 and differ at the N-terminus but have similar behavior (Casin et al., 1998). Conversely variant AAC(6′)-Ib11, found in a class 1 integron in S. Typhimurium, exhibits a two amino acids difference with AAC(6′)-Ib at positions 118 and 119 that results in an extended resistance spectrum that would merit the definition of a new subclass (Casin et al., 2003). Another variation to the nomenclature used only once is the addition of a prime symbol. The Pseudomonas fluorescens BM2687 AAC(6′)-Ib’ is encoded by a gene that has a Ser instead of a Leu residue at position 90, a substitution previously recognized as responsible for changing the resistance profile from subclass I to II (Lambert et al., 1994b; Rather et al., 1992). Besides the addition of a prime symbol, the name of this enzyme is also unusual, although not unique, in that in spite of having a AAC(6′)-II phenotype is called as if belonging to subclass AAC(6′)-I. AAC(6′)-Ib’ also exist as a fusion protein with a nucleotidyltransferase identified as ANT(3″)-Ii/AAC(6′)-IId in a S. marcescens integron that includes a group II intron (Centron and Roy, 2002). Considering the total identity between AAC(6′)-IId portion of the S. marcescens enzyme and AAC(6′)-Ib’, the name of this latter enzyme should be changed to AAC(6′)-IId. Other modifications to the nomenclature include removal of the roman number that identifies the subclass and the addition of a number, e.g. AAC(6′)-29a, AAC(6′)-29b, AAC(6′)-31, AAC(6′)-32, or AAC(6′)-33 (Gutierrez et al., 2007; Mendes et al., 2007; Poirel et al., 2001; Viedma et al., 2009); or the substitution of the low case letter for a number as in the S. enterica AAC(6′)-I30 enzyme (Mulvey et al., 2004). Other recent variations to the nomenclature consist on the addition of whole words or acronyms such as AAC(6′)-Ib-Suzhou (Huang et al., 2008) or AAC(6′)-Isa (Hamano et al., 2004). The monumental number of identified genes together with the de facto lack of a unified and agreed nomenclature for AAC(6′) enzymes make it extremely difficult to get a clear nomenclature landscape about these enzymes. The AAC(6′)-Id protein has been mentioned several times in the literature, but the accession number provided (X12618) does not currently correspond to an acetyltransferase and for that reason it has not been included in Table 1.
AAC(6′) enzymes can exist as fusion proteins occupying the N or C terminal region of the composite protein (Zhang et al., 2009). These fused aac(6′) genes are usually found within integrons and they can be the result of integrase-mediated recombination events (Centron and Roy, 2002). Interestingly, proteins containing AAC(6′)-I activities have been found fused to APH, ANT, a different AAC, and another AAC(6′)-I activities. AAC(6′)-Ie is located to the amino terminal end of a bifunctional Enterococcus faecalis and Staphylococcus aureus enzyme with AAC(6′) and APH(2″) activities (Boehr et al., 2004; Ferretti et al., 1986). The aac(6′)-aph(2″) gene is usually present in Tn4001-like transposons (Culebras and Martinez, 1999). As described above, the AAC(6′)-IId is the carboxy terminal region of the protein fusion that also includes an ANT(3″)-I activity. Fusions of two AAC(6′)-I activities, AAC(6′)-30/AAC(6′)-Ib’, or two AAC belonging to different subclasses, AAC(3)-Ib and AAC(6′)-Ib’ were found in P. aeruginosa integrons (Dubois et al., 2002; Mendes et al., 2004).
Three phylogenetic subgroups have been recognized among AAC(6′)-I and AAC(6′)-II enzymes (Hannecart-Pokorni et al., 1997; Shaw et al., 1993; Shmara et al., 2001) but an alternative theory has been published that proposes that the three groups are less related than thought before and the 6′ acetylating activity has evolved independently at least three times (Salipante and Hall, 2003).
An immunochromatographic method based on the utilization of monoclonal antibodies against the AAC(6′)-Iae has recently been reported (Kitao et al., 2010). This enzyme was selected for these studies because aac(6′)-Iae is prevalent in Japan and appears linked to the metallo-!-lactamase gene blaIMP and ant(3″)-Ia in the integron In113, making the assay a useful tool to detect multiple drug resistance in P. aeruginosa in this country (Kitao et al., 2010). However, 37% of the negative isolates from Japan still showed a multiple drug resistance phenotype and 76% of these negative isolates include aac(6′)-Ib and the metallo-!-lactamase gene blaIMP-1. At present, the authors of this study are developing an immunochromatography assay targeting AAC(6′)-Ib and metallo-!-lactamase IMP to complement that one targeting AAC(6′)-Iae for a more complete diagnostics tool (Kitao et al., 2010).
AAC(6′)-Ib is probably the most clinically relevant acetyltransferase and is responsible for the resistance to amikacin and other aminoglycosides found in several gram-negatives belonging to the genus Acinetobacter and to the Enterobacteriaceae, Pseudomonadaceae, and Vibrionaceae (reviewed in Tolmasky, 2007a; Vakulenko and Mobashery, 2003). It is present in over 70% of AAC(6′)-I-producing gram-negative clinical isolates (Vakulenko and Mobashery, 2003) and, as mentioned above, some of its variants show an extended spectrum including resistance to gentamicin [AAC(6′)-Ib11] (Casin et al., 2003) or reduced susceptibility to quinolones [AAC(6′)-Ib-cr] (Robicsek et al., 2006). Since it was first identified, this latter enzyme has been detected in a large number of geographical regions in numerous genetic environments (Strahilevitz et al., 2009). It is usually found as a gene cassette in different integrons and associated to quinolone resistance genes such as qnrA1, qnrB2, qnrB4, qnrB6, qnrB10, qnrS1, qnrS2, and qepA or β-lactamase genes such as blaCTX-M-1, blaCTX-M-14, blaCTX-M-15, blaCTX-M-24, blaDHA-1, blaSHV-12, and blaKPC-2 (Strahilevitz et al., 2009).
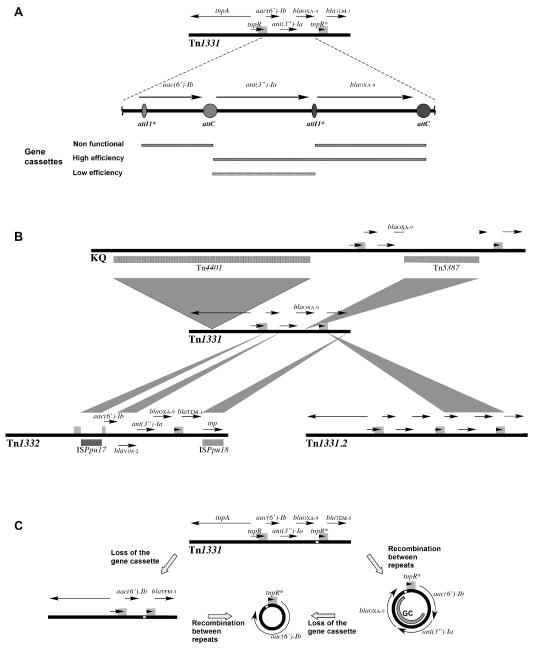
The prevalence of AAC(6′)-Ib together with its numerous variants attracted the interest of several research groups that studied them from different points of view. The aac(6′)-Ib gene is mostly found as a gene cassette within class 1 integrons or as a defective gene cassette within an unusual structure resembling the variable portion of the integrons but lacking the 5′ and 3′ conserved regions as in Tn1331 and its derivatives Tn1331.2, Tn1332 or the KQ element (Chamorro et al., 1990; Dery et al., 1997; Poirel et al., 2006; Rice et al., 2008; Sarno et al., 2002; Soler Bistue et al., 2008; Tolmasky et al., 1988; Tolmasky and Crosa, 1987). The structures of Tn1331 and the modifications occurred for the generation of Tn1331.2, Tn1332, and the KQ element are shown in Fig. 2. Interestingly, the aac(6′)-Ib environment found in these genetic elements has a number of particular characteristics. While in integrons the gene located at the 5′ end of the variable region is preceded by an attI recombination site located adjacent to the intI gene (Partridge et al., 2000), in these elements there is no attI upstream of the aac(6′)-Ib gene. Instead, an 8 bp sequence known as attI1* is found near the beginning of the structural gene at the location where a gene fusion between a blaTEM gene and a precursor of aac(6′)-Ib is believed to have occurred, incorporating the first six amino acids of the TEM β-lactamase at the N-terminus of this version of AAC(6′)-Ib (Fig. 2A) (Ramirez et al., 2008; Tolmasky, 1990). These features define an imperfect gene cassette with attI1* at the 5′ end within the aac(6′)-Ib structural gene (Fig. 2A). IntI1 integrase-mediated excision of this imperfect gene cassette could not be detected in cells harboring a recombinant clone with intI1 under the control of the Ptac promoter (Ramirez et al., 2008). Products of evolution of the Tn1331 transposon by insertion of DNA fragments or duplications have been found and are shown in Fig. 2B. This version of the aac(6′)-Ib gene was also found in the chromosome of a P. mirabilis isolate as part of a mosaic structure containing several resistance genes (Zong et al., 2009). In this case the upstream region of the gene is derived from Tn1331 but it shows a different organization, it is preceded by the region located downstream of blaOXA-9 in Tn1331. The authors of this report proposed that homologous recombination events between the duplicated regions of Tn1331 could have led to formation of a circular molecule that could have then been integrated into the chromosome (Fig. 2C) (Zong et al., 2009).
Fig. 2.
A. Genetic map of the Tn1331 transposon with the region including genes aac(6′)-Ib, aadA1 and blaOXA-9 amplified. Circles and ovals represent attC and attI1* loci respectively. For clarity the points of potential crossover reactions are not indicated but they can be found in Ramirez et al. (Ramirez et al., 2008). Regions with a gene cassette structure are indicated below the genetic map by bars of different patterns. Their functionality as determined in recombination assays in the presence of IntI1 expressed from a recombinant clone harboring intI1 under the control of the Ptac promoter is shown. Directly repeated regions are shown as gray boxes on the sequences. B. Genetic maps of Tn1331, Tn1331.2, Tn1332, and the KQ element. Shadowed areas show the fragments inserted within the Tn1331 sequence that generated the other three genetic elements. C. Model for generation of a circular molecule containing aac(6′)-Ib (Zong et al., 2009). The white box indicates the DNA region that is found upstream of the gene in P. mirabilis JIE273. GC, gene cassette. Circular molecules are not drawn to scale.
The translation of the aac(6′)-Ib7 gene cassette has been studied in some detail. This is one of about 20% of gene cassettes that lack a discernible translation initiation region. Instead, a short open reading frame is located immediately upstream of the structural gene that significantly enhances translation through translational coupling (Hanau-Bercot et al., 2002; Jacquier et al., 2009).
A large number of variants of AAC(6′)-Ib have been found that differ at the N-terminal end, a phenomenon that may be a consequence of the high mobility of the gene. The fact that most of all of these variants are active shows a high flexibility in the structural requirements at this portion of the protein. This property has been proposed to be a contributing factor to the successful distribution and predominance among aminoglycoside resistant Enterobacteriaceae (Casin et al., 1998).
The AAC(6′)-Ib protein has been the subject of numerous mutagenesis as well as structural and mechanistic studies (Casin et al., 2003; Chavideh et al., 1999; Kim et al., 2007; Maurice et al., 2008; Panaite and Tolmasky, 1998; Pourreza et al., 2005; Rather et al., 1992; Shmara et al., 2001; Vetting et al., 2004; Vetting et al., 2008). Significant progress in understanding AAC(6′)-Ib and its variants has been achieved recently after the elucidation of the crystal structures of AAC(6′)-Ib and the extended spectrum AAC(6′)-Ib11 in conjunction with the construction of a molecular model of AAC(6′)-Ib-cr (Vetting et al., 2008). These studies showed that unlike AAC(6′)-Ii and AAC(6′)-Iy, which are dimers (Draker et al., 2003; Vetting et al., 2004; Wybenga-Groot et al., 1999), AAC(6′)-Ib and AAC(6′)-Ib-cr exist as a monomer while AAC(6′)-Ib11 shows monomer/dimer equilibrium (Maurice et al., 2008). Structural features behind the ability of AAC(6′)-Ib to catalyze acetylation of semisynthetic aminoglycosides, as well as the ordered kinetic mechanism could be explained (Maurice et al., 2008). Furthermore, a flexible flap was identified in AAC(6′)-Ib11 that might explain its ability to utilize as substrate both amikacin and gentamicin (Maurice et al., 2008). The modeling of AAC(6′)-Ib-cr, which has the substitutions D179Y and W102R with respect to AAC(6′)-Ib, permitted to determine that the Asp179Tyr substitution produces the greatest structural effect that results in an enhanced binding to the antibiotic molecule and the W102R acts by stabilizing the positioning of the Y179 (Robicsek et al., 2006; Strahilevitz et al., 2009). This attractive model explains the effects of each individual substitution. While D179Y is enough to confer a partial resistance phenotype, the effect of W102R is hardly detectable. Another model that emphasizes plasticity in the active site has also been suggested (Maurice et al., 2008). Quick methods for identification and genotyping of aac(6′)-Ib-cr have recently been published (Bell et al., 2010; Hidalgo-Grass and Strahilevitz, 2010).
Detailed subcellular localization studies of the AAC(6′)-Ib encoded by Tn1331 using physical separation methods together with gene fusions to phoA in which the signal peptide coding sequence has been removed, and fluorescence microscopy in which the gene was fused to the cyan fluorescent protein demonstrated that the enzyme is evenly distributed within the cytoplasmic compartment of E. coli (Dery et al., 2003). Care should be taken when determining the subcellular location of aminoglycoside modifying enzymes. In the past there were contradictory reports indicating that they are located in the periplasmic space or the cytosol (Franklin and Clarke, 2001; Perlin and Lerner, 1981; Tolmasky, 2007a; Vakulenko and Mobashery, 2003; Vliegenthart et al., 1991a). These contradictory findings could be due to the fact that in osmotically shocked E. coli, proteins are released through a molecular sieve formed by the damaged cell envelope (Vazquez-Laslop et al., 2001). As a consequence, cytoplasmic proteins small in native size tend to be released after osmotic shock treatment while larger proteins or protein complexes remain inside the cells (Vazquez-Laslop et al., 2001). We confirmed this by extracting the periplasmic proteins by spheroplast formation under different conditions and found that while the controls behaved as expected under all conditions, when we used mild conditions the AAC(6′)-Ib signal was present in the cytosolic extract but when we used harsher conditions a considerable fraction of the total AAC(6′)-Ib was found in the periplasmic extract (Dery et al., 2003; Tolmasky, 2007a).
Two shorter proteins of this class, AAC(6′)-29a and AAC(6′)-29b, have been identified from a multidrug-resistant clinical isolate of P. aeruginosa (Magnet et al., 2003; Poirel et al., 2001). The 131-amino acids AAC(‘6)-29b protein was studied in more detail and it was found that it does not mediate resistance by enzymatic modification but rather by tightly binding aminoglycoside molecules, a result that led to the conclusion that the mechanism of aminoglycoside resistance mediated by this protein is by sequestering the drug as a result of tight binding to the molecule (Magnet et al., 2003).
3.3. Aminoglycoside modifying enzymes: aminoglycoside O-nucleotidyltransferases (ANTs)
ANTs mediate inactivation of aminoglycosides by catalyzing the transfer of an AMP group from the donor substrate ATP to and hydroxyl group in the aminoglycoside molecule. There are five classes of ANTs that catalyze adenylylation at the 6 [ANT(6)], 9 [ANT(9)], 4′ [ANT(4′)], 2″ [ANT(2″)], and 3″ [ANT(3″)] positions, of which only ANT(4′) includes two subclasses, I and II (Fig. 1 and Table 2).
