Abstract
During inflammatory bowel disease, TNFα is the major pro-inflammatory cytokine mainly secreted from macrophages and dendritic cells. Here, we have demonstrated that TNFα siRNA/polyethyleneimine loaded into polylactide at an optimal concentration of 20 g/L nanoparticles covered with polyvinyl alcohol are efficiently taken up by inflamed macrophages and inhibit TNFα secretion by the macrophages. Those nanoparticles have a diameter of ~380 nm and zeta potential of −8 mV at pH 7.2, and are non-cytotoxic. Complexation, interactions and protection from RNAse between TNFα siRNA and polyethyleneimine were higher than those using chitosan. Importantly, complexation between TNFα siRNA and polyethyleneimine facilitated higher rates of siRNA loading into nanoparticles, compared to Chi or free siRNA mixed with Lipofectamine. Oral administration of encapsulated TNFα-loaded nanoparticles specifically reduced the TNFα expression/secretion in colonic tissue in LPS-treated mice. In conclusion, we have shown: (1) that proposed siRNA TNFα-loaded NPs are prepared via a non-denaturing synthetic process; (2) a high encapsulation rate of TNFα siRNA complexed to polyethyleneimine into NPs; (3) effective enzymatic protection of TNFα siRNA by polyethyleneimine; (4) non-cytotoxicity and biodegradability of nanoparticles loaded with polyethyleneimine/TNFα siRNA; and (5) in vitro and in vivo significant anti-inflammatory effects at low TNFα siRNA dose that is specific and restricted to the colonic cells. Our results collectively indicate that polyethyleneimine/TNFα siRNA nanocomplexes represent an efficient therapeutic option for diseases such as IBD.
Keywords: Poly (lactic acid) nanoparticle, macrophages, transfection, polyethyleimine complex, small interfering RNA, mice colon
Introduction
Inflammatory bowel disease (IBD), which includes ulcerative colitis and Crohn’s disease, is a chronic debilitating inflammatory condition for which existing effective and targeted treatments are largely limited due to significant systemic side effects. Until a decade ago, the treatment options for IBD limited to anti-inflammatory medications (such as 5-aminosalicylic acid, steroids) or immunosuppressants. Despite the efficacy of these drugs, their use is restricted due to non-specific effects on the immune system, which result in short- and long-term debilitating side effects. Furthermore, anti-inflammatory drugs that are locally active with minimal systemic absorption (5-aminosalicylates) require frequent intake at high doses to exert measurable clinical activity. Sustained drug-release devices, such as pellets, capsules or tablets designed to deliver drugs specifically to the colon for longer periods, have been developed. However, these drugs have limited therapeutic efficacy, and are effective in only a subset of IBD patients.
Tumor necrosis factor alpha (TNFα) plays a central role in the pathogenesis of IBD, as evidenced by the successful treatment of patients treated with anti-TNFα antibodies in multiple clinical trials. [1–3] Although these antibodies are highly effective in general, nearly 25% of patients administered the monoclonal antibody infliximab experienced at least one serious adverse event, including pneumonia, cancer, and acute inflammation [4] largely due to systemic TNFα suppression. Macrophages (MPs) and dendritic cells are the two main cell lines that secrete TNFα in the colon. Thus, it seems reasonable to hypothesize that direct and local inhibition of TNFα secretion from MPs/dendritic cells would be highly effective not only to prevent intestinal inflammation but also to reduce systemic side-effects.
Small-interfering RNA (siRNA)-mediated knockdown of pro-inflammatory cytokines at the messenger RNA (mRNA) level (termed RNA interference) [5] is an attractive therapeutic strategy to overcome inflammatory conditions. One of the major obstacles in siRNA therapy is low penetration of naked siRNA across cell membranes. [6] Several delivery systems have been investigated to overcome this problem. Recent efforts in developing tissue-targetable nucleic acid delivery systems based on synthetic reagents have yielded promising results. [4, 7–13] Among the drug carriers, nanoparticles (NPs) (biodegradable or otherwise) have shown significant potential in binding and delivering siRNA. [14] Indeed, NPs protect siRNA against degradation in vitro and in vivo, and markedly increase their pharmacological activity under both cell culture and physiological conditions. [4] In the context of IBD, a non-cytotoxic system seems the most relevant. A biodegradable polymeric envelope provides protection and transport of siRNA into the cytosol, and facilitates efficiency of siRNA activity in vivo. [15] Here, we investigated the use of biodegradable, non-cytotoxic NPs for targeting of TNFα siRNA, with a view to inhibiting TNFα secretion from MPs, the main source of the cytokine during intestinal inflammation. Finally we will investigate the specificity of oral administration of encapsulated TNFα-loaded NPs to the colonic tissue of lipopolysaccharides (LPS)- treated mice.
Experimental Section
Materials
Branched PEI (Mn=1800g/mol, Mw=2000g/mol), PLA (Mw=75–120 kg/mol), Chi (high molecular weight, viscosity 800,000 cps and >75% deacethylation) and lipopolysaccharides from Salmonella enteric serotype typhimurium were purchased from Aldrich Chemistry, St Louis, MO, USA. Alexa Fluor 568 phalloidin (Mw=1590), 4′,6-diamidino-2-phenyl-indole dihydrochloride (DAPI) and fluorescently tagged siRNA (Block-it fluorescent control) were obtained from Invitrogen, Eugene, OR, USA. The cell proliferation reagent, WST-1, was purchased from Roche Diagnostics (Indianapolis, IN, USA) and LDH from Clonetech Laboratories (Mountain View, CA, USA). PVA (86–89% hydrolyzed, low molecular weight) was purchased from Alfa Aesar (Ward Hill, MA, USA). Silencer negative control and Tnf Silencer pre-designed siRNA were acquired from Ambion, Austin, TX, USA. The mouse TNF-α Elisa kit was from eBioscience, San Diego, CA, USA.
Preparation of TNFα siRNA/PEI or Chi loaded NPs covered with PVA
NPs were synthesized via double emulsion/solvent evaporation, as described previously. [16] Briefly, an internal phase (see details below) containing the drug was mixed with 5, 10, 15 or 20 g/L of PLA in dichloromethane to generate a water-in-oil (W/O) emulsion after 2 min of vortexing (Maxi Mix II, Thermodyne, Dubuque, Iowa, USA) and 1 min of sonication with 50% active cycles at 70% power (Pmax=400 W) (Digital Sonifier 450, Branson, Danbury, CT, USA). This first emulsion was dropped in a second water phase containing 0.3g/L of PVA to generate a water/oil/water emulsion (W/O/W).