Table 2.
Aminoglycoside O-nucleotydyltransferases
| ANTs | gene names | Genetic location |
Accession number |
Host | References |
|---|---|---|---|---|---|
| ANT(6)-Ia | ant(6)-Ia, ant6, aadE | Plasmid, chromosome |
NC_006663, NC_012924, GQ900487 |
Staphylococcus
epidermidis, E. faecium, Streptococcus suis, S. aureus |
(Gill et al., 2005; Holden et al., 2009) |
| ant6 | Plasmid | AB247327 | E. faecalis | ||
| aadE | Chromosome | NC_013853 | Streptococcus mitis | ||
| aadK | Chromosome | M26879 | B. subtilis, Bacillus spp. | (Noguchi et al., 1993; Ohmiya et al., 1989) |
|
| aadE | Plasmid | AJ489618 | C. jejuni | ||
| aad(6) | Plasmid |
NC_008445, AY712687 |
E. faecalis, Streptococcus
oralis |
(Cerda et al., 2007; Schwarz et al., 2001) |
|
| ANT(6)-Ib | ant(6)-Ib | Transferable pathogenicity island |
FN594949, NZ_ABDU0 1000081 |
C. fetus subsp. fetus, B.
subtilis |
(Abril et al., 2010) |
| ANT(9)-Ia | ant(9)-Ia, aad(9), spc | Plasmid, transposon |
X02588, GU235985 |
S. aureus, Enterococcus spp., Sathylococcus sciuri |
(Murphy, 1985) |
| ANT(9)-Ib | ant(9)-Ib, aad(9), spc | Plasmid | M69221 | E. faecalis | (LeBlanc et al., 1991) |
| ANT(4′)-Ia C |
ant(4′)-Ia, aadD2, aadD,
ant(4′,4″)-I |
Plasmid |
U35229, M19465 |
S. epidermidis, S. aureus, Enterococcus spp., Bacillus spp. |
(McKenzie et al., 1986; Santanam and Kayser, 1978) |
| ANT(4′)-IIa | ant(4′)-IIa | Plasmid | M98270 |
P. aeruginosa,
Enterobacteriaceae |
(Jacoby et al., 1990) |
| ANT(4′)-IIb | ant(4′)-IIb | Transposon | AY114142 | P. aeruginosa | (Sabtcheva et al., 2003) |
| ANT(2″)-Ia | ant(2″)-Ia, aadB | Plasmid, integron |
X04555 |
P. aeruginosa, K.
pneumoniae , Morganella morganii, E. coli, S. typhimurium, C. freundii, A. baumannii |
(Cameron et al., 1986) |
| ANT(3″)-Ia |
ant(3″)-Ia, aadA, aadA1,
aad(3″)(9) |
Plasmid, transposon, integron |
X02340 |
Enterobacteriaceae, A.
baumannii, P. aeruginosa, Vibrio cholerae |
(Hollingshead and Vapnek, 1985; Tolmasky, 1990) |
| aadA2 | Plasmid, integron |
NC_010870 |
K. peneumoniae, Salmonella spp., Corynebacterium glutamicum, C. freundii, Aeromonas spp. |
(Chen et al., 2007) | |
| aadA3 | Plasmid, transposon, integron |
AF047479 | E. coli | (Parent and Roy, 1992) | |
| aadA4 | Plasmid, chromosome |
NC_002928, NC_010558 |
Bordetella parapertussis,
E. coli |
(Parkhill et al., 2003; Perichon et al., 2008) |
|
| aadA5 | Plasmid, transposon, integron |
AF137361 |
E. coli, K. pneumoniae,
Kluyvera georgiana, P. aeruginosa, E. cloacae |
(Sandvang, 1999) | |
| aadA6 | Integron | AM087411 | P. aeruginosa | (Fiett et al., 2006) | |
| aadA7 | Integron | AB114632 |
V. fluvialis, P. aeruginosa,
E. coli, V. cholerae, S. enterica |
(Ahmed et al., 2004) | |
| aadA8 | Plasmid, integron |
AY139603 |
V. cholerae, K.
pneumoniae, Bacillus endophyticus |
(Tennstedt et al., 2003) | |
| aadA9 | Plasmid | NC_003227 | C. glutamicum | (Tauch et al., 2002) | |
| aadA10 | Plasmid, integron |
AM087405 | P. aeruginosa, E. coli. | (Fiett et al., 2006; Partridge et al., 2002) |
|
| aadA11 | Integron |
AJ567827, AY758206 |
E. coli, P. aeruginosa. | (Llanes et al., 2006) | |
| aadA12 | Integron | FJ381668 |
E. coli, Yersinia
enterocolitica, S. enterica |
(Ajiboye et al., 2009) | |
| aadA13 | Plasmid, integron |
NC_010643 |
Pseudomonas rettgeri, P.
aeruginosa, Y. enterocolitica, E. coli |
(Revilla et al., 2008) | |
| aadA14 | Plasmid | AJ884726 | Pasteurella multocida | (Kehrenberg et al., 2005) | |
| aadA15 | Integron | DQ393783 | P. aeruginosa | (Yan et al., 2006) | |
| aadA16 | Plasmid, integron |
EU675686 |
E. coli, V. cholerae, K.
pneumoniae |
(Wei et al., 2009) | |
| aadA17 | Integron | FJ460181 | Aeromonas media | ||
| aadA21 | Integron | AY171244 | Salmonella spp. | (Faldynova et al., 2003) | |
| aadA22 | Plasmid, integron |
AM261837 | S. enterica, E. coli | (Herrero et al., 2008) | |
| aadA23 | Integron | AJ809407 | S. enterica | (Michael et al., 2005) | |
| aadA24 | Integron | DQ677333 | Salmonella spp. | (Egorova et al., 2007) | |
| aadA6/aadA10 | Integron | AM087405 | P. aeruginosa | (Fiett et al., 2006) |
Only representative hosts, references and accession numbers are shown.
C, three dimensional structure has been resolved. ANT(4′)-Ia pdb id: 1KNY (Pedersen et al., 1995).
3.3.1. ANT(6)
Genes coding for enzymes with related amino acid sequences have been named ant(6)-Ia, ant6, ant(6), and aadE. They all exhibit the same substrate profile (resistance to streptomycin) and therefore belong into the same subclass, but they are not identical. These genes are highly widespread among gram-positive bacteria (Tolmasky, 2007a; Vakulenko and Mobashery, 2003). Two genes called aadE with 87% identity at the amino acid level were found in the E. faecalis plasmid pRE25 and in C. jejuni (Schwarz et al., 2001). Genes coding for ANT enzymes are found in plasmids, transposons, and chromosomes. The ant(6) gene is often found in a cluster ant(6)-sat4-aph(3′)-III that specifies resistance to aminoglycosides and streptothricin (Cerda et al., 2007). This cluster is part of Tn5405 and other related transposons, which are distributed among Staphylococci and Enterococci (Werner et al., 2003) and are located in plasmids and chromosomes. Another gene originally found in Bacillus subtilis was named aadK (Noguchi et al., 1993) and was subsequently found in other species of Bacillus (Vakulenko and Mobashery, 2003). The protein encoded by this gene shows 58% identity and 74% similarity with one encoded by an aadE gene (Vakulenko and Mobashery, 2003). A novel ant(6) gene, named ant(6)-Ib, was recently identified in Campylobacter fetus subsp. fetus within a transferable pathogenicity island (Abril et al., 2010). This gene is identical to that called aad(6) in a contig of an unfinished Clostridium genome (accession number NZ_ABDU01000081).
3.3.2. ANT(9)
Two enzymes with the ANT(9) characteristics have been described, ANT(9)-Ia and ANT(9)-Ib, both mediating resistance to spectinomycin. The genes coding for these enzymes were called as ant(9)-Ia and ant(9)-Ib, but unfortunately they have both also been called spc or aad(9) facilitating confusion. The amino acid sequences of ANT(9)-Ia and ANT(9)-Ib share 39% identity. ANT(9)-Ia was first described in S. aureus and then also in Enterococcus avium, E. faecium, and E. faecalis. In all four bacteria the gene was part of Tn554 (Mahbub Alam et al., 2005; Murphy, 1985). Our BLAST analysis showed a protein with 100% identity to ANT(9)-Ia present as part of a novel transposon, Tn6072 (Chen et al., 2010). However, although the gene is correctly named as spc it is described as a streptomycin 3′-adenyltransferase. ANT(9)-Ib was found in a plasmid from E. faecalis (LeBlanc et al., 1991).
3.3.3. ANT(4′)
ANT(4′)-Ia is found in plasmids of gram-positives such as Staphylococci, Enterococci, and Bacillus spp., and the gene has been also named aadD, aadD2, and ant(4′,4″)-I (Bozdogan et al., 2003; Kobayashi et al., 2001; Muller et al., 1986; Perez-Vazquez et al., 2009). This latter name is due to the fact that this enzyme was found to modify 4′ and 4″ groups, which makes it capable of conferring resistance to dibekacin, an aminoglycoside that lacks a 4′ target (Santanam and Kayser, 1978). Both subclasses, I and II, confer resistance to tobramycin, amikacin, isepamicin, but subclass I also codifies resistance to dibekacin. ANT(4′)-Ia is the only ANT enzyme for which the three dimensional structure has been resolved (Pedersen et al., 1995). An ANT(4′) was also the subject of NMR studies to clarify aspects of the process of the recognition of the substrate (Revuelta et al. 2008). Two ANT(4′)-II enzymes have been described in gram-negative bacilli. These enzymes do not modify dibekacin, and therefore they must be unable to use the position 4″ as target. ANT(4′)-IIa was identified in plasmids of Pseudomonas and Enterobacteriaceae (Jacoby et al., 1990), and ANT(4′)-IIb was identified more recently in a P. aeruginosa transposon (Coyne et al., 2010).
3.3.4. ANT(2″)
This class consists only of ANT(2″)-Ia (Cameron et al., 1986), an enzyme that is widely distributed as a gene cassette in class 1 and 2 integrons (Ramirez et al., 2005; Vakulenko and Mobashery, 2003) and mediates resistance to gentamicin, tobramycin, dibekacin, sisomicin, and kanamycin. Therefore it is commonly encoded by plasmids and transposons. This enzyme, encoded by a gene more commonly called aadB, is present in enterobacteria and non-fermentative gram-negative bacilli.
3.3.5. ANT(3″)
These are the most commonly found ANT enzymes, they specify resistance to spectinomycin and streptomycin, and the coding genes are most commonly named aadA (Hollingshead and Vapnek, 1985). At least 22 highly related gene versions are found in GenBank, that are identified as aadA1 through aadA24, but some numbers are missing. The alternative nomenclature for the protein coded for by aadA1 is ANT(3″)-Ia. Another name used to identify ANT(3″)-Ia is AAD(3″)(9). The aadA genes exist as gene cassettes and are part of a large number of integrons, plasmids and transposons. They can be part of unusual gene cassettes and exist as gene fusions as described in the following paragraphs.
In Tn1331, the aadA1 [ant(3″)-Ia] gene is present within two unusual gene cassette structures (see Fig. 2A). At the 3′ end of the gene, instead of the usual attC site, there is a copy of attI1*, which may have been formed by an illegitimate recombination event between the attC site located 3′ of aadA1 of an integron and the attI1 locus located 5′ of blaOXA-9 of another integron in which the blaOXA-9 gene cassette is adjacent to the 5′-conserved sequence (Sarno et al., 2002). The resulting structure defines a gene cassette consisting of aadA1-attI1* but that lacks the usual attC site, and another gene cassette that includes two genes aadA1-attI1*-blaOXA-9–attC (see Fig. 2A) (Ramirez et al., 2008; Sarno et al., 2002; Tolmasky, 1990; Tolmasky and Crosa, 1993). While the aadA1-attI1* gene cassette is excised by the IntI1 integrase at a very low frequency the gene cassette that includes both genes is fully functional (Ramirez et al., 2008).
The aadA genes are also found fused to other resistance enzymes, e.g., in a P. aeruginosa class 1 integron aadA15 is fused 3′ of blaOXA-10 (Yan et al., 2006) and aadA6 is fused to aadA10 in another P. aeruginosa class 1 integron (Fiett et al., 2006). The aadA1 and aadA4 genes were also found disrupted by insertion of IS26 (Adrian et al., 2000; Han et al., 2008).
The ant(3″)-Ia gene is part of numerous transposons, some of them exhaustively studied such as: a) Tn21 and other related transposons of what is known as the Tn21 subfamily. These transposons are widely disseminated probably as a result of the association of an integron and a gene conferring resistance to a toxic metal within the same mobile element (Liebert et al., 1999); b) Tn1331, already described above; and c) Tn7, which includes in its structure a class 2 integron (Hansson et al., 2002).
3.4 Aminoglycoside modifying enzymes: aminoglycoside O-phosphotransferases (APHs)
APHs catalyze the transfer of a phosphate group to the aminoglycoside molecule (Wright and Thompson, 1999). The classes and subclasses are: APH(4)-I, APH(6)-I, APH(9)-I, APH(3′)-I through VII, APH(2″)-I through IV, APH(3″)-I, APH(7″)-I (Fig. 1 and Table 3).
3.4.1. APH(4)
There are two enzymes within the only subclass defined in this group: APH(4)-Ia (Kaster et al., 1983) and APH(4)-Ib (Zalacain et al., 1986), whose genes have also been named hph and hyg, respectively. These enzymes mediate resistance to hygromycin and are not clinically relevant. These genes have been used in the construction of cloning vehicles for both prokaryotes and eukaryotes (Abhyankar et al., 2009; Gritz and Davies, 1983).
3.4.2. APH(6)
There are 4 enzymes in the only described subclass of APH(6)s, which confer resistance to streptomycin. The aph(6)-Ia, also known as aphD and strA, was originally found in the chromosome of Streptomyces griseus (Distler et al., 1987). The aph(6)-Ib was also named sph and was found in Streptomyces glaucescens (Vogtli and Hutter, 1987). The gene coding for APH(6)-Ic is one of three resistance genes present in Tn5, a composite transposon found in gram-negatives (Steiniger-White et al., 2004). Although this transposon is not widely distributed, it has been extensively studied and modified as tool for molecular genetics (Steiniger-White et al., 2004). The aph(6)-Id gene, also denominated strB and orfI, was first found in the plasmid RSF1010, a 8,684 bp broad host range multicopy plasmid RSF1010 that can replicate in most gram-negative bacteria and also in gram-positive actinomyces, and is also known as R300B and R1162 (Meyer, 2009). This plasmid was also the first source identified for another APH, aph(3″)-Ib (see below), which is contiguous to aph(6)-Id. These genes are part of a fragment that includes the genes repA, repC, sul2, aph(3″)-Ib, and aph(6)-Id that has been found, complete or in part, within plasmids, integrative conjugative elements, and chromosomal genomic islands (Daly et al., 2005; Gordon et al., 2008). As a consequence of the dissemination of this DNA fragment, the aph(6)-Id and aph(3″)-Ib genes are found in both gram-positives and gram-negatives.
3.4.3. APH(9)
The aph(9)-Ia gene was first found in Legionella pneumophila (Suter et al., 1997). BLAST analysis of this nucleotide sequence also showed that there is a gene with 87% homology within the genome of L. pneumophila strain Lens that is identified as aph (Cazalet et al., 2004). The APH(9)-Ia has been the subject of detailed analysis. The enzyme was overproduced and purified, and it was determined that it does not bind to any tested aminoglycoside other than spectinomycin (Thompson et al., 1998). The Km and kcat values were also determined and the reaction product was purified and characterized by mass spectrometry and 1H, 13C, and 31P NMR (Thompson et al., 1998). Further studies led to determination of the crystal structures of APH(9)-Ia in its apo form, its binary complex with the nucleotide, AMP, and its ternary complex bound with ADP and spectinomycin (Fong et al., 2010). These structures showed that APH(9)-Ia presents similar folding to APH(3′) and APH(2″) enzymes but differs significantly in its substrate binding area and in undergoing a conformation change upon ligand binding (Fong et al., 2010).
The phosphotransferase APH(9)-Ib isolated from Streptomyces flavopersicus (Str. netropsis) has also been called SpcN and it has no significant homology to that of L. pneumophila. A BLAST analysis of this aph(9)-Ib gene nucleotide sequence showed 78-79% identity with genes from 3 Stretomyces spectabilis strains (Lyutzkanova et al., 1997). Despite of the differences these genes are also called spcN in GenBank.
3.4.4. APH(3′)
The APH(3′)-I subclass shows a resistance profile including kanamycin, neomycin, paromomycin, ribostamycin, lividomycin, is composed of three enzymes that are widely distributed mainly among gram-negatives within wide host range plasmids and transposons (Vakulenko and Mobashery, 2003). The aph(3′)-Ia gene, also known as aphA-1, is part of the well known Tn903 transposon (Bernardi and Bernardi, 1991) and it is commonly used as marker gene in cloning vehicles. The aph(3′)-Ib gene is part of the wide host range conjugative RP4 plasmid (Pansegrau et al., 1987). This gene was originally named aphA. The aph(3′)-Ic gene, also called aphA7 and aphA1-Iab, is part of plasmids and transposons and its wide distribution includes Corynebacterium spp. (Tauch et al., 2000; Vakulenko and Mobashery, 2003). This gene has also been included in cloning vehicles.
The APH(3′)-II subclass includes three isozymes that specify resistance to kanamycin, neomycin, butirosin, paromomycin, and ribostamycin. The APH(3′)-IIa, also known as aphA-2 is one of the three resistance genes encoded by Tn5 (Steiniger-White et al., 2004) (see above) and it is used as resistance marker in cloning vectors for both prokaryotes and eukaryotes (Wright and Thompson, 1999). The enzyme coded by this gene has been characterized in detail and its crystal structure in complex with kanamycin has been resolved (Nurizzo et al., 2003; Siregar et al., 1994). The aph(3′)-IIb gene was identified in the P. aeruginosa chromosome (Winsor et al., 2005) and the third member of this subclass, aph(3′)-IIc, was recently defined in S. maltophilia but an accession number is not available (Okazaki and Avison, 2007).
The APH(3′)-IIIa is highly disseminated within gram-positives, confers resistance to kanamycin, neomycin, lividomycin, paromomycin, livostamycin, butirosin, amikacin, and isepamicin, and the epidemiological data has been extensively reviewed by Vakulenko and Mobashery (Vakulenko and Mobashery, 2003). Its crystal structure in complex with ADP has been resolved and it shows a close resemblance to kinases from eukaryotes (Hon et al., 1997). An interesting property of this enzyme is that it is competitively inhibited by tobramycin, which one would expect not to be substrate because it lacks a free 3′-hydroxyl group (McKay et al., 1994). However, other aminoglycosides that also lack a free 3′-hydroxyl group like the case of lividomycin can be phosphorylated at the position 5″ (Thompson et al., 1996). This enzyme has also the capability to di-phosphorylate aminoglycosides such as butirosin and neomycin B that have free 3′- and 5″-hydroxyl groups (Hon et al., 1997; Wright and Thompson, 1999). A recent study showed that APH(3′)-IIIa uses only ATP as donor substrate (Shakya and Wright, 2010).
The APH(3′)-IVa coding gene is present in the chromosome of Bacillus circulans (Herbert et al., 1983) and those coding for APH(3′)-Va through c are found in the chromosome of actinomycetes (Wright and Thompson, 1999). The resistance profile for this subclass includes neomycin, paromomycin, and ribostamycin. The aph(3′)-VIa, also known as aphA-6, was described in A. baumannii (Martin et al., 1988), and aph(3′)-VIb was described in K. pneumoniae and S. marcescens but an accession number for this gene is not available (Gaynes et al., 1988). The resistance profile specified by this subclass includes kanamycin, neomycin, paromomycin, ribostamycin, butirosin, amikacin, and isepamycin. The aph(3′)-VIIa, also known as aphA-7, was described in C. jejuni and confers resistance to kanamycin and neomycin (Tenover et al., 1989).
3.4.5. APH(2″)
The APH(2″) plays an important role in resistance to gentamicin in gram-positives. There were originally five APH(2″)-I enzymes described in the literature. However, Toth et al. (Toth et al., 2009) recently performed a detailed analysis of the resistance profiles, regiospecificity, and donor substrate preferences of these enzymes and concluded that on the basis of the aminoglycoside recipient substrate profiles presented by APH(2″)-Ib, APH(2″)-Ic, and APH(2″)-Id they should be reclassified as belonging to subclass II. A novel consideration in renaming these enzymes is the inclusion of the donor substrate as a criterion. Contrary to what it was believed at the time of that work, only APH(2!)-Ib among the four APH(2!) enzymes included in Toth et al. study showed a clear preference for ATP as donor of the phosphate group. APH(2!)-Ia and APH(2!)-Ic utilize GTP as the most efficient donor substrate, and APH(2!)-Id shows similar catalytic efficiencies with ATP or GTP (Toth et al., 2009). A contradictory result was obtained with a derivative of APH(2″)-Ib that includes a His6-tag at the N-terminus, this protein showed a slight selectivity for GTP over ATP but was still able to utilize both NTPs as donor substrates (Shakya and Wright, 2010). Therefore, at least in the case of phosphotransferases of the 2″ class, the subclass nomenclature now considers the recipient as well as the donor substrate profile. In consequence, these authors proposed to change the names of APH(2!)-Ib, -Ic, and -Id to APH(2!)-IIa, -IIIa, and IVa, respectively. The three dimensional structures of these enzymes have been resolved (Smith et al., 2010; Toth et al., 2010a; Toth et al., 2010b). The APH(2″)-Ie has not been included in this analysis and for now it has not been renamed.
The APH(2″)-Ia exists as a fusion to AAC(6′)-Ie, which is located at the N-terminal portion (Ferretti et al., 1986). Cloning both regions as separate genes resulted in active proteins (Ferretti et al., 1986) suggesting that the natural gene arose by gene fusion. However, it is of interest that although the domains do not functionally interact, they are structurally linked in a manner that is important for their stability and conformation and disruption of these interactions results in a negative impact for both activities (Boehr et al., 2004).
The aph(2″)-Ie gene was found downstream of a tnpA gene in an Enterococcus casseliflavus plasmid (Chen et al., 2006).
3.4.6. APH(3″)
The only subclass of APH(3″) enzymes mediates resistance to streptomycin. The APH(3″)-Ia and Ic coding genes were isolated from the chromosomes of S. griseus and M. fortuitum, respectively (Ramon-Garcia et al., 2006; Trower and Clark, 1990). The aph(3″)-Ia gene is also known as aphE and aphD2. The APH(3″)-Ib coding gene was originally found within the plasmid RSF1010 (Scholz et al., 1989) and then in a large number of plasmids, transposons, integrative conjugative elements, and at least one chromosome (chromosome 1 of V. cholerae MJ-1236, accession number CP001485). The gene can also be found named as strA (Scholz et al., 1989).
3.4.7. APH(7″)
The APH(7″)-Ia, which mediates resistance to hygromycin, was isolated from S. hygroscopicus and the gene has been cloned and engineered to be used in molecular genetic analysis of Chlamydomonas reinhardtii (Berthold et al., 2002).
4. Strategies to overcome the effect of aminoglycoside modifying enzymes
The development of new aminoglycosides, which is being pursued using numerous different approaches, is an obvious path to overcome the action of aminoglycoside modifying enzymes. Strategies and perspectives for the generation of novel aminoglycosides or aminoglycoside derivatives such as dimers or conjugates to small molecules have been recently reviewed (see Green et al., 2010; Houghton et al., 2010; Tolmasky, 2007a; Welch et al., 2005). ACHN-490, a novel aminoglycoside named neoglycoside proved to be a promising alternative for treating multiple drug resistance K. pneumoniae including those producing KPC !-lactamase (Endimiani et al. 2009). Other approaches such as the utilization of enzymatic inhibitors or strategies to interfere with gene expression could, if successful, reduce or eliminate the need of discarding aminoglycosides due to the broad dissemination of modifying enzymes. All these strategies together have the potential of increasing the armamentarium against the growing threat of multiresistant infections. A summary of the efforts to develop strategies to inhibit the action or biosynthesis of aminoglycoside modifying enzymes follows.
4.1 Inhibitors of aminoglycoside modifying enzymes
Compounds consisting of both substrates covalently linked, known as bisubstrates, are potential tools to inhibit enzymatic reactions that involve the initial formation of a ternary complex through ordered or random binding of the substrates. An aminoglycoside-CoA bisubstrate was first shown to inhibit the activity of AAC(3)-I in vitro but not in vivo, probably due to the inability of the compound to penetrate the cell wall (Williams and Northrop, 1979). Further research led to synthesis of other bisubstrates of smaller size by using truncated aminoglycosides or CoA. One of the compounds, showed a synergistic effect with kanamycin on the growth of E. faecium harboring AAC(6′)-Ii, an enzyme that catalyzes acetylation through an ordered mechanism (Draker et al., 2003; Gao et al., 2005; Gao et al., 2006). Subsequent kinetic and structural studies using AAC(6′)-Iy, which binds the substrates on a random manner, as a target found that the bisubstrates analyzed bind to this enzyme with much lower affinity (Magalhaes et al., 2008). Aminoglycoside–CoA bisubstrates containing sulfonamide, sulfoxide, or sulfone groups were recently synthesized. Only the sulfone- and sulfoxide-containing bisubstrates showed inhibition of AAC(6)-Ii at nanomolar concentrations (Gao et al., 2008).
In another study, cationic antimicrobial peptides were tested as inhibitors of APH(3!)-IIIa, AAC(6!)-Ii, and AAC(6!)-APH(2!). The results showed that the bovine peptide indolicidin and analogs have an inhibitory effect against both aminoglycoside phosphotransferases and aminoglycoside acetyltransferases, albeit by different mechanisms (Boehr et al., 2003). These peptides were the first example of broad-spectrum inhibitors of aminoglycoside resistance enzymes. However, although the research shows enormous potential for therapeutic purposes none of the peptides showed inhibitory effect in vivo (Boehr et al., 2003).
Two non-carbohydrate diamine derivatives with inhibitory activity were isolated from a library of compounds. One of these compounds, N-cyclohexyl-N-(3-dimethylamino-propyl)-propane-1,3-diamine, was active against ANT(2″), and the other, N-[2-(3,4-dimethoxyphenyl)-ethyl]-N’-(3-dimethylamino-propyl)-propane-1,3-diamine was active against APH(3′) and ANT(2″) (Welch et al., 2005).
In the case of APHs it has been shown that it is possible to take advantage of the structural relation found between these enzymes and eukaryotic protein kinases. Burk et al. recently reviewed possible strategies to inhibit aminoglycoside phosphotransferases (Burk and Berghuis, 2002). Known inhibitors of eukaryotic protein kinases were tested to determine if they had also activity against two aminoglycoside phosphotransferases, APH(3!)-IIIa and the fusion protein AAC(6!)-APH(2″). The results showed that several of the tested compounds were inhibitors of these enzymes. Compounds belonging to the isoquinolinesulfonamide group were the most active in these experiments (Daigle et al., 1997). Compounds that act as inhibitors can target the antibiotic binding region, the ATP-binding site, or the bridged nature of the active site, which binds both the aminoglycoside and the donor nucleotide. Compounds that target the aminoglycoside binding site would have the potential of showing a broader spectrum by being able to bind the pocket of more than one kind of aminoglycoside modifying enzyme.
Liu et al. synthesized bisubstrate compounds consisting of adenosine tethered covalently to neamine using methylene groups as linkers. Compounds including linkers of 5 – 8 carbons in length acted as competitive inhibitors of APH(3′)-Ia and APH(3′)-IIIa (Liu et al., 2000).
A compound that exhibited a modest level of inhibition of AAC(6′)-Ib has been constructed using non-aminoglycoside-like fragments (Lombes et al., 2008). This could be a first step towards generating a strong inhibitor of this clinically important enzyme.
An interesting approach to beat the activity of modifying enzymes without inhibiting their activity is that proposed by Haddad et al. in which an aminoglycoside is chemically unstable after phosphorylation and spontaneously sheds the phosphate self-regenerating the antibiotic (Haddad et al., 1999). The authors prepared an analog of kanamycin A, whose hydrated variant undergoes spontaneous, non-enzymatic elimination of the phosphate donated by ATP via APH(3′) catalysis (Haddad et al., 1999).
4.2 Inhibition of expression of aminoglycoside modifying enzymes
Inhibition of gene expression by antisense oligonucleotides or oligonucleotide analogs can be achieved by a variety of strategies. A number of them have been explored in bacteria with therapeutic purposes, mainly to target essential genes and inhibit growth. A detailed description of these attempts can be found in recent reviews (Hebert et al., 2008; Lundblad and Altman, 2010; Rasmussen et al., 2007; Woodford and Wareham, 2009). In a few instances the targets of antisense inhibition of gene expression were genes specifying antibiotic resistance rather than essential or virulence genes. Antisense oligonucleotides targeting regulatory regions of the multiple antibiotic resistance operon (marORAB) in E. coli increased susceptibility to multiple antibiotics and nuclease-resistant phosphorothioate oligonucleotides enhanced the killing effect of nor! oxacin after introduction into competent cells by chemical transformation or electroporation (White et al., 1997). Whether inhibition of resistance occurred by the action of RNase H or steric hindrance has not been determined.
A recombinant clone containing an engineered gene consisting of a vanH promoter driving expression of a vanA antisense inhibited vancomycin resistance in E. faecalis by a dual mechanism. The phosphorylated VanR, a transcriptional activator, is sequestered by the native vanH promoter present in the recombinant plasmid reducing expression of the vanHAX genes. In addition expression of a vanA RNA antisense placed under the control the recombinant plasmid’s vanH promoter prevents translation of any vanA mRNA that is transcribed by forming a duplex followed by degradation (Torres Viera et al., 2001).
Phosphorothioate deoxyribozymes have been used to target genes coding for mecR1 or blaR1 in methicillin resistant S. aureus and restore susceptibility to methicillin or oxacillin, respectively, after they were delivered inside the cells by electroporation (Hou et al., 2007a; Hou et al., 2007b). Subsequently liposome-encapsulated antisense compounds targeting the mecA gene were delivered into untreated S. aureus. The compounds induced a reduction of the MICs of commonly used antibiotics for methicillin resistant S. aureus clinical isolates and improved the survival rate when administered together with oxacillin to infected mice (Meng et al., 2009).