The W/O/W emulsion was dropped in a dispersing phase of 0.1g/L PVA, and stirred at 45ºC under a vacuum to remove dichloromethane. As each synthesis made around 50 mg of dry NPs, each group of NPs is the accumulation of 3 independent syntheses. NPs were then centrifuged at 9953g and freeze-dried overnight at −50ºC under 0.1 mbar pressure. As the second emulsion allowed PVA to be grafted on the surface by hydrophobic interaction with the PLA matrix, NPs are coated with PVA that prevent from aggregations through electrostatic repulsions. To determine the physicochemical characteristics of NPs, PVA absorbed on NPs were measured. Doses of absorbed PVA in NPs and total PVA introduced during the NP synthesis process were employed, as specified by Zambaux et al. [17] Briefly, initial PVA (total PVA) was assayed with boric acid and iodine solution and 20 mg of nanoparticles (absorbed PVA) hydrolyzed overnight in 5 ml of 1N NaOH. The solution was neutralized with 1N HCl. We added 3.7% boric acid solution (w/v) and iodine solution (1.66% potassium iodide and 1.27% iodine in distilled water) (w/v) to each sample. Absorbance was measured at 620 nm against buffer alone. A solution containing known amounts of PVA was used as the reference.
Preparation of the internal phase
The internal phase has a typical N/P ratio of the number of negative charges of siRNA (TNFα siRNA or FITC-TNFα siRNA) (P as the phosphorous charge) and positive charges of Chi or PEI (N as the ammonium charge). We employed N/P ratios of 30 for PEI and 100 for chitosan. A mixture of siRNA/PEI: 29 μL TNFα siRNA (5 μM) was combined with 18 μL PEI (5mM), and incubated for 10 min at room temperature for complexation. Similarly, a mixture of siRNA/Chi: 60 μL TNFα siRNA (5 μM) was combined with 13 μL Chi (4 mg/mL), and incubated for 10 min at room temperature for complexation. After 10 min, a polyplex was formed, and 750 μL bovine serum albumin (BSA, 50g/L) added, generating the first emulsion with dichloromethane.
TNFα siRNA/PEI and siRNA/Chi polyplex formation
Formation of each polyplex was assessed via examination of electrophoretic mobility on a 4% (w/v) agarose gel for 45 min at 100V in 0.5X TBE buffer (40 mM Tris/HCl, 445mM boric acid, 1mM ETDA). Ethidium bromide was used to stain siRNA.
TNFα siRNA/PEI and siRNA/Chi polyplex protection from RNAse A
Each polyplex was incubated with 4 μL of RNAse A (0.04 mg/mL) at 37ºC for 2 h and 4 h. Polyplex and siRNA integrity were assessed via electrophoretic mobility analysis. To evaluate siRNA integrity after polyplex exposure to RNAse A, we added 4μL of concentrated NaOH solution.
Cytotoxicity tests
WST-1
To assess the potential toxicity of TNFα siRNA/PEI or TNFα siRNA/Chi-loaded NPs, a WST-1 assay was performed with long-time exposure (3 days). As described previously, [18] MPs were seeded in 96-well plates at a density of 5×104 cells per well and exposed to 1 mg/mL of loaded NPs with or without TNFα siRNA/PEI or TNFα siRNA/Chi-loaded NPs for 72 hours. The WST-1 assay measures cleavage of the soluble red tetrazolium salt, WST-1 (4-[3-(4- iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate), by dehydrogenase present in intact mitochondria, which leads to the formation of dark red formazan crystals. WST-1 proliferation reagent (10 μL) was added to MPs (10 μL/well) and incubated for 1–2 hours at 37°C. The wavelength for measuring absorbance of the formazan product was 440 nm.
LDH
The Lactate dehydrogenase (LDH) test is based on LDH secretion in the medium culture as a marker of dead or damaged cells. In total, 10,000 MPs were seeded and exposed to 1 mg/mL of loaded NPs with or without TNFα siRNA/PEI or TNFα siRNA/Chi-loaded NPs or Lipofectamine for 24 h. An aliquot (100 μL) of culture medium was sampled and added to 100 μL of the LDH reagent. The wavelength for measuring absorbance of the formazan product was 492 nm.
Intracellular NP uptake visualization
Confocal microscopy was employed to visualize the intracellular uptake of NPs (Zeiss LSM510 Meta confocal). Cells (104) were seeded in four-chamber tissue culture glass slides (BD Falcon, Bedford, MA, USA). FITC-siRNA/PEI or FITC-siRNA/Chi-loaded NPs covered with PVA (800 μg/mL, NPs were loaded with 7.3 ng FITC-siRNA/PEI or FITC-siRNA/Chi) or FITC-TNFα siRNA (7.3 ng) in the presence of Lipofectamine (concentration) were added to medium at pH 7.2. For siRNA uptake, MPs were exposed to load NPs for 30 min, 1 h or 2 h. Next, MPs were washed 3 times with medium and 3 times with PBS to remove extracellular NPs, and subsequently fixed for 20 min in 4% paraformaldehyde. Alexa Fluor 568 phalloidin and DAPI were successively added, and diluted 60 and 10,000 times for staining cells for 45 min and 5 min, respectively. To obtain comparable data, all photomultipliers used were adjusted to the same sensitivity (gain/offset).
Intracellular NP uptake quantification
For intracellular uptake quantification, MPs (104) were seeded in 24-well plates in a similar manner to that for visualization. The concentration of loaded NPs or FITC siRNA in the presence of Lipofectamine and exposure time used were analogous to those for intracellular NP uptake visualization. Cell lysis buffer (RIPA buffer [150 mM NaCl, 0.5% sodium deoxycholate, 50 mM Tris HCl (pH 8.0), 0.1% SDS, 0.1% Nonidet P-40] supplemented with protease inhibitors (Roche Diagnostics, Mannheim, Germany)) (200 μL) was added to each well for 1 hour to release total tagged siRNA from NPs taken up by cells. Fluorescence (λex=494 nm, λem=519 nm) was measured using a SpectraMax M2 (Molecular Devices, Sunnyvale, CA).
NP release
For determination of total loading (TNFα siRNA or FITC-tagged scrambled siRNA), 90 mg of NP was treated with 4 mL of acetic acid (pH=2) for 2 h at 25°C. The solution pH was increased to 6 with concentrated NaOH and diluted to 6 mL with 20 mM acetate buffer, pH 6. The release kinetic study was performed by sampling 100 μL of 10 mg/mL NP suspension in PBS buffer at pH=7.4 corresponding to the intracellular pH of MPs at all time-points.
NPs Size and potential zeta measurements
Diameters (nm) and potential zeta (mV) of NPs were measured by light scattering using 90 Plus/BI-MAS (Multi angle particle sizing) or light scattering after applying an electric field using a ZetaPlus (Zeta potential analyzer, Brookhaven Instruments Corporation, Holtsville, NY, USA). The average and standard deviations of the diameters (nm) or zeta potential (mV) were calculated using 3 runs. Each run is an average of 10 measurements.
Calculation of Association constants
Association and dissociation constants of interactions between TNFα siRNA and PEI were calculated using the BI-2000 Biosensing Instrument (Tempe, AZ, USA), based on the surface plasmon resonance (SPR) theory. KD (parameter expressed in mol. L−1), representing 50% of siRNA adsorption on PEI covered gold chip, is commonly used to describe the affinity between two molecules that is how tightly a ligand binds to a particular protein. Ligand-protein affinities are influenced by non-covalent intermolecular interactions between the two molecules such as hydrogen bonding, electrostatic interactions, hydrophobic and Van Der Waals forces. Briefly, after coating the chip with PEI, increasing concentrations of siRNA solutions were passed on. As a result, a two-step interaction curve was obtained. The first step involved adsorption of siRNA on PEI until maximum siRNA adsorption was represented. In the second step, when the passing siRNA concentration reached null concentration, the running buffer released “unspecific” adsorption. The kinetics of the adsorption curve decreased to a plateau at an intermediate level above the initial baseline. The amplitude of siRNA binding to PEI represents the difference between the initial and final levels. KD is then determined by analyzing binding curves at different concentration of siRNA.
Scattering and transmission electron microscopy of NPs
Suspensions of NPs on plots were fixed overnight with glutaraldehyde containing 2.5% cacodylate buffer (0.1mol/L, pH 7) at 4°C. Subsequently, samples were rinsed and dehydrated via 2-minute rinses with a series of increasing alcohol concentrations (30%, 50%, 70%, 80%, 90%, 100% vol/vol), incubated for 5 minutes with hexamethyldisilazane, and dried under a range hood. Plots were fixed on an aluminum support with carbon-adhesive glue and coated with gold palladium (Spotter Coater SC7640). Samples were observed under a scattering or transmission electron microscope (Cambridge Instruments Stereoscan S).
In vitro anti-inflammation effect of TNFα siRNA-loaded NPs
For measurement of TNFα secretion, MPs (104 cells) were seeded in 24-well plates. Empty NPs or loaded with TNFα siRNA or loaded with a scrambled siRNA were added for 24 h. As a control, MPs transfected with TNFα siRNA using Lipofectamine were used at the same concentration of siRNA loaded in NPs. After 24 hours, cells were stimulated with LPS (10 μg/mL) for 1 h. TNFα secretion in the medium was measured with an eBioscience kit (mouse TNFα ELISA kit, eBioscience, San Diego, CA, USA).
In vivo anti-inflammation effect of encapsulated TNFα siRNA-loaded NPs
To deliver the TNFα-loaded NPs to C57BL/6 mice colonic lumen, we encapsulated them into a biomaterial comprised of alginate and chitosan at a ratio of 7/3 (wt/wt). We have previously shown that the biomaterial collapses in intestinal solution at pH 5 or 6, which is the colonic pH under inflamed and non-inflamed states. C57BL/6 mice (8 per group) were gavaged daily for 4 days with encapsulated TNFα siRNA-loaded, scrambled siRNA or empty NPs. After 4 days of gavage, mice were treated with an intra-peritoneal injection of LPS (100μg LPS/kg mice). After one hour, colon tissue, liver tissue and blood were collected. Colon and liver pieces were cultured as described by Siegmund B. at al. [19] In brief, 1-sq cm of liver and 1 cm segments of proximal colons were washed in HBSS supplemented with penicillin and streptomycin (Cellgro). These segments were cultured in 24-well flat-bottom culture plates (Costar) in serum-free RPMI 1640 medium (Cellgro) supplemented with penicillin and streptomycin, L-glutamine, and nonessential amino acids. After 24 hours, supernatant was collected and centrifuged at 13,000 g for 10 minutes at 4°C and stored at –20°C until analyzed. Samples were processed as described in the in vitro part by an ELISA protocol.
Statistical analysis
Data are presented as average values and standard deviations from experiments performed in triplicate (n=3), except for cytotoxicity tests and in vivo experiments (n=8). ANOVA tests were performed to obtain statistical comparisons between samples.
Results and Discussion
Complexation/protection of TNFα siRNA by PEI
Polyethyleneimine (PEI) or chitosan (Chi) was used for condensing negatively charged TNFα siRNA (Figure 1A). Electrostatic interactions are formed between the positive charges of PEI (or Chi) and negative charges of siRNA. For Chi, positive charges are induced via overnight stirring of Chi solution (4 mg/mL) in acetic acid (0.6% v/v). N/P represents the ratio between the number of negative charges of siRNA (P is the negative phosphorous charge) and positive charges of PEI or Chi (N is the positive ammonium charge). The N/P ratio was set at 30 for PEI and 100 for Chi. Due to the small size and high quantity of available nitrogen, the PEI ratio is lower as a result of higher charge density and higher efficiency of complexation at the same N/P.
Figure 1. Complexation and protection of TNFα siRNA by polyethylenimine (PEI).
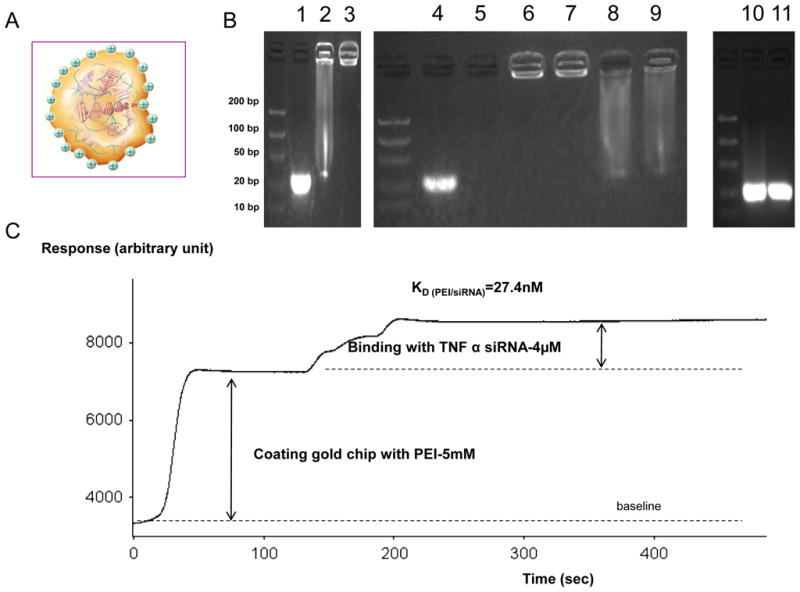
TNFα siRNA was complexed and protected from RNAse A digestion using PEI or chitosan (Chi) as electrostatic binding molecule. A-Schematic of a polyplex of TNFα siRNA negatively charged (red) and PEI positively charged (green) B- Electrophoretic migration (100V) of TNFα siRNA in a 4% agarose gel containing 0.4μg/mL (w/v) free TNFα siRNA (1) and complex formed with Chi (2) or PEI (3). Protection study was performed by electrophoretic migration (100V) of a free TNFα siRNA alone (lane 4) or at 37°C with RNAse A during 2h digestion (lane 5) and complex formed with PEI (lane 6 during 2h of RNAse, lane 7 during 4h of RNAse) or Chi (lane 8 during 2h of RNAse, lane 9 during 4h of RNAse). Decomplexation of polyplexes of lane 7 (PEI) and lane 9 (Chi), respectively, showed in lane 10 (PEI) and lane 11 (Chi) with a concentrated NaOH (0.1M, 1min) solution. C- Kinetic curves of adsorption of siRNA (4μM) on PEI (5mM) linked on gold chip. Surface plasmon resonance (SPR) measurement was performed after immobilizing the PEI onto a carboxyl-terminated self-assembled monolayer (carboxydextran) formed on the gold-coated chip using N-hydroxysuccinimide (NHS) and N-ethyl-N9-(3 diethylaminopropyl) carbodiimide (EDC) coupling (Frey and Corn, 1996). KD (nM) is calculated by SPR.