To inhibit resistance to aminoglycosides mediated by the aac(6′)-Ib gene present in Tn1331 a series of oligodeoxynucleotides targeting mRNA regions identified by RNase H mapping in combination with computer generated secondary structures were synthesized and tested. At least three oligodeoxynucleotides were identified that induced in vitro degradation of mRNA, inhibit in vitro synthesis of the enzyme, and upon delivery by electroporation significantly reduced the number of cells surviving after exposure to amikacin (Sarno et al., 2003). Although it has not been determined, the most probable mechanism of inhibition in vivo is through mRNA degradation by RNase H. However, steric hindrance is a possibility that cannot be discarded at this time. Another approach to reduce or silence expression of aac(6′)-Ib consisted of the design of external guide sequences, short antisense RNA molecules that elicit RNase P-mediated degradation of the mRNA, encoded by recombinant plasmids (Guerrier-Takada et al., 1997). This approach had been already used to reverse resistance to ampicillin and chloramphenicol (Guerrier-Takada et al., 1997). Recombinant clones coding for the selected sequences under an inducible promoter were introduced into E. coli harboring aac(6′)-Ib, and the transformant strains were tested to determine their resistance to amikacin. Two external guide sequences that showed strong binding to the mRNA in vitro induced inhibition of expression of the resistance phenotype in cells harboring the aac(6′)-Ib gene (Soler Bistue et al., 2007). Although these results were an indication that the use of external guide sequences could be a viable strategy to preserve the efficacy of aminoglycosides, as it is the case with other antisense approaches there are several problems that must be addressed. A crucial one is to find nuclease resistant oligonucleotide analogs that still induce inhibition of gene expression, in this case the analog must behave as RNA with respect to eliciting RNase P degradation of the target mRNA while being impervious to RNases. A survey of a variety of oligoribonucleotide analogs including phosphorothioate oligodeoxynucleotides, 2!-O-methyl oligoribonucleotides, phosphorodiamidate morpholino oligomers, or locked nucleic acids (LNA)/DNA co-oligomers showed that selected LNA/DNA co-oligomers elicited RNase P-mediated cleavage of mRNA in vitro. Analyses of isosequential LNA/DNA co-oligomers with different numbers and locations of LNA substitutions suggested that different configurations must be tested to identify an oligomer that promotes high enough levels of RNase P cleavage, it is specific, and it is resistant to the action of nucleases. As a results of these assays a configuration of LNA/DNA residues with the desired properties was found for the particular case of inhibition of expression of aac(6′)-Ib. Administration of 50 nM of an LNA/DNA co-oligomer to the hyperpermeable E. coli AS19 harboring aac(6′)-Ib inhibited growth in the presence of amikacin suggesting that the oligoribonucleotide analog induced RNase P-mediated inhibition of expression of the gene (Soler Bistue et al., 2009).
5. Concluding remarks
Inactivation by enzymatic modification is the most prevalent mechanism of resistance to aminoglycoside antibiotics in the clinical setting. The raise and dissemination of aminoglycoside modifying enzymes has reduced the efficacy of these antibiotics and in some cases rendered them virtually unusable. There are three kinds of aminoglycoside modifying enzymes, nucleotidyltranferases, phosphotransferases, or acetyltransferases, which catalyze the modification at different −OH or −NH2 groups in the antibiotic molecule. The large number and ability of the genes coding for these enzymes to evolve, as well as the numerous mobile elements where they are located, results in a high adaptability by these enzymes to utilize new antibiotics as substrates and to efficiently disseminate among bacteria. As a consequence virtually all bacteria of medical interest can support enzymatic resistance to aminoglycosides. Two nomenclature schemes have been proposed in the past, but the dizzying rate of discovery of new genes together with the appearance of enzymes with new characteristics superseded the criteria defined. We suggest that members of the community should engage in a debate to come up with a consensus new nomenclature. We suggest that returning to a simpler nomenclature with the support of an internet repository site could facilitate the naming of the genes, avoid duplications, and facilitate further changes when new enzymes with new, and may be unexpected, characteristics are discovered. The fight to keep aminoglycosides as useful tools in the armamentarium against bacterial infectious diseases includes the development of new aminoglycosides that must be refractory to as many as possible modifying enzymes, the development of inhibitors of aminoglycoside modifying enzymes, and inhibitors of their expression by the action of antisense oligonucleotide analogs.
Acknowledgements
The authors’ work cited in this review article was funded by Public Health Service grant 2R15AI047115 (to M.E.T.) from the National Institutes of Health. M.S.R. is a research career member of C.O.N.I.C.E.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abhyankar MM, Hochreiter AE, Connell SK, Gilchrist CA, Mann BJ, Petri WA., Jr. Development of the Gateway system for cloning and expressing genes in Entamoeba histolytica. Parasitol Int. 2009;58:95–97. doi: 10.1016/j.parint.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abril C, Brodard I, Perreten V. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob Agents Chemother. 2010;54:3052–3055. doi: 10.1128/AAC.00304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol. 2008;190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian PV, Thomson CJ, Klugman KP, Amyes SG. New gene cassettes for trimethoprim resistance, dfr13, and Streptomycin-spectinomycin resistance, aadA4, inserted on a class 1 integron. Antimicrob Agents Chemother. 2000;44:355–361. doi: 10.1128/aac.44.2.355-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AM, Nakagawa T, Arakawa E, Ramamurthy T, Shinoda S, Shimamoto T. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J Antimicrob Chemother. 2004;53:947–951. doi: 10.1093/jac/dkh221. [DOI] [PubMed] [Google Scholar]
- Ainsa JA, Perez E, Pelicic V, Berthet FX, Gicquel B, Martin C. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol Microbiol. 1997;24:431–441. doi: 10.1046/j.1365-2958.1997.3471717.x. [DOI] [PubMed] [Google Scholar]
- Aires JR, Kohler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajiboye RM, Solberg OD, Lee BM, Raphael E, Debroy C, Riley LW. Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin Infect Dis. 2009;49:365–371. doi: 10.1086/600301. [DOI] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Allmansberger R, Brau B, Piepersberg W. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol Gen Genet. 1985;198:514–520. doi: 10.1007/BF00332949. [DOI] [PubMed] [Google Scholar]
- Azucena E, Mobashery S. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist Updat. 2001;4:106–117. doi: 10.1054/drup.2001.0197. [DOI] [PubMed] [Google Scholar]
- Bakker EP. Aminoglycoside and aminocyclitol antibiotics: hygromycin B is an atypical bactericidal compound that exerts effects on cells of Escherichia coli characteristics for bacteriostatic aminocyclitols. J Gen Microbiol. 1992;138:563–569. doi: 10.1099/00221287-138-3-563. [DOI] [PubMed] [Google Scholar]
- Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Bell JM, Turnidge JD, Andersson P. aac(6′)-Ib-cr genotyping by simultaneous high-resolution melting analyses of an unlabeled probe and full-length amplicon. Antimicrob Agents Chemother. 2010;54:1378–1380. doi: 10.1128/AAC.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousoff MJ, Graham B, Spiccia L, Tor Y. Cleavage of RNA oligonucleotides by aminoglycosides. Org Biomol Chem. 2009;7:30–33. doi: 10.1039/b813252f. [DOI] [PubMed] [Google Scholar]
- Benet LZ, Hoener BA. Changes in plasma protein binding have little clinical relevance. Clin Pharmacol Ther. 2002;71:115–121. doi: 10.1067/mcp.2002.121829. [DOI] [PubMed] [Google Scholar]
- Bernardi F, Bernardi A. Inter- and intramolecular transposition of Tn903. Mol Gen Genet. 1991;227:22–27. doi: 10.1007/BF00260701. [DOI] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Daigle DM, Wright GD. Domain-domain interactions in the aminoglycoside antibiotic resistance enzyme AAC(6′)-APH(2″) Biochemistry. 2004;43:9846–9855. doi: 10.1021/bi049135y. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Draker KA, Koteva K, Bains M, Hancock RE, Wright GD. Broad-spectrum peptide inhibitors of aminoglycoside antibiotic resistance enzymes. Chem Biol. 2003;10:189–196. doi: 10.1016/s1074-5521(03)00026-7. [DOI] [PubMed] [Google Scholar]
- Bozdogan B, Galopin S, Gerbaud G, Courvalin P, Leclercq R. Chromosomal aadD2 encodes an aminoglycoside nucleotidyltransferase in Bacillus clausii. Antimicrob Agents Chemother. 2003;47:1343–1346. doi: 10.1128/AAC.47.4.1343-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brau B, Pilz U, Piepersberg W. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol Gen Genet. 1984;193:179–187. doi: 10.1007/BF00327434. [DOI] [PubMed] [Google Scholar]
- Brossier F, Veziris N, Aubry A, Jarlier V, Sougakoff W. Detection by Genotype MTBDRsl test of complex resistance mechanisms to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010;48:1683–1689. doi: 10.1128/JCM.01947-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnemann SR, Segal JL. Amikacin serum protein binding in spinal cord injury. Life Sci. 1991;49:PL1–5. doi: 10.1016/0024-3205(91)90030-f. [DOI] [PubMed] [Google Scholar]
- Bryan L, van der Elzen H. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother. 1977;12:163–177. doi: 10.1128/aac.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan LE, Kowand SK, Van Den Elzen HM. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob Agents Chemother. 1979;15:7–13. doi: 10.1128/aac.15.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryskier A. Antibiotics and antibacterial agents: classifications and structure-activity relationships. In: Bryskier A, editor. Antimicrobial agents. ASM Press; Washington, DC: 2005. pp. 13–38. [Google Scholar]
- Bucke WE, Leitzke S, Diederichs JE, Borner K, Hahn H, Ehlers S, Muller RH. Surface-modified amikacin-liposomes: organ distribution and interaction with plasma proteins. J Drug Target. 1998;5:99–108. doi: 10.3109/10611869808995863. [DOI] [PubMed] [Google Scholar]
- Bunny KL, Hall RM, Stokes HW. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob Agents Chemother. 1995;39:686–693. doi: 10.1128/AAC.39.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DL, Berghuis AM. Protein kinase inhibitors and antibiotic resistance. Pharmacol Ther. 2002;93:283–292. doi: 10.1016/s0163-7258(02)00197-3. [DOI] [PubMed] [Google Scholar]
- Burk DL, Ghuman N, Wybenga-Groot LE, Berghuis AM. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 2003;12:426–437. doi: 10.1110/ps.0233503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk DL, Hon WC, Leung AK, Berghuis AM. Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase. Biochemistry. 2001;40:8756–8764. doi: 10.1021/bi010504p. [DOI] [PubMed] [Google Scholar]
- Burk DL, Xiong B, Breitbach C, Berghuis AM. Structures of aminoglycoside acetyltransferase AAC(6′)-Ii in a novel crystal form: structural and normal-mode analyses. Acta Crystallogr D Biol Crystallogr. 2005;61:1273–1279. doi: 10.1107/S0907444905021487. [DOI] [PubMed] [Google Scholar]
- Busse HJ, Wostmann C, Bakker EP. The bactericidal action of streptomycin: membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane of Escherichia coli and subsequent caging of the antibiotic inside the cells due to degradation of these proteins. J Gen Microbiol. 1992;138:551–561. doi: 10.1099/00221287-138-3-551. [DOI] [PubMed] [Google Scholar]
- Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother. 2010;54:590–596. doi: 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron FH, Groot Obbink DJ, Ackerman VP, Hall RM. Nucleotide sequence of the AAD(2″) aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986;14:8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casin I, Bordon F, Bertin P, Coutrot A, Podglajen I, Brasseur R, Collatz E. Aminoglycoside 6′-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob Agents Chemother. 1998;42:209–215. doi: 10.1128/aac.42.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casin I, Hanau-Bercot B, Podglajen I, Vahaboglu H, Collatz E. Salmonella enterica serovar Typhimurium bla(PER-1)-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob Agents Chemother. 2003;47:697–703. doi: 10.1128/AAC.47.2.697-703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- Centron D, Roy PH. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Iq from the integron of a natural multiresistance plasmid. Antimicrob Agents Chemother. 1998;42:1506–1508. doi: 10.1128/aac.42.6.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centron D, Roy PH. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda P, Goni P, Millan L, Rubio C, Gomez-Lus R. Detection of the aminoglycosidestreptothricin resistance gene cluster ant(6)-sat4-aph(3 ′)-III in commensal viridans group streptococci. Int Microbiol. 2007;10:57–60. [PubMed] [Google Scholar]
- Chamorro RM, Actis LA, Crosa JH, Tolmasky ME. Dissemination of plasmid-mediated amikacin resistance among pathogenic Klebsiella pneumoniae. Medicina (B Aires) 1990;50:543–547. [PubMed] [Google Scholar]
- Chavideh R, Sholly S, Panaite D, Tolmasky ME. Effects of F171 mutations in the 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] enzyme on susceptibility to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2811–2812. doi: 10.1128/aac.43.11.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts RB, Robinson DA, Kreiswirth BN. Identification of a novel transposon (Tn6072) and a truncated SCCmec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother. 2010 doi: 10.1128/AAC.00001-10. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Qu TT, Yu YS, Zhou JY, Li LJ. Insertion sequence ISEcp1-like element connected with a novel aph(2″) allele [aph(2″)-Ie] conferring high-level gentamicin resistance and a novel streptomycin adenylyltransferase gene in Enterococcus. J Med Microbiol. 2006;55:1521–1525. doi: 10.1099/jmm.0.46702-0. [DOI] [PubMed] [Google Scholar]
- Chen YT, Lauderdale TL, Liao TL, Shiau YR, Shu HY, Wu KM, Yan JJ, Su IJ, Tsai SF. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 β- lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2007;51:3004–3007. doi: 10.1128/AAC.00167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother. 2009;53:1235–1237. doi: 10.1128/AAC.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JW, Kak V, You I, Kao SJ, Petrin J, Clewell DB, Lerner SA, Miller GH, Shaw KJ. Aminoglycoside resistance genes aph(2″)-Ib and aac(6′)-Im detected together in strains of both Escherichia coli and Enterococcus faecium. Antimicrob Agents Chemother. 2001;45:2691–2694. doi: 10.1128/AAC.45.10.2691-2694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JW, Zervos MJ, Lerner SA, Thal LA, Donabedian SM, Jaworski DD, Tsai S, Shaw KJ, Clewell DB. A novel gentamicin resistance gene in Enterococcus. Antimicrob Agents Chemother. 1997;41:511–514. doi: 10.1128/aac.41.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37:1896–1903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne S, Courvalin P, Galimand M. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology. 2010;156:1448–1458. doi: 10.1099/mic.0.033639-0. [DOI] [PubMed] [Google Scholar]
- Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culebras E, Martinez JL. Aminoglycoside resistance mediated by the bifunctional enzyme 6′-N-aminoglycoside acetyltransferase-2″-O-aminoglycoside phosphotransferase. Front Biosci. 1999;4:D1–8. doi: 10.2741/culebras. [DOI] [PubMed] [Google Scholar]
- Dabertrand F, Mironneau J, Henaff M, Macrez N, Morel JL. Comparison between gentamycin and exon skipping treatments to restore ryanodine receptor subtype 2 functions in mdx mouse duodenum myocytes. Eur J Pharmacol. 2010;628:36–41. doi: 10.1016/j.ejphar.2009.11.034. [DOI] [PubMed] [Google Scholar]
- Dahmen S, Bettaieb D, Mansour W, Boujaafar N, Bouallegue O, Arlet G. Characterization and molecular epidemiology of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae in a Tunisian university hospital. Microb Drug Resist. 2010;16:163–170. doi: 10.1089/mdr.2009.0108. [DOI] [PubMed] [Google Scholar]
- Daigle DM, McKay GA, Wright GD. Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J Biol Chem. 1997;272:24755–24758. doi: 10.1074/jbc.272.40.24755. [DOI] [PubMed] [Google Scholar]
- Daly M, Villa L, Pezzella C, Fanning S, Carattoli A. Comparison of multidrug resistance gene regions between two geographically unrelated Salmonella serotypes. J Antimicrob Chemother. 2005;55:558–561. doi: 10.1093/jac/dki015. [DOI] [PubMed] [Google Scholar]
- Davies JE. Aminoglycosides: ancient and modern. J Antibiot (Tokyo) 2006;59:529–532. doi: 10.1038/ja.2006.73. [DOI] [PubMed] [Google Scholar]
- Davis BB. The lethal action of aminoglycosides. J Antimicrob Chemother. 1988;22:1–3. doi: 10.1093/jac/22.1.1. [DOI] [PubMed] [Google Scholar]
- Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD. Non-specific membrane permeability and aminoglycoside action. J Antimicrob Chemother. 1989;24:77–78. doi: 10.1093/jac/24.1.77. [DOI] [PubMed] [Google Scholar]
- Del Campo R, Galan JC, Tenorio C, Ruiz-Garbajosa P, Zarazaga M, Torres C, Baquero F. New aac(6′)-I genes in Enterococcus hirae and Enterococcus durans: effect on β-lactam/aminoglycoside synergy. J Antimicrob Chemother. 2005;55:1053–1055. doi: 10.1093/jac/dki138. [DOI] [PubMed] [Google Scholar]
- Dery KJ, Chavideh R, Waters V, Chamorro R, Tolmasky LS, Tolmasky ME. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid. 1997;38:97–105. doi: 10.1006/plas.1997.1303. [DOI] [PubMed] [Google Scholar]
- Dery KJ, Soballe B, Witherspoon MS, Bui D, Koch R, Sherratt DJ, Tolmasky ME. The aminoglycoside 6′-N-acetyltransferase type Ib encoded by Tn1331 is evenly distributed within the cell’s cytoplasm. Antimicrob Agents Chemother. 2003;47:2897–2902. doi: 10.1128/AAC.47.9.2897-2902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler J, Ebert A, Mansouri K, Pissowotzki K, Stockmann M, Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987;15:8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- Doi Y, Wachino J, Yamane K, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. Spread of novel aminoglycoside resistance gene aac(6′)-Iad among Acinetobacter clinical isolates in Japan. Antimicrob Agents Chemother. 2004;48:2075–2080. doi: 10.1128/AAC.48.6.2075-2080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet B, Weill FX, Fabre L, Chaslus-Dancla E, Cloeckaert A. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar newport. Antimicrob Agents Chemother. 2004;48:3806–3812. doi: 10.1128/AAC.48.10.3806-3812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker KA, Northrop DB, Wright GD. Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6′)-Ii from Enterococcus faecium: evidence of dimer subunit cooperativity. Biochemistry. 2003;42:6565–6574. doi: 10.1021/bi034148h. [DOI] [PubMed] [Google Scholar]
- Dubois V, Arpin C, Coulange L, Andre C, Noury P, Quentin C. TEM-21 extended-spectrum β-lactamase in a clinical isolate of Alcaligenes faecalis from a nursing home. J Antimicrob Chemother. 2006;57:368–369. doi: 10.1093/jac/dki450. [DOI] [PubMed] [Google Scholar]
- Dubois V, Arpin C, Dupart V, Scavelli A, Coulange L, Andre C, Fischer I, Grobost F, Brochet JP, Lagrange I, Dutilh B, Jullin J, Noury P, Larribet G, Quentin C. β-lactam and aminoglycoside resistance rates and mechanisms among Pseudomonas aeruginosa in French general practice (community and private healthcare centres) J Antimicrob Chemother. 2008;62:316–323. doi: 10.1093/jac/dkn174. [DOI] [PubMed] [Google Scholar]
- Dubois V, Poirel L, Marie C, Arpin C, Nordmann P, Quentin C. Molecular characterization of a novel class 1 integron containing bla(GES-1) and a fused product of aac3-Ib/aac6′-Ib’ gene cassettes in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2002;46:638–645. doi: 10.1128/AAC.46.3.638-645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley MN, Loutit J, Griffith DC. Aerosol antibiotics: considerations in pharmacological and clinical evaluation. Curr Opin Biotechnol. 2008;19:637–643. doi: 10.1016/j.copbio.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Egorova S, Kaftyreva L, Grimont PA, Weill FX. Prevalence and characterization of extended-spectrum cephalosporin-resistant nontyphoidal Salmonella isolates in adults in Saint Petersburg, Russia (2002-2005) Microb Drug Resist. 2007;13:102–107. doi: 10.1089/mdr.2007.712. [DOI] [PubMed] [Google Scholar]
- Eliopoulos GM. Synergism and antagonism. Infect Dis Clin North Am. 1989;3:399–406. [PubMed] [Google Scholar]
- Endimiani A, Hujer KM, Hujer AM, Armstrong ES, Choudhary Y, Aggen JB, Bonomo RA. ACHN-490, a neoglycoside with potent in vitro activity against multidrug-resistant Klebsiella pneumoniae isolates. Antimicrob Agents Chemother. 2009;53:4504–4507. doi: 10.1128/AAC.00556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faldynova M, Pravcova M, Sisak F, Havlickova H, Kolackova I, Cizek A, Karpiskova R, Rychlik I. Evolution of antibiotic resistance in Salmonella enterica serovar typhimurium strains isolated in the Czech Republic between 1984 and 2002. Antimicrob Agents Chemother. 2003;47:2002–2005. doi: 10.1128/AAC.47.6.2002-2005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, Gilmore KS, Courvalin P. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2″-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J Bacteriol. 1986;167:631–638. doi: 10.1128/jb.167.2.631-638.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiett J, Baraniak A, Mrowka A, Fleischer M, Drulis-Kawa Z, Naumiuk L, Samet A, Hryniewicz W, Gniadkowski M. Molecular epidemiology of acquired-metallo-β-lactamase-producing bacteria in Poland. Antimicrob Agents Chemother. 2006;50:880–886. doi: 10.1128/AAC.50.3.880-886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong DH, Berghuis AM. Structural basis of APH(3′)-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics. Antimicrob Agents Chemother. 2009;53:3049–3055. doi: 10.1128/AAC.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong DH, Berghuis AM. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 2002;21:2323–2331. doi: 10.1093/emboj/21.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong DH, Lemke CT, Hwang J, Xiong B, Berghuis AM. Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila. J Biol Chem. 2010;285:9545–9555. doi: 10.1074/jbc.M109.038364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K, Clarke AJ. Overexpression and characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob Agents Chemother. 2001;45:2238–2244. doi: 10.1128/AAC.45.8.2238-2244.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother. 2005;49:2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yan X, Baettig OM, Berghuis AM, Auclair K. Regio- and chemoselective 6′-N-derivatization of aminoglycosides: bisubstrate inhibitors as probes to study aminoglycoside 6′-N-acetyltransferases. Angew Chem Int Ed Engl. 2005;44:6859–6862. doi: 10.1002/anie.200501399. [DOI] [PubMed] [Google Scholar]
- Gao F, Yan X, Shakya T, Baettig OM, Ait-Mohand-Brunet S, Berghuis AM, Wright GD, Auclair K. Synthesis and structure-activity relationships of truncated bisubstrate inhibitors of aminoglycoside 6′-N-acetyltransferases. J Med Chem. 2006;49:5273–5281. doi: 10.1021/jm060732n. [DOI] [PubMed] [Google Scholar]
- Gao F, Yan X, Zahr O, Larsen A, Vong K, Auclair K. Synthesis and use of sulfonamide-, sulfoxide-, or sulfone-containing aminoglycoside-CoA bisubstrates as mechanistic probes for aminoglycoside N-6′-acetyltransferase. Bioorg Med Chem Lett. 2008;18:5518–5522. doi: 10.1016/j.bmcl.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynes R, Groisman E, Nelson E, Casadaban M, Lerner SA. Isolation, characterization, and cloning of a plasmid-borne gene encoding a phosphotransferase that confers high-level amikacin resistance in enteric bacilli. Antimicrob Agents Chemother. 1988;32:1379–1384. doi: 10.1128/aac.32.9.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gionechetti F, Zucca P, Gombac F, Monti-Bragadin C, Lagatolla C, Tonin E, Edalucci E, Vitali LA, Dolzani L. Characterization of antimicrobial resistance and class 1 integrons in Enterobacteriaceae isolated from Mediterranean herring gulls (Larus cachinnans) Microb Drug Resist. 2008;14:93–99. doi: 10.1089/mdr.2008.0803. [DOI] [PubMed] [Google Scholar]
- Gomez-Luis R, Vergara Y, Lopez L, Castillo J, Rubio M. 1-N-Aminoglycoside acetyltransferase [AAC(1)] in clinical isolates of Campylobacter spp.. Interscience Conference on Antimicrobial Agents and Chemotherapy; San Francisco, CA. 1999. [Google Scholar]
- Gordon L, Cloeckaert A, Doublet B, Schwarz S, Bouju-Albert A, Ganiere JP, Le Bris H, Le Fleche-Mateos A, Giraud E. Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J Antimicrob Chemother. 2008;62:65–71. doi: 10.1093/jac/dkn166. [DOI] [PubMed] [Google Scholar]
- Gordon RC, Regamey C, Kirby WM. Serum protein binding of the aminoglycoside antibiotics. Antimicrob Agents Chemother. 1972;2:214–216. doi: 10.1128/aac.2.3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KD, Chen W, Houghton JL, Fridman M, Garneau-Tsodikova S. Exploring the substrate promiscuity of drug-modifying enzymes for the chemoenzymatic generation of N-acylated aminoglycosides. Chembiochem. 2010;11:119–126. doi: 10.1002/cbic.200900584. [DOI] [PubMed] [Google Scholar]
- Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Salavati R, Altman S. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc Natl Acad Sci U S A. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie OW. Aminoglycoside induced ototoxicity. Toxicology. 2008;249:91–96. doi: 10.1016/j.tox.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Gutierrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, Perez JL, Oliver A. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother. 2007;51:4329–4335. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad J, Vakulenko SB, Mobashery S. An antibiotic cloaked by its own resistance enzyme. J. Am. Chem. Soc. 1999;121:11922–11923. [Google Scholar]
- Hamano Y, Hoshino Y, Nakamori S, Takagi H. Overexpression and characterization of an aminoglycoside 6′-N-acetyltransferase with broad specificity from an epsilon-poly-L-lysine producer, Streptomyces albulus IFO14147. J Biochem. 2004;136:517–524. doi: 10.1093/jb/mvh146. [DOI] [PubMed] [Google Scholar]
- Han HL, Jang SJ, Park G, Kook JK, Shin JH, Shin SH, Kim DM, Cheon JS, Moon DS, Park YJ. Identification of an atypical integron carrying an IS26-disrupted aadA1 gene cassette in Acinetobacter baumannii. Int J Antimicrob Agents. 2008;32:165–169. doi: 10.1016/j.ijantimicag.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Hanau-Bercot B, Podglajen I, Casin I, Collatz E. An intrinsic control element for translational initiation in class 1 integrons. Mol Microbiol. 2002;44:119–130. doi: 10.1046/j.1365-2958.2002.02843.x. [DOI] [PubMed] [Google Scholar]
- Hancock RE. Aminoglycoside uptake and mode of action--with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J Antimicrob Chemother. 1981;8:249–276. doi: 10.1093/jac/8.4.249. [DOI] [PubMed] [Google Scholar]
- Hannecart-Pokorni E, Depuydt F, de wit L, van Bossuyt E, Content J, Vanhoof R. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Im [corrected] associated with a sulI-type integron. Antimicrob Agents Chemother. 1997;41:314–318. doi: 10.1128/aac.41.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson K, Sundstrom L, Pelletier A, Roy PH. IntI2 integron integrase in Tn7. J Bacteriol. 2002;184:1712–1721. doi: 10.1128/JB.184.6.1712-1721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert CG, Valdes JJ, Bentley WE. Beyond silencing-engineering applications of RNA interference and antisense technology for altering cellular phenotype. Curr Opin Biotechnol. 2008;19:500–505. doi: 10.1016/j.copbio.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Hegde SS, Javid-Majd F, Blanchard JS. Overexpression and mechanistic analysis of chromosomally encoded aminoglycoside 2′-N-acetyltransferase (AAC(2′)-Ic) from Mycobacterium tuberculosis. J Biol Chem. 2001;276:45876–45881. doi: 10.1074/jbc.M108810200. [DOI] [PubMed] [Google Scholar]
- Heinze A, Holzgrabe U. Determination of the extent of protein binding of antibiotics by means of an automated continuous ultrafiltration method. Int J Pharm. 2006;311:108–112. doi: 10.1016/j.ijpharm.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Herbert CJ, Giles IG, Akhtar M. The sequence of an antibiotic resistance gene from an antibiotic-producing bacterium. Homologies with transposon genes. FEBS Lett. 1983;160:67–71. doi: 10.1016/0014-5793(83)80937-5. [DOI] [PubMed] [Google Scholar]
- Hermann T. Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell Mol Life Sci. 2007;64:1841–1852. doi: 10.1007/s00018-007-7034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A, Rodicio MR, Echeita MA, Mendoza MC. Salmonella enterica serotype Typhimurium carrying hybrid virulence-resistance plasmids (pUO-StVR): a new multidrug-resistant group endemic in Spain. Int J Med Microbiol. 2008;298:253–261. doi: 10.1016/j.ijmm.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Heuer H, Krogerrecklenfort E, Wellington EM, Egan S, Elsas JD, Overbeek L, Collard JM, Guillaume G, Karagouni AD, Nikolakopoulou TL, Smalla K. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol Ecol. 2002;42:289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Grass C, Strahilevitz J. High Resolution Melt Curve Analysis for Identification of Single Nucleotide Mutations in the Quinolone Resistance Gene aac(6′)-Ib-cr. Antimicrob Agents Chemother. 2010;54:3509–3511. doi: 10.1128/AAC.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D, Vogne C, El Garch F, Vejux A, Gotoh N, Lee A, Lomovskaya O, Plesiat P. MexXY-OprM efflux pump is necessary for a adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 2003;47:1371–1375. doi: 10.1128/AAC.47.4.1371-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C, Rabbinowitsch E, Sharp S, Croucher NJ, Chieu TB, Mai NT, Diep TS, Chinh NT, Kehoe M, Leigh JA, Ward PN, Dowson CG, Whatmore AM, Chanter N, Iversen P, Gottschalk M, Slater JD, Smith HE, Spratt BG, Xu J, Ye C, Bentley S, Barrell BG, Schultsz C, Maskell DJ, Parkhill J. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 2009;4:e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- Hon WC, McKay GA, Thompson PR, Sweet RM, Yang DS, Wright GD, Berghuis AM. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell. 1997;89:887–895. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- Hoshiko S, Nojiri C, Matsunaga K, Katsumata K, Satoh E, Nagaoka K. Nucleotide sequence of the ribostamycin phosphotransferase gene and of its control region in Streptomyces ribosidificus. Gene. 1988;68:285–296. doi: 10.1016/0378-1119(88)90031-5. [DOI] [PubMed] [Google Scholar]