To investigate siRNA complexation with positive molecules, electrophoretic migration on a 4% agarose gel was performed for 45 min under 100 V. As shown in Figure 1B, the free siRNA band at 21 bp (Figure 1B, lane 1) was barely visible when complexed with Chi (Figure 1B, lane 2) and not visible after complexation with PEI (Figure 1B, lane 3), indicating strong affinity for both PEI and Chi at the N/P ratio analyzed. Due to complexation of siRNA in each polyplex, band brightness of complexed siRNA (Figure 1B, lanes 2 and 3) was slightly decreased for the same amount of siRNA, compared with that of non-complexed siRNA (Figure 1B, lane 1). This is explained by the fact that ethidium bromide (EtBr) integrates between the two siRNA chains and introduction of PEI or Chi leads to steric modifications, making interactions between siRNA and EtBr more difficult. As shown in Figure 1B (lanes 2 and 3), at each N/P ratio, Chi complexed with siRNA to a lower extent than PEI due to slight degradation of siRNA (smear observed in Figure 1B, lane 2, where siRNA is complexed to Chi, but not Figure 1B, lane 3, where siRNA is complexed to PEI). Electrophoretic migration data obtained under similar conditions revealed that both polyplexes protected siRNA from RNAse A (0.04 mg/mL) degradation at 37ºC during 2- and 4-h incubations (Figure 1B lanes 2 and 3). Free siRNA was fully degraded after 2-h digestion with RNAse A (Figure 1B; lane 5), whereas no degradation was observed in lanes 6 and 7 representing siRNA within a polyplex composed of PEI for 2 h (lane 6) and PEI for 4 h (lane 7). Data in Figure 1C (lanes 8 and 9) show that the integrity of siRNA complexed with Chi was maintained, but not as well as that complexed with PEI (smear observed). Finally, siRNA integrity after RNAse A exposure was assessed following polyplex decomplexation by neutralizing the positive charges on PEI and Chi using a NaOH-concentrated solution for a short time preventing siRNA hydrolyzation. Migration patterns of the polyplex (siRNA/PEI) in alkaline solution revealed that the siRNA remained intact, as evident from the bright band at 21 bp (lane 10 and 11). Brightness decreased upon complexation (Figure 1B lane 1 versus lane 3) and was reversed by disruption of the siRNA/PEI complex (lane 10 and 11). Based on these results, we conclude that the degree of complexation/protection of siRNA with PEI is higher than that with Chi.
Direct force measurement between TNFα siRNA and PEI using surface plasmon resonance
The association and dissociation constants of interactions between PEI with siRNA were calculated using a technique based on surface plasmon resonance (SPR). KD calculation, representing 50% of siRNA adsorption on PEI, is the main value of a SPR study. Our data show that TNFα siRNA has a low KD=27.4nM for binding to PEI indicating a strong binding (Figure 1E). Low KD was predictable according to the very weak desorption corresponding to the second phase of siRNA binding on PEI (Figure 1E). It is likely that electrostatic represent most interactions between TNFα siRNA/PEI. This finding indicates that TNFα siRNA displays high complexation/stability index with PEI.
Characterization of TNFα siRNA-loaded NPs covered with PVA
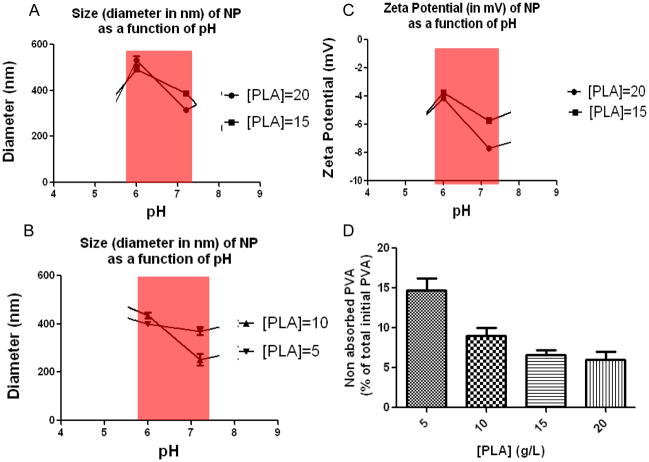
In view of the excellent physicochemical parameters of TNFα siRNA/PEI complexation, we initiated the process of loading NP with TNFα siRNA/PEI. TNFα siRNA/PEI loaded into NPs with different concentrations of polylactide (PLA) (NPs matrix) and covered with polyvinyl alcohol (PVA) (NPs shell) were prepared as described in the Materials and Methods section. The sizes of these particles were further measured in solution at pH 6.2 and 7.2. These pH values were selected because they represent the physiological extracellular pH in the colonic lumen and physiological neutral intracellular pH (cytoplasma) or acidic intracellular pH (endosomes). The sizes of TNFα siRNA-loaded NPs (for all PLA concentration) covered with PVA decreased at neutral pH, compared with that at more acidic pH (Figure 2A and 2B). These observations indicate that the hydrodynamic radius is pH-dependent. The change in diameter is possibly determined by the PVA surface characteristics. Pure PVA, corresponding to 100% hydrolysis, is totally hydrophobic and therefore non-soluble in water or biological medium. Upon introduction of acetate groups (14% in this study), PVA became water soluble but, interestingly, retained a hydrophobic part that made it an emulsifier. The ester interchange of PVA led to reduced acetate function and increased hydrophobicity. Thus, the biological medium became a less effective solvent for PVA, which, in turn, had a direct effect by reducing the hydrodynamic radius of the molecule. Because PVA represents the NP shell, decreasing the hydrodynamic radius of PVA automatically leads to a reduction in the NP diameter. Next, we investigated the zeta potential of NPs (Figure 2C); this measure represents a direct index of colloidal stability. The colloidal stability of particles is based on electrostatic repulsions between particles and aggregation phenomenon happens at low zeta potential. TNFα siRNA-loaded NPs covered with PVA did not provide a stable zeta potential at PLA concentrations of 5 and 10 g/L. The latter finding may be explained by incomplete and irregular PVA absorption by NPs at PLA concentrations of 5 and 10 g/L, which would consequently render the NPs surface charges unstable (Figure 2D). In contrast, TNFα siRNA-loaded NPs covered with PVA generate a stable zeta potential and maximal PVA absorption at PLA concentrations of 15 and 20g/L, and display optimal activity at pH 7 (Figure 2C). The interdependence between pH and zeta potential is directly correlated to PVA absorbed on the NPs surfaces. PVA is 86–89% hydrolyzed, meaning that 11–14% of the remaining functions are acetate groups. Between pH 6 to 7.2, the number of hydroxide groups is 10-fold higher and acetate groups of PVA are hydrolyzed via ester interchange. These reactions increase deprotonation of hydroxyl function and lead to more negative NPs zeta potentials. The negative zeta potential of TNFα siRNA-loaded NPs covered with PVA at PLA concentrations of 15 and 20 g/L (Figure 2C) are indicative of negatively charge. A negative zeta potential (−5 mV) is required for strong cell/NP interactions due to the positive cell charges. [20, 21] However, internalization of negatively charged nanoparticles is believed to occur through nonspecific binding and clustering of the particles on cationic sites on the plasma membrane and their subsequent endocytosis. Also, NPs within this size range are taken up by macrophages cells. [22] To illustrate, SEM and TEM (Figure 3A-left and right) images of NPs with 20g/L PLA loaded with TNFα siRNA/PEI confirmed diameters of ~530 nm measured at pH=6 by using the light scattering theory. Both images reveal diameters of about 500 nm and unimodal distribution of NP sizes (Figure 3A-center). Interestingly, TEM images (Figure 3A-right) allowed observation of differences in electron densities of the hydrophobic core (PLA in black) and NP shells made of PVA (PVA in white) assessing a effective adsorption of PVA on NPs made with 20g/L of PLA
Figure 2. Size and charge characterization of TNFα siRNA-loaded nanoparticles (NPs) covered with polyvinyl alcohol (PVA).
Diameter (nm) of high (A) and low (B) concentration of D,L polylactide (PLA) NPs loaded with TNFα siRNA measured by photon correlation spectroscopy (PCS). C-Zeta potential (mV) of high PLA concentration of NPs loaded with TNFα siRNA measured by zeta potential analyzer. D- Percentage of non-absorbed PVA after centrifugation for NPs groups calculated for NPs made with [PLA]=20, 15, 10 and 5 g/L.
Figure 3. Visualization and cytotoxicity assessment on macrophages of TNFα siRNA-loaded nanoparticles (NPs) covered with polyvinyl alcohol (PVA).
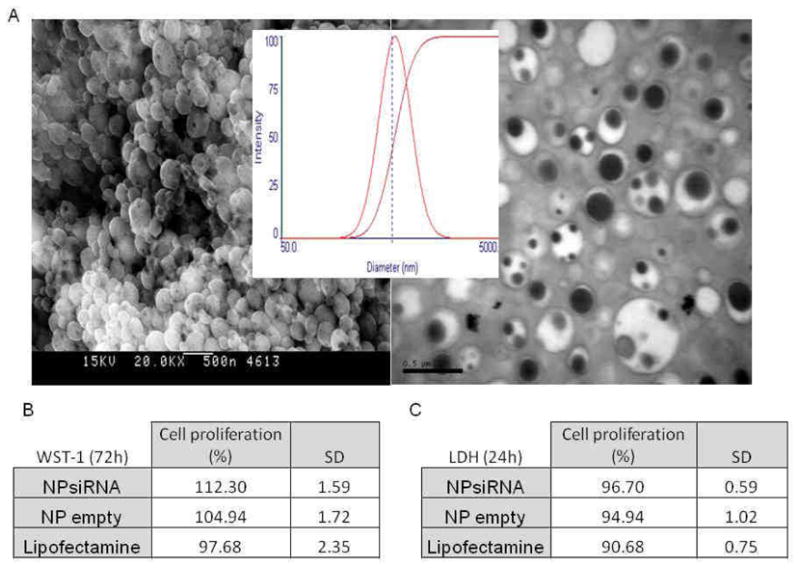
A- Scanning electronic microscopy (SEM) (left), size distribution measured by light scattering (center) and transmission electronic microscopy (TEM) (right) of 500μg/mL TNFα siRNA-loaded NPs suspension. B- WST-1 test was used to determine the cytotoxicity of TNFα siRNA NPs (1 mg/ml) and compared to empty NPs (1mg/mL) and lipofectamine on RAW 264.7 cells (mouse macrophages) after 72h of exposure (N=8). C- As another measure of cytotoxicity, lactate dehydrogenase (LDH) level was measured after treating 1mg/mL suspension of TNFα siRNA NPs on RAW 264.7 cells (mouse macrophage) and compared to empty NPs (1 mg/ml) and lipofectamine for 24h (N=8).
Non-cytotoxicity measurement of TNFα siRNA-loaded NPs covered with PVA
Cytotoxicity assessments need to be performed for all biomaterials. Accordingly, we performed simple cytotoxicity tests (WST-1 and LDH assays). As shown in Figure 3B and 3C, TNFα siRNA-loaded NPs at a concentration of 1 mg/mL did not induce cellular death of MPs over a short (24-h period) or a longer (72-h period) exposure time, compared with control materials (empty NPs). Notably, Lipofectamine, an agent commonly used to transfect cells, significantly affects LDH release from MPs (Figure 3B and 3C). These observations collectively demonstrate that our TNFα siRNA-loaded NPs do not affect cell viability compared with Lipofectamine, and may be used to safely transfect MPs.
Uptake and kinetic of FITC-tagged scrambled siRNA/PEI loaded NPs by macrophages
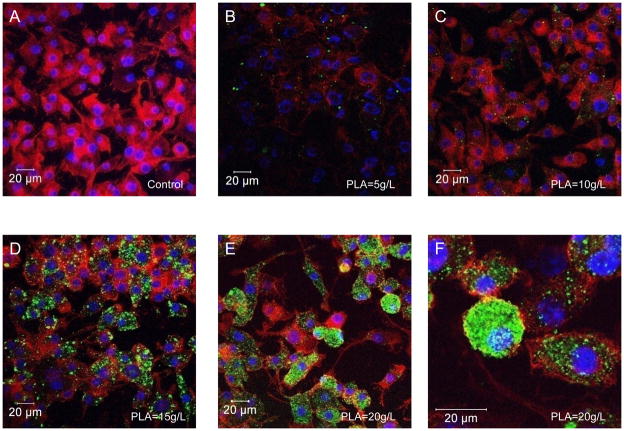
Because modulation of PLA concentrations modified the physicochemical properties of NPs, such as final size and electrostatic charges (Figure 2), we predicted that these changes directly affect loaded NP uptake by MPs. FITC-tagged scrambled siRNA/PEI-loaded NPs at different PLA concentrations (respectively 5, 10, 15 and 20g/L) and covered with PVA were added to MPs in vitro in the extracellular medium at pH 7.2 for 24 hours. Next, intracellular uptake was visualized using confocal microscopy. As shown in Figure 4A, B, C, D, E the increasing fluorescence signal was correlated with the increase of the PLA concentration. 20g/L of D, L PLA in the organic phase was the optimal concentration for maximal fluorescent NP uptake.