- Hotta K, Sunada A, Ishikawa J, Mizuno S, Ikeda Y, Kondo S. The novel enzymatic 3″-N-acetylation of arbekacin by an aminoglycoside 3-N- acetyltransferase of Streptomyces origin and the resulting activity. J Antibiot (Tokyo) 1998;51:735–742. doi: 10.7164/antibiotics.51.735. [DOI] [PubMed] [Google Scholar]
- Hou Z, Meng JR, Niu C, Wang HF, Liu J, Hu BQ, Jia M, Luo XX. Restoration of antibiotic susceptibility in methicillin-resistant Staphylococcus aureus by targeting mecR1 with a phosphorothioate deoxyribozyme. Clin Exp Pharmacol Physiol. 2007a;34:1160–1164. doi: 10.1111/j.1440-1681.2007.04705.x. [DOI] [PubMed] [Google Scholar]
- Hou Z, Meng JR, Zhao JR, Hu BQ, Liu J, Yan XJ, Jia M, Luo XX. Inhibition of β-lactamase-mediated oxacillin resistance in Staphylococcus aureus by a deoxyribozyme. Acta Pharmacol Sin. 2007b;28:1775–1782. doi: 10.1111/j.1745-7254.2007.00646.x. [DOI] [PubMed] [Google Scholar]
- Houghton JL, Green KD, Chen W, Garneau-Tsodikova S. The future of aminoglycosides: the end or renaissance? Chembiochem. 2010;11:880–902. doi: 10.1002/cbic.200900779. [DOI] [PubMed] [Google Scholar]
- Huang ZM, Shan H, Mi ZH, Yang HY, Wu L, Chu QJ, Qin L. [Analysis on 168 rRNA methylase genes and aminoglycoside modifying enzymes genes in Enterobacter cloacae in China] Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29:369–373. [PubMed] [Google Scholar]
- Hurwitz C, Braun CB, Rosano CL. Role of ribosome recycling in uptake of dihydrostreptomycin by sensitive and resistant Escherichia coli. Biochim Biophys Acta. 1981;652:168–176. doi: 10.1016/0005-2787(81)90220-3. [DOI] [PubMed] [Google Scholar]
- Ishikawa J, Sunada A, Oyama R, Hotta K. Identification and characterization of the point mutation which affects the transcription level of the chromosomal 3-N-acetyltransferase gene of Streptomyces griseus SS-1198. Antimicrob Agents Chemother. 2000;44:437–440. doi: 10.1128/aac.44.2.437-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA, Blaser MJ, Santanam P, Hachler H, Kayser FH, Hare RS, Miller GH. Appearance of amikacin and tobramycin resistance due to 4′-aminoglycoside nucleotidyltransferase [ANT(4′)-II] in gram-negative pathogens. Antimicrob Agents Chemother. 1990;34:2381–2386. doi: 10.1128/aac.34.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier H, Zaoui C, Sanson-le Pors MJ, Mazel D, Bercot B. Translation regulation of integrons gene cassette expression by the attC sites. Mol Microbiol. 2009;72:1475–1486. doi: 10.1111/j.1365-2958.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- Jana S, Deb JK. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol. 2006;70:140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- Teran F. Javier, Alvarez M, Suarez JE, Mendoza MC. Characterization of two aminoglycoside-(3)-N-acetyltransferase genes and assay as epidemiological probes. J Antimicrob Chemother. 1991;28:333–346. doi: 10.1093/jac/28.3.333. [DOI] [PubMed] [Google Scholar]
- Kao SJ, You I, Clewell DB, Donabedian SM, Zervos MJ, Petrin J, Shaw KJ, Chow JW. Detection of the high-level aminoglycoside resistance gene aph(2″)-Ib in Enterococcus faecium. Antimicrob Agents Chemother. 2000;44:2876–2879. doi: 10.1128/aac.44.10.2876-2879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaster KR, Burgett SG, Rao RN, Ingolia TD. Analysis of a bacterial hygromycin B resistance gene by transcriptional and translational fusions and by DNA sequencing. Nucleic Acids Res. 1983;11:6895–6911. doi: 10.1093/nar/11.19.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Catry B, Haesebrouck F, de Kruif A, Schwarz S. Novel spectinomycin/streptomycin resistance gene, aadA14, from Pasteurella multocida. Antimicrob Agents Chemother. 2005;49:3046–3049. doi: 10.1128/AAC.49.7.3046-3049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R. Translational readthrough induction of pathogenic nonsense mutations. Eur J Med Genet. 2006;49:445–450. doi: 10.1016/j.ejmg.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Kim C, Villegas-Estrada A, Hesek D, Mobashery S. Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6′)-Ib’ from Pseudomonas aeruginosa. Biochemistry. 2007;46:5270–5282. doi: 10.1021/bi700111z. [DOI] [PubMed] [Google Scholar]
- Kingsley JD, Dou H, Morehead J, Rabinow B, Gendelman HE, Destache CJ. Nanotechnology: a focus on nanoparticles as a drug delivery system. J Neuroimmune Pharmacol. 2006;1:340–350. doi: 10.1007/s11481-006-9032-4. [DOI] [PubMed] [Google Scholar]
- Kitao T, Miyoshi-Akiyama T, Kirikae T. AAC(6′)-Iaf, a novel aminoglycoside 6′-N-acetyltransferase from multidrug-resistant Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2009;53:2327–2334. doi: 10.1128/AAC.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao T, Miyoshi-Akiyama T, Shimada K, Tanaka M, Narahara K, Saito N, Kirikae T. Development of an immunochromatographic assay for the rapid detection of AAC(6′)-Iae-producing multidrug-resistant Pseudomonas aeruginosa. J Antimicrob Chemother. 2010;65:1382–1386. doi: 10.1093/jac/dkq148. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Alam M, Nishimoto Y, Urasawa S, Uehara N, Watanabe N. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol Infect. 2001;126:197–204. doi: 10.1017/s0950268801005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl A, Amstutz P, Parizek P, Binz HK, Briand C, Capitani G, Forrer P, Pluckthun A, Grutter MG. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure. 2005;13:1131–1141. doi: 10.1016/j.str.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Lambert T, Gerbaud G, Courvalin P. Characterization of the chromosomal aac(6′)-Ij gene of Acinetobacter sp. 13 and the aac(6′)-Ih plasmid gene of Acinetobacter baumannii. Antimicrob Agents Chemother. 1994a;38:1883–1889. doi: 10.1128/aac.38.9.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert T, Gerbaud G, Galimand M, Courvalin P. Characterization of Acinetobacter haemolyticus aac(6′)-Ig gene encoding an aminoglycoside 6′-N-acetyltransferase which modifies amikacin. Antimicrob Agents Chemother. 1993;37:2093–2100. doi: 10.1128/aac.37.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert T, Ploy MC, Courvalin P. A spontaneous point mutation in the aac(6′)-Ib’ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol Lett. 1994b;115:297–304. doi: 10.1111/j.1574-6968.1994.tb06654.x. [DOI] [PubMed] [Google Scholar]
- Lambert T, Ploy MC, Denis F, Courvalin P. Characterization of the chromosomal aac(6′)-Iz gene of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1999;43:2366–2371. doi: 10.1128/aac.43.10.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc DJ, Lee LN, Inamine JM. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Hopkins JD, Syvanen M. Direct involvement of IS26 in an antibiotic resistance operon. J Bacteriol. 1990;172:3229–3236. doi: 10.1128/jb.172.6.3229-3236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings RS, Partridge SR, Lightfoot D, Hall RM, Djordjevic SP. New integron-associated gene cassette encoding a 3-N-aminoglycoside acetyltransferase. Antimicrob Agents Chemother. 2005;49:1238–1241. doi: 10.1128/AAC.49.3.1238-1241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Haddad J, Azucena E, Kotra LP, Kirzhner M, Mobashery S. Tethered bisubstrate derivatives as probes for mechanism and as inhibitors of aminoglycoside 3′-phosphotransferases. J Org Chem. 2000;65:7422–7431. doi: 10.1021/jo000589k. [DOI] [PubMed] [Google Scholar]
- Llanes C, Neuwirth C, El Garch F, Hocquet D, Plesiat P. Genetic analysis of a multiresistant strain of Pseudomonas aeruginosa producing PER-1 β-lactamase. Clin Microbiol Infect. 2006;12:270–278. doi: 10.1111/j.1469-0691.2005.01333.x. [DOI] [PubMed] [Google Scholar]
- Lombes T, Begis G, Maurice F, Turcaud S, Lecourt T, Dardel F, Micouin L. NMR-guided fragment-based approach for the design of AAC(6′)-Ib ligands. Chembiochem. 2008;9:1368–1371. doi: 10.1002/cbic.200700677. [DOI] [PubMed] [Google Scholar]
- Lopez-Cabrera M, Perez-Gonzalez JA, Heinzel P, Piepersberg W, Jimenez A. Isolation and nucleotide sequencing of an aminocyclitol acetyltransferase gene from Streptomyces rimosus forma paromomycinus. J Bacteriol. 1989;171:321–328. doi: 10.1128/jb.171.1.321-328.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering AM, White LO, Reeves DS. AAC(1): a new aminoglycoside-acetylating enzyme modifying the Cl aminogroup of apramycin. J Antimicrob Chemother. 1987;20:803–813. doi: 10.1093/jac/20.6.803. [DOI] [PubMed] [Google Scholar]
- Lundblad EW, Altman S. Inhibition of gene expression by RNase P. N Biotechnol. 2010;27:212–221. doi: 10.1016/j.nbt.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Lyutzkanova D, Distler J, Altenbuchner J. A spectinomycin resistance determinant from the spectinomycin producer Streptomyces flavopersicus. Microbiology. 1997;143(Pt 7):2135–2143. doi: 10.1099/00221287-143-7-2135. [DOI] [PubMed] [Google Scholar]
- Mabilat C, Lourencao-Vital J, Goussard S, Courvalin P. A new example of physical linkage between Tn1 and Tn21: the antibiotic multiple-resistance region of plasmid pCFF04 encoding extended-spectrum β-lactamase TEM-3. Mol Gen Genet. 1992;235:113–121. doi: 10.1007/BF00286188. [DOI] [PubMed] [Google Scholar]
- MacLeod DL, Nelson LE, Shawar RM, Lin BB, Lockwood LG, Dirk JE, Miller GH, Burns JL, Garber RL. Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J Infect Dis. 2000;181:1180–1184. doi: 10.1086/315312. [DOI] [PubMed] [Google Scholar]
- Magalhaes ML, Vetting MW, Gao F, Freiburger L, Auclair K, Blanchard JS. Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6′-N-acetyltransferase. Biochemistry. 2008;47:579–584. doi: 10.1021/bi701957c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- Magnet S, Courvalin P, Lambert T. Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J Bacteriol. 1999;181:6650–6655. doi: 10.1128/jb.181.21.6650-6655.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Courvalin P, Lambert T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother. 2001;45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S, Smith TA, Zheng R, Nordmann P, Blanchard JS. Aminoglycoside resistance resulting from tight drug binding to an altered aminoglycoside acetyltransferase. Antimicrob Agents Chemother. 2003;47:1577–1583. doi: 10.1128/AAC.47.5.1577-1583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. Mahbub, Kobayashi N, Ishino M, Sumi A, Kobayashi K, Uehara N, Watanabe N. Detection of a novel aph(2″) allele (aph[2″]-Ie) conferring high-level gentamicin resistance and a spectinomycin resistance gene ant(9)-Ia (aad 9) in clinical isolates of enterococci. Microb Drug Resist. 2005;11:239–247. doi: 10.1089/mdr.2005.11.239. [DOI] [PubMed] [Google Scholar]
- Majumder K, Wei L, Annedi S, Kotra LP. Aminoglycoside antibiotics. In: Bonomo RA, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press; Washington, DC: 2007. pp. 7–20. [Google Scholar]
- Martin P, Jullien E, Courvalin P. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol Microbiol. 1988;2:615–625. doi: 10.1111/j.1365-2958.1988.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Salgado C, Lopez-Hernandez FJ, Lopez-Novoa JM. Glomerular nephrotoxicity of aminoglycosides. Toxicol Appl Pharmacol. 2007;223:86–98. doi: 10.1016/j.taap.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Matsuhashi Y, Murakami T, Nojiri C, Toyama H, Anzai H, Nagaoka K. Mechanisms of aminoglycoside-resistance of Streptomyces harboring resistant genes obtained from antibiotic-producers. J Antibiot (Tokyo) 1985;38:279–282. doi: 10.7164/antibiotics.38.279. [DOI] [PubMed] [Google Scholar]
- Maurice F, Broutin I, Podglajen I, Benas P, Collatz E, Dardel F. Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep. 2008;9:344–349. doi: 10.1038/embor.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier P, Cossart P, Giraud E, Gasser F. Completion of the nucleotide sequence of the central region of Tn5 confirms the presence of three resistance genes. Nucleic Acids Res. 1985;13:195–205. doi: 10.1093/nar/13.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay GA, Thompson PR, Wright GD. Broad spectrum aminoglycoside phosphotransferase type III from Enterococcus: overexpression, purification, and substrate specificity. Biochemistry. 1994;33:6936–6944. doi: 10.1021/bi00188a024. [DOI] [PubMed] [Google Scholar]
- McKenzie T, Hoshino T, Tanaka T, Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986;15:93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Mehta R, Champney WS. Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr Microbiol. 2003;47:237–243. doi: 10.1007/s00284-002-3945-9. [DOI] [PubMed] [Google Scholar]
- Mendes R, Toleman M, Ribeiro J, Sader H, Jones R, Walsh T. Integron carrying a novel metallo-b-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistance gene aac(6′)-30/aac(6′)-Ib: report from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. 2004;48:4693–4702. doi: 10.1128/AAC.48.12.4693-4702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes RE, Castanheira M, Toleman MA, Sader HS, Jones RN, Walsh TR. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac(6′)-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob Agents Chemother. 2007;51:2611–2614. doi: 10.1128/AAC.00838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Wang H, Hou Z, Chen T, Fu J, Ma X, He G, Xue X, Jia M, Luo X. Novel anion liposome-encapsulated antisense oligonucleotide restores susceptibility of methicillin-resistant Staphylococcus aureus and rescues mice from lethal sepsis by targeting mecA. Antimicrob Agents Chemother. 2009;53:2871–2878. doi: 10.1128/AAC.01542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies D, Benedetti A, Paydar A, Martin I, Royce S, Pai M, Vernon A, Lienhardt C, Burman W. Effect of duration and intermittency of rifampin on tuberculosis treatment outcomes: a systematic review and meta-analysis. PLoS Med. 2009;6:e1000146. doi: 10.1371/journal.pmed.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael GB, Cardoso M, Schwarz S. Class 1 integron-associated gene cassettes in Salmonella enterica subsp. enterica serovar Agona isolated from pig carcasses in Brazil. J Antimicrob Chemother. 2005;55:776–779. doi: 10.1093/jac/dki081. [DOI] [PubMed] [Google Scholar]
- Mikkelsen NE, Brannvall M, Virtanen A, Kirsebom LA. Inhibition of RNase P RNA cleavage by aminoglycosides. Proc Natl Acad Sci U S A. 1999;96:6155–6160. doi: 10.1073/pnas.96.11.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnier P, Casin I, Bouthors AT, Collatz E. Novel OXA-10-derived extended-spectrum β-lactamases selected in vivo or in vitro. Antimicrob Agents Chemother. 1998a;42:3113–3116. doi: 10.1128/aac.42.12.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnier P, Podglajen I, Goldstein FW, Collatz E. Carbapenems as inhibitors of OXA-13, a novel, integron-encoded β-lactamase in Pseudomonas aeruginosa. Microbiology. 1998b;144(Pt 4):1021–1031. doi: 10.1099/00221287-144-4-1021. [DOI] [PubMed] [Google Scholar]
- Muir ME, van Heeswyck RS, Wallace BJ. Effect of growth rate on streptomycin accumulation by Escherichia coli and Bacillus megaterium. J Gen Microbiol. 1984;130:2015–2022. doi: 10.1099/00221287-130-8-2015. [DOI] [PubMed] [Google Scholar]
- Muller RE, Ano T, Imanaka T, Aiba S. Complete nucleotide sequences of Bacillus plasmids pUB110dB, pRBH1 and its copy mutants. Mol Gen Genet. 1986;202:169–171. doi: 10.1007/BF00330534. [DOI] [PubMed] [Google Scholar]
- Mulvey M, Boyd D, Baker L, Mykytczuk O, Resi E, Asensi M, Rodrigues D, Ng L. Characterization of a Salmonella enterica serovar Agona strain harbouring a class 1 integron containing novel OXA-type ß-lactamase (blaOXA-53) and 6′-N-aminoglycoside acetyltransferase genes [aac(6′)-I30] J Antimicrob Chemother. 2004;54:354–359. doi: 10.1093/jac/dkh347. [DOI] [PubMed] [Google Scholar]
- Murphy E. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3″) (9) Mol Gen Genet. 1985;200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- Nakashima T, Teranishi M, Hibi T, Kobayashi M, Umemura M. Vestibular and cochlear toxicity of aminoglycosides-a review. Acta Otolaryngol. 2000;120:904–911. doi: 10.1080/00016480050218627. [DOI] [PubMed] [Google Scholar]
- Nichols WW. The enigma of streptomycin transport. J Antimicrob Chemother. 1989;23:673–676. doi: 10.1093/jac/23.5.673. [DOI] [PubMed] [Google Scholar]
- Nichols WW, Dorrington SM, Slack MP, Walmsley HL. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother. 1988;32:518–523. doi: 10.1128/aac.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WW, Young SN. Respiration-dependent uptake of dihydrostreptomycin by Escherichia coli. Its irreversible nature and lack of evidence for a uniport process. Biochem J. 1985;228:505–512. doi: 10.1042/bj2280505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, Tolmasky ME, Crosa LM, Crosa JH. Sequencing and expression of the 6′-N-acetyltransferase gene of transposon Tn1331 from Klebsiella pneumoniae. J Bacteriol. 1988;170:3769–3773. doi: 10.1128/jb.170.8.3769-3773.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi N, Sasatsu M, Kono M. Genetic mapping in Bacillus subtilis 168 of the aadK gene which encodes aminoglycoside 6-adenylyltransferase. FEMS Microbiol Lett. 1993;114:47–52. doi: 10.1016/0378-1097(93)90140-w. [DOI] [PubMed] [Google Scholar]
- Novick RP, Clowes RC, Cohen SN, Curtiss R, 3rd, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976;40:168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurizzo D, Shewry SC, Perlin MH, Brown SA, Dholakia JN, Fuchs RL, Deva T, Baker EN, Smith CA. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J Mol Biol. 2003;327:491–506. doi: 10.1016/s0022-2836(03)00121-9. [DOI] [PubMed] [Google Scholar]
- O’Connor M, De Stasio EA, Dahlberg AE. Interaction between 16S ribosomal RNA and ribosomal protein S12: differential effects of paromomycin and streptomycin. Biochimie. 1991;73:1493–1500. doi: 10.1016/0300-9084(91)90183-2. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Jr., Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ohmiya K, Tanaka T, Noguchi N, O’Hara K, Kono M. Nucleotide sequence of the chromosomal gene coding for the aminoglycoside 6-adenylyltransferase from Bacillus subtilis Marburg 168. Gene. 1989;78:377–378. doi: 10.1016/0378-1119(89)90241-2. [DOI] [PubMed] [Google Scholar]
- Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Okazaki A, Avison MB. Aph(3′)-IIc, an aminoglycoside resistance determinant from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2007;51:359–360. doi: 10.1128/AAC.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JF, Silva CA, Barbieri CD, Oliveira GM, Zanetta DM, Burdmann EA. Prevalence and risk factors for aminoglycoside nephrotoxicity in intensive care units. Antimicrob Agents Chemother. 2009;53:2887–2891. doi: 10.1128/AAC.01430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J, Navarro C, Cercenado E, Delgado-Iribarren A, Wilhelmi I, Orden B, Garcia C, Miguelanez S, Perez-Vazquez M, Garcia-Cobos S, Aracil B, Bautista V, Campos J. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J Clin Microbiol. 2006;44:2359–2366. doi: 10.1128/JCM.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Over U, Gur D, Unal S, Miller GH. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin Microbiol Infect. 2001;7:470–478. doi: 10.1046/j.1198-743x.2001.00284.x. [DOI] [PubMed] [Google Scholar]
- Overhage J, Bains M, Brazas MD, Hancock RE. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol. 2008;190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaite DM, Tolmasky ME. Characterization of mutants of the 6′-N-acetyltransferase encoded by the multiresistance transposon Tn1331: effect of Phe171-to-Leu171 and Tyr80-to-Cys80 substitutions. Plasmid. 1998;39:123–133. doi: 10.1006/plas.1997.1330. [DOI] [PubMed] [Google Scholar]
- Pansegrau W, Miele L, Lurz R, Lanka E. Nucleotide sequence of the kanamycin resistance determinant of plasmid RP4: homology to other aminoglycoside 3′-phosphotransferases. Plasmid. 1987;18:193–204. doi: 10.1016/0147-619x(87)90062-x. [DOI] [PubMed] [Google Scholar]
- Parent R, Roy PH. The chloramphenicol acetyltransferase gene of Tn2424: a new breed of cat. J Bacteriol. 1992;174:2891–2897. doi: 10.1128/jb.174.9.2891-2897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeno-Tarraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O’Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Partridge SR, Collis CM, Hall RM. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob Agents Chemother. 2002;46:2400–2408. doi: 10.1128/AAC.46.8.2400-2408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge SR, Recchia GD, Scaramuzzi C, Collis CM, Stokes HW, Hall RM. Definition of the attI1 site of class 1 integrons. Microbiology. 2000;146(Pt 11):2855–2864. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- Pedersen LC, Benning MM, Holden HM. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry. 1995;34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- Perez-Vazquez M, Vindel A, Marcos C, Oteo J, Cuevas O, Trincado P, Bautista V, Grundmann H, Campos J. Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of the aminoglycoside-modifying enzyme gene ant(4′)-Ia and the efflux pump genes msrA/msrB. J Antimicrob Chemother. 2009;63:21–31. doi: 10.1093/jac/dkn430. [DOI] [PubMed] [Google Scholar]
- Perichon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob Agents Chemother. 2008;52:2581–2592. doi: 10.1128/AAC.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin MH, Lerner SA. Localization of an amikacin 3′-phosphotransferase in Escherichia coli. J Bacteriol. 1981;147:320–325. doi: 10.1128/jb.147.2.320-325.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Alphandary H, Andremont A, Couvreur P. Targeted delivery of antibiotics using liposomes and nanoparticles: research and applications. Int J Antimicrob Agents. 2000;13:155–168. doi: 10.1016/s0924-8579(99)00121-1. [DOI] [PubMed] [Google Scholar]
- Poirel L, Cabanne L, Collet L, Nordmann P. Class II transposon-borne structure harboring metallo-β-lactamase gene blaVIM-2 in Pseudomonas putida. Antimicrob Agents Chemother. 2006;50:2889–2891. doi: 10.1128/AAC.00398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Lambert T, Turkoglu S, Ronco E, Gaillard J, Nordmann P. Characterization of Class 1 integrons from Pseudomonas aeruginosa that contain the bla(VIM-2) carbapenem-hydrolyzing b-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob Agents Chemother. 2001;45:546–552. doi: 10.1128/AAC.45.2.546-552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Newmark J, Sorto N, Sherratt DJ, Tolmasky ME. Sublethal concentrations of the aminoglycoside amikacin interfere with cell division without affecting chromosome dynamics. Antimicrob Agents Chemother. 2007;51:252–256. doi: 10.1128/AAC.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourreza A, Whiterspoon M, Fox J, Newmark J, Bui D, Tolmasky ME. Mutagenesis analysis of a conserved region involved in acetyl coenzyme A binding in the aminoglycoside 6′-N-acetyltransferase type Ib encoded by the plasmid pJHCMW1. Antimicrob Agents Chemother. 2005;49:2979–2982. doi: 10.1128/AAC.49.7.2979-2982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MS, Parenteau TR, Centron D, Tolmasky ME. Functional characterization of Tn1331 gene cassettes. J Antimicrob Chemother. 2008;62:669–673. doi: 10.1093/jac/dkn279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MS, Quiroga C, Centron D. Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:5179–5181. doi: 10.1128/AAC.49.12.5179-5181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Garcia S, Otal I, Martin C, Gomez-Lus R, Ainsa JA. Novel streptomycin resistance gene from Mycobacterium fortuitum. Antimicrob Agents Chemother. 2006;50:3920–3922. doi: 10.1128/AAC.00223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb Cell Fact. 2007;6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather PN, Mann PA, Mierzwa R, Hare RS, Miller GH, Shaw KJ. Analysis of the aac(3)-VIa gene encoding a novel 3-N-acetyltransferase. Antimicrob Agents Chemother. 1993a;37:2074–2079. doi: 10.1128/aac.37.10.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather PN, Munayyer H, Mann PA, Hare RS, Miller GH, Shaw KJ. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J Bacteriol. 1992;174:3196–3203. doi: 10.1128/jb.174.10.3196-3203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather PN, Orosz E, Shaw KJ, Hare R, Miller G. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J Bacteriol. 1993b;175:6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla C, Garcillan-Barcia MP, Fernandez-Lopez R, Thomson NR, Sanders M, Cheung M, Thomas CM, de la Cruz F. Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother. 2008;52:1472–1480. doi: 10.1128/AAC.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revuelta J, Vacas T, Torrado M, Corzana F, Gonzalez C, Jiménez-Barbero J, Menendez M, Bastida A, Asensio L. NMR-based analysis of aminoglycoside recognition by the resistance enzyme ANT(4!): the pattern of OH/NH3+ substitution determines the preferred antibiotic binding mode and is critical for drug inactivation. J Am Chem Soc. 2008;130:5086–5103. doi: 10.1021/ja076835s. [DOI] [PubMed] [Google Scholar]
- Riccio ML, Docquier JD, Dell’Amico E, Luzzaro F, Amicosante G, Rossolini GM. Novel 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ic, from a Pseudomonas aeruginosa integron. Antimicrob Agents Chemother. 2003;47:1746–1748. doi: 10.1128/AAC.47.5.1746-1748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LB, Carias LL, Hutton RA, Rudin SD, Endimiani A, Bonomo RA. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2008;52:3427–3429. doi: 10.1128/AAC.00493-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich DP, Anderson MP, Gregory RJ, Cheng SH, Paul S, Jefferson DM, McCann JD, Klinger KW, Smith AE, Welsh MJ. Expression of cystic fibrosis transmembrane conductance regulator corrects defective chloride channel regulation in cystic fibrosis airway epithelial cells. Nature. 1990;347:358–363. doi: 10.1038/347358a0. [DOI] [PubMed] [Google Scholar]
- Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- Rosenberg EY, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch DA, Byrne ME, Kong YC, Skurray RA. The aacA-aphD gentamicin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J Gen Microbiol. 1987;133:3039–3052. doi: 10.1099/00221287-133-11-3039. [DOI] [PubMed] [Google Scholar]
- Rudant E, Bourlioux P, Courvalin P, Lambert T. Characterization of the aac(6′)-Ik gene of Acinetobacter sp. 6. FEMS Microbiol Lett. 1994;124:49–54. doi: 10.1016/0378-1097(94)90331-x. [DOI] [PubMed] [Google Scholar]
- Rudant E, Bouvet P, Courvalin P, Lambert T. Phylogenetic analysis of proteolytic Acinetobacter strains based on the sequence of genes encoding aminoglycoside 6′-N-acetyltransferases. Syst Appl Microbiol. 1999;22:59–67. doi: 10.1016/S0723-2020(99)80028-9. [DOI] [PubMed] [Google Scholar]
- Sabtcheva S, Galimand M, Gerbaud G, Courvalin P, Lambert T. Aminoglycoside resistance gene ant(4′)-IIb of Pseudomonas aeruginosa BM4492, a clinical isolate from Bulgaria. Antimicrob Agents Chemother. 2003;47:1584–1588. doi: 10.1128/AAC.47.5.1584-1588.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salauze D, Perez-Gonzalez JA, Piepersberg W, Davies J. Characterisation of aminoglycoside acetyltransferase-encoding genes of neomycin-producing Micromonospora chalcea and Streptomyces fradiae. Gene. 1991;101:143–148. doi: 10.1016/0378-1119(91)90237-6. [DOI] [PubMed] [Google Scholar]
- Salipante SJ, Hall BG. Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol Biol Evol. 2003;20:653–659. doi: 10.1093/molbev/msg074. [DOI] [PubMed] [Google Scholar]
- Sandvang D. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob Agents Chemother. 1999;43:3036–3038. doi: 10.1128/aac.43.12.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santanam P, Kayser FH. Purification and characterization of an aminoglycoside inactivating enzyme from Staphylococcus epidermidis FK109 that nucleotidylates the 4′- and 4″-hydroxyl groups of the aminoglycoside antibiotics. J Antibiot (Tokyo) 1978;31:343–351. doi: 10.7164/antibiotics.31.343. [DOI] [PubMed] [Google Scholar]
- Sarno R, Ha H, Weinsetel N, Tolmasky ME. Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob Agents Chemother. 2003;47:3296–3304. doi: 10.1128/AAC.47.10.3296-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tolmasky ME. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother. 2002;46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione F, Dugnani S, Demartini G, Arcidiacono MM, Cocuzza CE, Fraschini F. Bactericidal kinetics of an in vitro infection model of once-daily ceftriaxone plus amikacin against gram-positive and gram-negative bacteria. Chemotherapy. 1995;41:239–246. doi: 10.1159/000239351. [DOI] [PubMed] [Google Scholar]
- Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Schwarz FV, Perreten V, Teuber M. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid. 2001;46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- Schwocho LR, Schaffner CP, Miller GH, Hare RS, Shaw KJ. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1790–1796. doi: 10.1128/aac.39.8.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Fujino T, Kobayashi I, Morita K, Kikuchi Y, Kuratsuji T, Kirikae T. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob Agents Chemother. 2005;49:3734–3742. doi: 10.1128/AAC.49.9.3734-3742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya T, Wright GD. Nucleotide selectivity of antibiotic kinases. Antimicrob Agents Chemother. 2010;54:1909–1913. doi: 10.1128/AAC.01570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Cramer CA, Rizzo M, Mierzwa R, Gewain K, Miller GH, Hare RS. Isolation, characterization, and DNA sequence analysis of an AAC(6′)-II gene from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989;33:2052–2062. doi: 10.1128/aac.33.12.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Rather PN, Sabatelli FJ, Mann P, Munayyer H, Mierzwa R, Petrikkos GL, Hare RS, Miller GH, Bennett P, et al. Characterization of the chromosomal aac(6′)-Ic gene from Serratia marcescens. Antimicrob Agents Chemother. 1992;36:1447–1455. doi: 10.1128/aac.36.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmara A, Weinsetel N, Dery KJ, Chavideh R, Tolmasky ME. Systematic analysis of a conserved region of the aminoglycoside 6′-N-acetyltransferase type Ib. Antimicrob Agents Chemother. 2001;45:3287–3292. doi: 10.1128/AAC.45.12.3287-3292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siregar JJ, Lerner SA, Mobashery S. Purification and characterization of aminoglycoside 3′-phosphotransferase type IIa and kinetic comparison with a new mutant enzyme. Antimicrob Agents Chemother. 1994;38:641–647. doi: 10.1128/aac.38.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Toth M, Frase H, Byrnes L, Vakulenko S. Kinetic and structural characterization of the enterococcal aminoglycoside phosphotransferases from the APH(2″) family. 110th General Meeting of the American Society for Microbiology; San Diego, CA. 2010. [Google Scholar]