Figure 4. Effects of different concentrations of D,L polylactide (PLA) for a maximum uptake of FITC-tagged scrambled siRNA/polyethylenimine loaded NPs by macrophages.
Cells were treated for 24h with NPs (500μg/mL) loaded with FITC-tagged scrambled siRNA (green) and processed for confocal immunofluorescence staining as described in the Methods section. Fixed cells were stained with rhodamine/phalloidin and DAPI to visualize actin (red) and nuclei (purple), respectively. Intracellular FITC-tagged scrambled siRNA in NPs made with variable PLA concentration was determined. A- Untreated control cells. B-C-D-E- Macrophages exposed to 24h of nanoparticles loaded with FITC-tagged scrambled siRNA (green) made of variable PLA concentration (5 (B), 10 (C), 15 (D) and 20 g/L (E and F).
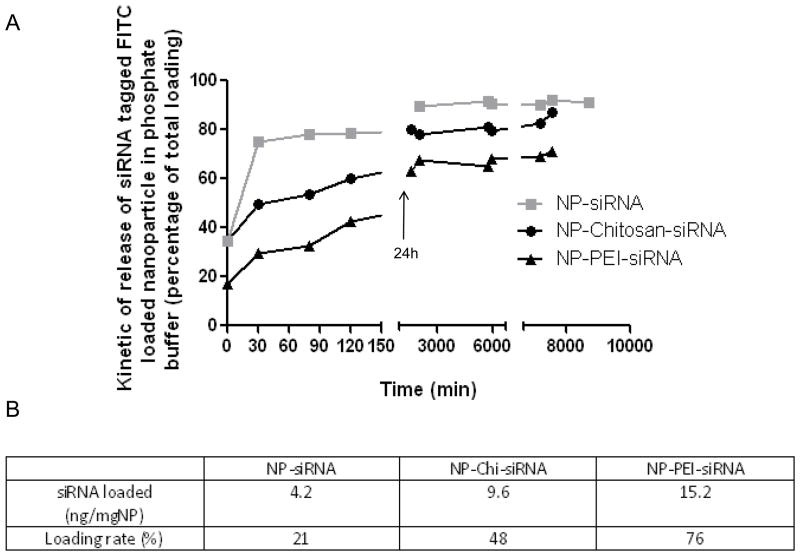
According to those observations at 20 g/L of PLA for NPs, we decided to measure the loading and the kinetics characteristic of NPs made with siRNA alone or complexed with Chi or PEI. Interestingly, the formation of the polyplex quantitatively changed the “shape” of the kinetic release curve made in PBS buffer at pH=6 and 37ºC (Figure 5-A). Without any complex, a typical burst effect of siRNA released from NPs is observed. After 30 min of suspension in PBS buffer at pH=6, 80% of the total siRNA is already released out of the NP. Complexation of siRNA with Chi significantly decreased this burst effect allowing more than 50% of NPs to remain loaded with siRNA after 30 min of release. Optimal results were obtained for PEI/siRNA complex that allowed about 70% of the siRNA drug to remain loaded after 30 min of release in PBS buffer. In our model of synthesis of NPs, siRNA complexed by PEI or Chi significantly increased the NPs value not only by protecting the loaded siRNA but also by increasing the final amount of loaded siRNA and beneficially changing the kinetic release curve to insure high delivery of siRNA in the intracellular domain of mouse MPs. In the table above (Figure 5-B), PEI complexation lead to higher effective loading (15.2 ng/mg of NPs) compared to siRNA naked or complexed with Chi respectively 4.2 and 9.6 ng/mg of NPs.
Figure 5. Kinetics of release of FITC-tagged scrambled siRNA/polyethylenimine (PEI) loaded nanoparticles (NPs) covered with polyvinyl alcohol (PVA).
Kinetics of release of FITC-tagged scrambled siRNA loaded in NPs in phosphate buffer, pH=6. ( non complexed siRNA,
non complexed siRNA,  siRNA complexed with Chi and
siRNA complexed with Chi and  siRNA complexed with PEI). As the total amount of FITC-tagged scrambled siRNA loaded is different in each group of NPs, siRNA release curves were normalized to their specific total siRNA loaded. B- Effective loaded amount of TNFα siRNA in NPs (ng/mg NP) and loading rate (% initial TNFα siRNA introduced in the synthesis process) of TNFα siRNA loaded NPs.
siRNA complexed with PEI). As the total amount of FITC-tagged scrambled siRNA loaded is different in each group of NPs, siRNA release curves were normalized to their specific total siRNA loaded. B- Effective loaded amount of TNFα siRNA in NPs (ng/mg NP) and loading rate (% initial TNFα siRNA introduced in the synthesis process) of TNFα siRNA loaded NPs.
Effects of siRNA complexation/protection on NP uptake by macrophages
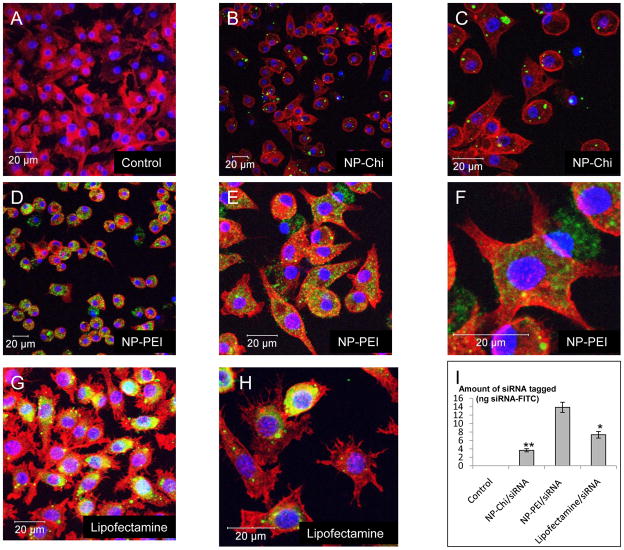
We showed that the degree of complexation/protection/loading of siRNA by PEI is greater than that by Chi. FITC-tagged scrambled siRNA/PEI-loaded or siRNA/Chi-loaded NPs with 20g/L PLA and covered with PVA were added to MPs in the extracellular medium at pH 7.2. Exposure time of NPs with MPs was set at 24 hours. As shown in Figure 6, MPs displayed greater uptake of FITC-tagged scrambled siRNA/PEI loaded NPs (Figure 6D, 6E and 6F) compared with FITC-tagged scrambled siRNA/Chi-loaded NPs. As a positive control, MPs were transfected with free FITC-tagged scrambled siRNA at the same concentration (7.3 ng) as with NPs using classical Lipofectamine technology (Figure 6G and 6H). To compare the transfection efficiencies of the different experimental approaches, the amount of FITC-tagged scrambled siRNA was quantified using fluorometry. As shown in Figure 6I, FITC-tagged scrambled siRNA/PEI-loaded NPs exhibited higher transfection efficiency than FITC-tagged scrambled siRNA with Lipofectamine and FITC-tagged scrambled siRNA/Chiloaded NPs. Finally, MPs transfected with Lipofectamine, but not with loaded NPs, induced numerous filiform and convoluted dendritic processes (Figure 6H versus Figure 6A or 6F). This typical shape, observed previously, is a characteristic marker of MPs or dendritic cell activation. [23, 24] This finding indicates that in contrast to Lipofectamine, loaded NPs do not stimulate MPs to an inflamed state.
Figure 6. Complexation using PEI results in improved uptake of FITC-tagged scrambled siRNA compared to chitosan (Chi).
Cells were treated with NPs (500μg/mL) loaded with FITC-tagged scrambled siRNA (green) for 24 h and processed for confocal immunofluorescence staining as described in the Methods section. Fixed cells were stained with rhodamine/phalloidin and DAPI to visualize actin (red) and nuclei (purple), respectively. Images show intracellular FITC-tagged scrambled siRNA in NPs (20g/L of PLA) made with variable polyplex. A-Untreated control cells. B-C- FITC-tagged scrambled siRNA complexed with Chi. D-E-F- FITC-tagged scrambled siRNA complexed with PEI. G-H- FITC-tagged siRNA scrambled transfected by lipofectamine. I- Fluorescence quantification (ng FITC tagged scrambled siRNA) of A, B, D and G. (** p<0.01, *p<0.05, ANOVA test)
Kinetic of uptake of FITC-tagged scrambled siRNA/PEI loaded NPs by macrophages
We also demonstrate that FITC-tagged scrambled siRNA/PEI-loaded NPs covered with PVA are taken up by MPs within 30 min after addition (Figure 7-B). Intracellular loading of these molecules is maximal at 1 hour after NP addition because no increase in the fluorescence signal is observed after this time-point (Figure 7B and 7C). In contrast, a weak fluorescence signal was observed after 2 hour (nothing after 1h) when using Lipofectamine to transfect free FITC-tagged scrambled siRNA (Figure 7A). MPs observations also showed that PEI had not any toxicity on MPs as previously described for transfection of siRNA or plasmids using PEI. [25] These findings demonstrate that NPs act as an efficient vehicle that is rapidly deliver siRNA into the cytoplasm and prevent siRNA exposure to a hostile extracellular environment.
Figure 7. Time course and uptake of FITC-tagged scrambled siRNA/polyethylenimine (PEI) loaded nanoparticles (NPs) by macrophages.
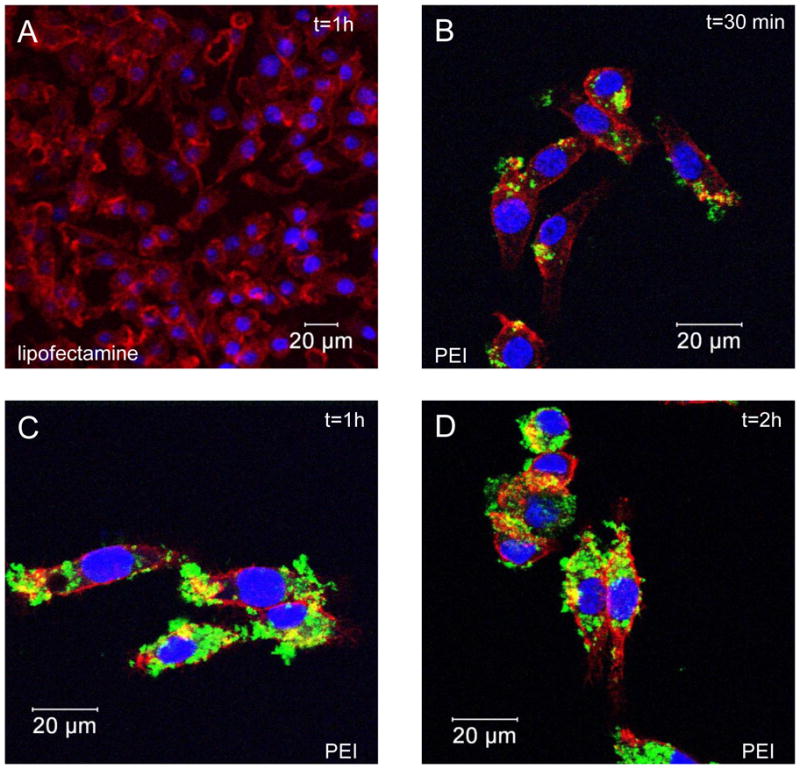
Cells were treated for 30 min, 1h and 2h with NPs (500μg/mL) loaded with FITC-tagged scrambled siRNA (green) and processed for confocal immunofluorescence staining as described in the Methods section. Fixed cells were stained with rhodamine/phalloidin and DAPI to visualize actin (red) and nuclei (purple), respectively. Picture of FITC-tagged scrambled siRNA intracellular accumulation with lipofectamine (1h exposure) (A) and kinetics of FITC-tagged scrambled siRNA complexed with polyethylenimine after 30 min (B), 1h (C) and 2h (D).