- Bistue A.J. Soler, Birshan D, Tomaras AP, Dandekar M, Tran T, Newmark J, Bui D, Gupta N, Hernandez K, Sarno R, Zorreguieta A, Actis LA, Tolmasky ME. Klebsiella pneumoniae multiresistance plasmid pMET1: similarity with the Yersinia pestis plasmid pCRY and integrative conjugative elements. PLoS One. 2008;3:e1800. doi: 10.1371/journal.pone.0001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistue A.J. Soler, Ha H, Sarno R, Don M, Zorreguieta A, Tolmasky ME. External guide sequences targeting the aac(6′)-Ib mRNA induce inhibition of amikacin resistance. Antimicrob Agents Chemother. 2007;51:1918–1925. doi: 10.1128/AAC.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bistue A.J. Soler, Martin FA, Vozza N, Ha H, Joaquin JC, Zorreguieta A, Tolmasky ME. Inhibition of aac(6′)-Ib-mediated amikacin resistance by nuclease-resistant external guide sequences in bacteria. Proc Natl Acad Sci U S A. 2009;106:13230–13235. doi: 10.1073/pnas.0906529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotts CR, Stanier RY. Mechanism of streptomycin action on bacteria: a unitary hypothesis. Nature. 1961;192:633–637. doi: 10.1038/192633a0. [DOI] [PubMed] [Google Scholar]
- Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition. Curr Opin Struct Biol. 2004;14:50–57. doi: 10.1016/j.sbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunada A, Nakajima M, Ikeda Y, Kondo S, Hotta K. Enzymatic 1-N-acetylation of paromomycin by an actinomycete strain #8 with multiple aminoglycoside resistance and paromomycin sensitivity. J Antibiot (Tokyo) 1999;52:809–814. doi: 10.7164/antibiotics.52.809. [DOI] [PubMed] [Google Scholar]
- Suter TM, Viswanathan VK, Cianciotto NP. Isolation of a gene encoding a novel spectinomycin phosphotransferase from Legionella pneumophila. Antimicrob Agents Chemother. 1997;41:1385–1388. doi: 10.1128/aac.41.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber HW, Mueller JP, Miller PF, Arrow AS. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauch A, Gotker S, Puhler A, Kalinowski J, Thierbach G. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid. 2002;48:117–129. doi: 10.1016/s0147-619x(02)00120-8. [DOI] [PubMed] [Google Scholar]
- Tauch A, Krieft S, Kalinowski J, Puhler A. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol Gen Genet. 2000;263:1–11. doi: 10.1007/pl00008668. [DOI] [PubMed] [Google Scholar]
- Tennstedt T, Szczepanowski R, Braun S, Puhler A, Schluter A. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol Ecol. 2003;45:239–252. doi: 10.1016/S0168-6496(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Tenover FC, Filpula D, Phillips KL, Plorde JJ. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J Bacteriol. 1988;170:471–473. doi: 10.1128/jb.170.1.471-473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Gilbert T, O’Hara P. Nucleotide sequence of a novel kanamycin resistance gene, aphA-7, from Campylobacter jejuni and comparison to other kanamycin phosphotransferase genes. Plasmid. 1989;22:52–58. doi: 10.1016/0147-619x(89)90035-8. [DOI] [PubMed] [Google Scholar]
- Teran FJ, Suarez JE, Mendoza MC. Cloning, sequencing, and use as a molecular probe of a gene encoding an aminoglycoside 6′-N-acetyltransferase of broad substrate profile. Antimicrob Agents Chemother. 1991;35:714–719. doi: 10.1128/aac.35.4.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CJ, Gray GS. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983;80:5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PR, Hughes DW, Cianciotto NP, Wright GD. Spectinomycin kinase from Legionella pneumophila. Characterization of substrate specificity and identification of catalytically important residues. J Biol Chem. 1998;273:14788–14795. doi: 10.1074/jbc.273.24.14788. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Hughes DW, Wright GD. Regiospecificity of aminoglycoside phosphotransferase from Enterococci and Staphylococci (APH(3′)-IIIa) Biochemistry. 1996;35:8686–8695. doi: 10.1021/bi960389w. [DOI] [PubMed] [Google Scholar]
- Tolmasky ME. Aminoglycoside-modifying enzymes: characteristics, localization, and dissemination. In: Bonomo RA, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press; Washington, DC: 2007a. pp. 35–52. [Google Scholar]
- Tolmasky ME. Bacterial resistance to aminoglycosides and β-lactams: the Tn1331 transposon paradigm. Front Biosci. 2000;5:D20–29. doi: 10.2741/tolmasky. [DOI] [PubMed] [Google Scholar]
- Tolmasky ME. Overview of dissemination mechanisms of genes coding for resistance to antibiotics. In: Bonomo RA, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press; Washington, DC: 2007b. pp. 267–270. [Google Scholar]
- Tolmasky ME. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid. 1990;24:218–226. doi: 10.1016/0147-619x(90)90005-w. [DOI] [PubMed] [Google Scholar]
- Tolmasky ME, Chamorro RM, Crosa JH, Marini PM. Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1988;32:1416–1420. doi: 10.1128/aac.32.9.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Crosa JH. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid. 1993;29:31–40. doi: 10.1006/plas.1993.1004. [DOI] [PubMed] [Google Scholar]
- Tolmasky ME, Crosa JH. Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1987;31:1955–1960. doi: 10.1128/aac.31.12.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Roberts M, Woloj M, Crosa JH. Molecular cloning of amikacin resistance determinants from a Klebsiella pneumoniae plasmid. Antimicrob Agents Chemother. 1986;30:315–320. doi: 10.1128/aac.30.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriya M, Sakakibara M, Matsushita K, Morohoshi T. Nucleotide sequence of aminoglycoside 6′-N-acetyltransferase [AAC(6′)] determinant from Serratia sp. 45. Chem Pharm Bull (Tokyo) 1992;40:2473–2477. doi: 10.1248/cpb.40.2473. [DOI] [PubMed] [Google Scholar]
- Viera C. Torres, Tsiodras S, Gold HS, Coakley EP, Wennersten C, Eliopoulos GM, Moellering RC, Jr., Inouye RT. Restoration of vancomycin susceptibility in Enterococcus faecalis by antiresistance determinant gene transfer. Antimicrob Agents Chemother. 2001;45:973–975. doi: 10.1128/AAC.45.3.973-975.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M, Chow JW, Mobashery S, Vakulenko SB. Source of phosphate in the enzymic reaction as a point of distinction among aminoglycoside 2″- phosphotransferases. J Biol Chem. 2009;284:6690–6696. doi: 10.1074/jbc.M808148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M, Frase H, Antunes NT, Smith CA, Vakulenko SB. Crystal structure and kinetic mechanism of aminoglycoside phosphotransferase-2″-IVa. Protein Sci. 2010a doi: 10.1002/pro.437. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M, Vakulenko S, Smith CA. Purification, crystallization and preliminary X-ray analysis of Enterococcus casseliflavus aminoglycoside-2″- phosphotransferase-IVa. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010b;66:81–84. doi: 10.1107/S1744309109050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nhieu G. Tran, Collatz E. Primary structure of an aminoglycoside 6′-N-acetyltransferase AAC(6′)-4, fused in vivo with the signal peptide of the Tn3-encoded β-lactamase. J Bacteriol. 1987;169:5708–5714. doi: 10.1128/jb.169.12.5708-5714.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5″-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Trower MK, Clark KG. PCR cloning of a streptomycin phosphotransferase (aphE) gene from Streptomyces griseus ATCC 12475. Nucleic Acids Res. 1990;18:4615. doi: 10.1093/nar/18.15.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SF, Zervos MJ, Clewell DB, Donabedian SM, Sahm DF, Chow JW. A new high-level gentamicin resistance gene, aph(2″)-Id, in Enterococcus spp. Antimicrob Agents Chemother. 1998;42:1229–1232. doi: 10.1128/aac.42.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 2003;16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Klundert JA, Vliegenthart JS. Nomenclature of aminoglycoside resistance genes: a comment. Antimicrob Agents Chemother. 1993;37:927–928. doi: 10.1128/aac.37.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoof R, Hannecart-Pokorni E, Content J. Nomenclature of genes encoding aminoglycoside-modifying enzymes. Antimicrob Agents Chemother. 1998;42:483. doi: 10.1128/aac.42.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoof R, Sonck P, Hannecart-Pokorni E. The role of lipopolysaccharide anionic binding sites in aminoglycoside uptake in Stenotrophomonas (Xanthomonas) maltophilia. J Antimicrob Chemother. 1995;35:167–171. doi: 10.1093/jac/35.1.167. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Lee H, Hu R, Neyfakh AA. Molecular sieve mechanism of selective release of cytoplasmic proteins by osmotically shocked Escherichia coli. J Bacteriol. 2001;183:2399–2404. doi: 10.1128/JB.183.8.2399-2404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting MW, Hegde SS, Javid-Majd F, Blanchard JS, Roderick SL. Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat Struct Biol. 2002;9:653–658. doi: 10.1038/nsb830. [DOI] [PubMed] [Google Scholar]
- Vetting MW, LP S.d.C., Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem Biol. 2004;11:565–573. doi: 10.1016/j.chembiol.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Vetting MW, Park CH, Hegde SS, Jacoby GA, Hooper DC, Blanchard JS. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6′)-Ib and its bifunctional, fluoroquinolone-active AAC(6′)-Ib-cr variant. Biochemistry. 2008;47:9825–9835. doi: 10.1021/bi800664x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyssier P, Bryskier A. Aminocyclitol aminoglycosides. In: Bryskier A, editor. Antimicrobial agents. ASM Press; Washington, DC: 2005. pp. 453–469. [Google Scholar]
- Vicens Q, Westhof E. Molecular recognition of aminoglycoside antibiotics by ribosomal RNA and resistance enzymes: an analysis of x-ray crystal structures. Biopolymers. 2003;70:42–57. doi: 10.1002/bip.10414. [DOI] [PubMed] [Google Scholar]
- Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother. 2009;53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vliegenthart JS, Ketelaar-van Gaalen PA, Eelhart J, van de Klundert JA. Localisation of the aminoglycoside-(3)-N-acetyltransferase isoenzyme II in Escherichia coli. FEMS Microbiol Lett. 1991a;65:101–105. doi: 10.1016/0378-1097(91)90479-t. [DOI] [PubMed] [Google Scholar]
- Vliegenthart JS, Ketelaar-van Gaalen PA, van de Klundert JA. Nucleotide sequence of the aacC3 gene, a gentamicin resistance determinant encoding aminoglycoside-(3)-N-acetyltransferase III expressed in Pseudomonas aeruginosa but not in Escherichia coli. Antimicrob Agents Chemother. 1991b;35:892–897. doi: 10.1128/aac.35.5.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtli M, Hutter R. Characterisation of the hydroxystreptomycin phosphotransferase gene (sph) of Streptomyces glaucescens: nucleotide sequence and promoter analysis. Mol Gen Genet. 1987;208:195–203. doi: 10.1007/BF00330442. [DOI] [PubMed] [Google Scholar]
- Wei Q, Jiang X, Yang Z, Chen N, Chen X, Li G, Lu Y. dfrA27, a new integron-associated trimethoprim resistance gene from Escherichia coli. J Antimicrob Chemother. 2009;63:405–406. doi: 10.1093/jac/dkn474. [DOI] [PubMed] [Google Scholar]
- Welch KT, Virga KG, Whittemore NA, Ozen C, Wright E, Brown CL, Lee RE, Serpersu EH. Discovery of non-carbohydrate inhibitors of aminoglycoside-modifying enzymes. Bioorg Med Chem. 2005;13:6252–6263. doi: 10.1016/j.bmc.2005.06.059. [DOI] [PubMed] [Google Scholar]
- Wenk M, Spring P, Vozeh S, Follath F. Multicompartment pharmacokinetics of netilmicin. Eur J Clin Pharmacol. 1979;16:331–334. doi: 10.1007/BF00605631. [DOI] [PubMed] [Google Scholar]
- Werner G, Hildebrandt B, Witte W. Linkage of erm(B) and aadE-sat4-aphA-3 in multiple-resistant Enterococcus faecium isolates of different ecological origins. Microb Drug Resist. 2003;9(Suppl 1):S9–16. doi: 10.1089/107662903322541847. [DOI] [PubMed] [Google Scholar]
- White DG, Maneewannakul K, von Hofe E, Zillman M, Eisenberg W, Field AK, Levy SB. Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob Agents Chemother. 1997;41:2699–2704. doi: 10.1128/aac.41.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Northrop DB. Synthesis of a tight-binding, multisubstrate analog inhibitor of gentamicin acetyltransferase I. J Antibiot (Tokyo) 1979;32:1147–1154. doi: 10.7164/antibiotics.32.1147. [DOI] [PubMed] [Google Scholar]
- Wilson NL, Hall RM. Unusual class 1 integron configuration found in Salmonella genomic island 2 from Salmonella enterica serovar Emek. Antimicrob Agents Chemother. 2010;54:513–516. doi: 10.1128/AAC.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor GL, Lo R, Sui SJ, Ung KS, Huang S, Cheng D, Ching WK, Hancock RE, Brinkman FS. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlleben W, Arnold W, Bissonnette L, Pelletier A, Tanguay A, Roy PH, Gamboa GC, Barry GF, Aubert E, Davies J, et al. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet. 1989;217:202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]
- Wolf E, Vassilev A, Makino Y, Sali A, Nakatani Y, Burley SK. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94:439–449. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- Woloj M, Tolmasky ME, Roberts MC, Crosa JH. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob Agents Chemother. 1986;29:315–319. doi: 10.1128/aac.29.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N, Wareham DW. Tackling antibiotic resistance: a dose of common antisense? J Antimicrob Chemother. 2009;63:225–229. doi: 10.1093/jac/dkn467. [DOI] [PubMed] [Google Scholar]
- Wright G, Berghuis A. Structural aspects of aminoglycoside-modifying enzymes. In: Bonomo RA, Tolmasky ME, editors. Enzyme-mediated resistance to antibiotics: mechanisms, dissemination, and prospects for inhibition. ASM Press; Washington, DC: 2007. pp. 21–33. [Google Scholar]
- Wright GD, Thompson PR. Aminoglycoside phosphotransferases: proteins, structure, and mechanism. Front Biosci. 1999;4:D9–21. doi: 10.2741/wright. [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot LE, Draker K, Wright GD, Berghuis AM. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure. 1999;7:497–507. doi: 10.1016/s0969-2126(99)80066-5. [DOI] [PubMed] [Google Scholar]
- Yan JJ, Hsueh PR, Lu JJ, Chang FY, Ko WC, Wu JJ. Characterization of acquired β-lactamases and their genetic support in multidrug-resistant Pseudomonas aeruginosa isolates in Taiwan: the prevalence of unusual integrons. J Antimicrob Chemother. 2006;58:530–536. doi: 10.1093/jac/dkl266. [DOI] [PubMed] [Google Scholar]
- Yao J, Moellering R. Antibacterial agents. In: Murray P, Baron E, Jorgensen J, Landry M, Pfaller M, editors. Manual of Clinical Microbiology. American Society for Microbiology Press; Washington, DC: 2007. pp. 1077–1113. [Google Scholar]
- Young PG, Walanj R, Lakshmi V, Byrnes LJ, Metcalf P, Baker EN, Vakulenko SB, Smith CA. The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2″-phosphotransferase-IIa [APH(2″)-IIa] provide insights into substrate selectivity in the APH(2″) subfamily. J Bacteriol. 2009;191:4133–4143. doi: 10.1128/JB.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalacain M, Gonzalez A, Guerrero MC, Mattaliano RJ, Malpartida F, Jimenez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986;14:1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fisher JF, Mobashery S. The bifunctional enzymes of antibiotic resistance. Curr Opin Microbiol. 2009;12:505–511. doi: 10.1016/j.mib.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Zingman LV, Park S, Olson TM, Alekseev AE, Terzic A. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin Pharmacol Ther. 2007;81:99–103. doi: 10.1038/sj.clpt.6100012. [DOI] [PubMed] [Google Scholar]
- Zong Z, Partridge SR, Iredell JR. A blaVEB-1 variant, blaVEB-6, associated with repeated elements in a complex genetic structure. Antimicrob Agents Chemother. 2009;53:1693–1697. doi: 10.1128/AAC.01313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]