LPS-induced TNFα secretion measurement in macrophages treated with TNFα siRNA/PEIloaded NPs
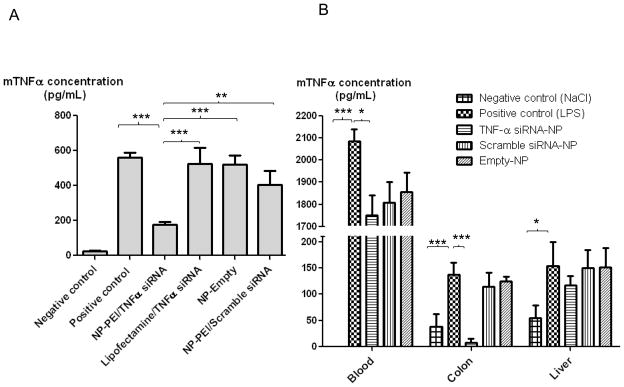
Although our experiments confirm that FITC-tagged scrambled siRNA/PEI is released in the cytosol of MPs, to ensure the effectiveness of TNFα siRNA, we assessed the anti-inflammatory effects of TNFα siRNA-loaded NPs in vitro on MPs stimulated with 10 μg/mL LPS. MPs were exposed to TNFα siRNA/PEI-loaded NPs, TNFα siRNA/Chi-loaded NPs or TNFα siRNA in the presence of Lipofectamine for 24 hours, followed by stimulation with LPS (10 μg/mL) for 1 h. As shown in Figure 8-A, exposure to LPS increased the mouse TNFα concentration in the supernatant 23-fold (559.8 pg/mL), compared with that in the negative control (Figure 8-A). TNFα siRNA/PEI-loaded NPs induced a significant decrease in TNFα secretion (175.9 pg/mL vs 559.8 pg/mL) from LPS-stimulated MPs. In contrast, siRNA/Chiloaded NPs and siRNA alone in the presence of Lipofectamine did not significantly affect TNFα secretion (siRNA in the presence of Lipofectamine: 523 pg/mL vs 559.8 pg/mL; siRNA/Chi loaded NPs: 523 pg/mL vs 520.0 pg/mL), indicating that siRNA TNFα/PEI loaded NPs have an anti-inflammatory effect on MPs. Appropriate controls were performed and showed that scrambled siRNA-loaded in NP or similar amount of siRNA with lipofectamine or free siRNA NPs had no anti-inflammatory effect on LPS-stimulated MPs (Figure 8-A). The biological effect showed that the high uptake of NPs into cells effectively release TNFα siRNA. Thus, we indirectly demonstrated that TNFα siRNA was released from the endocytosis vesicles. As already known, the strong buffer capacity, designed as proton sponge hypothesis, could be responsible for the fact that PEI-based delivery systems do not require endosome disruptive agents for lysosomal escape. [26]
Figure 8. TNFα siRNA/polyethylenimine (PEI) loaded nanoparticles (NPs) inhibit LPS-induced TNFα secretion in macrophages and in mice.
A- RAW 264-7 macrophages were pretreated with TNFα siRNA loaded NPs suspension (800μg/mL) for 24h. As a control, RAW 264-7 macrophages were pre-treated with empty NPs, scrambled siRNA loaded NPs, and lipofectamine with the same amount of TNFα siRNA compared to TNFα siRNA loaded NPs suspension. Cells were then treated with LPS (10mg/ml) for one hour (positive control cells were not pre-treated with NPs). Cells were lysed and TNFα protein measured as described in the Methods section. Data is SEM±S.E, n=6 (Significantly different from ANOVA test***P<0.001, **P<0.005).
B- C57BL/6 female wild type mice were gavaged with TNFα siRNA loaded NPs (5 mg/mL), scrambled siRNA NPs (5mg/mL) and empty NPs (5mg/mL) contained in a hydrogel (alginate and chitosan) daily for 4 consecutive days. Positive control mice were gavaged only with the hydrogel without NPs. Mice were then administered LPS (100μg. Kg−1 i.p.) and euthanized after one hour. The colon, spleen and blood were collected and TNFα expression was measured as described in the Materials and Methods section. Data is represented as SEM±S.E, n=8 (Significantly different from ANOVA test *P<0.05, ***P<0.001, ANOVA test).
LPS-induced colonic TNFα secretion measurement in mice treated with encapsulated TNFα siRNA/PEI-loaded NPs
Next, we investigated the effect of oral administration of encapsulated TNFα-loaded NPs on TNFα expression/secretion in colonic tissue in LPS-treated mice. To deliver the TNFα-loaded NPs to the colonic lumen, we encapsulated them into a biomaterial comprised of alginate and chitosan at a ratio of 7/3 (wt/wt). We previously shown that the biomaterial collapses in intestinal solution at pH 5 or 6, which is the colonic pH under inflamed and non-inflamed states respectively. Thus, NPs release from the hydrogel (and so TNFα release from NP) will occur mostly in the colonic lumen compared with other parts of the gastrointestinal tract such as the stomach and small intestine. [16] As shown in Figure 8-B, one hour after LPS administration by intra peritoneal injection, TNFα levels increased in blood, colonic tissue and liver when compared with untreated mice. Interestingly, when mice were pretreated for 4 days with the encapsulated TNFα siRNA/PEI-loaded NPs (oral administration of 5mg of NPs by mL of hydrogel), LPS-induced TNFα levels was significantly reduced in colonic tissue and blood but not in the liver tissue when compared to mice treated with empty NP (Blood: 1751.5 pg/mL vs 2084.5 pg/mL; Colon tissue: 7.5 pg/mL vs 136.2 pg/mL; Liver tissue: 117.1 pg/mL vs 154.2 pg/mL). Together these results demonstrated that TNFα siRNA/PEI-loaded NPs are delivered to the colonic tissue and that TNFα siRNA released from NPs efficiently inhibit LPS-induced colonic TNFα synthesis/secretion. These NP had no effect on LPS-induced TNF-α synthesis/secretion in the liver. These results suggest that NP can be used for site-specific delivery of therapies with minimal systemic effects. Given that the use of potent therapies (e.g. anti-TNF agents) in diseases such as inflammatory bowel disease is limited by profound systemic toxicity, our data provides a highly promising mode of delivery of anti-TNF agents to the site of inflammation and minimize systemic toxicity.
Conclusions
Here we demonstrated that siRNA-TNFα/PEI-loaded NPs covered with PVA effectively deliver siRNA-TNFα into the cytoplasm and consequently induce efficient gene silencing of endogenous TNFα in inflamed MPs. The physicochemical properties of the prepared NPs and complexation of siRNA TNFα with PEI play a crucial role in efficient delivery and TNFα gene silencing in inflamed MPs. Notably, the high loading efficiency of siRNA TNFα into the NPs and slow release from NPs are attractive in the context of gene silencing over a period of time for chronic diseases, such as IBD. Finally, we demonstrated that siRNA TNFα/PEI-loaded NPs covered with PVA are non-cytotoxic and thus applicable for therapeutic purposes. The next step is to specifically target siRNA TNFα/PEI-loaded NPs covered with PVA to MPs. Finally, we demonstrated effective and specific drug delivery of TNFα siRNA-loaded NPs to colonic tissue by oral administration of encapsulated TNFα siRNA/PEI-loaded NPs in mice.
Acknowledgments
This work was supported by grants from the Department of Veterans Affairs and the National Institutes of Health of Diabetes and Digestive and Kidney by the grants R24-DK-064399 (center grant), RO1-DK-071594 (to D.M), RO1-DK55850 (to S.S), K01-DK085222 (to A.L.T). G.D is a recipient of a research Fellowship Award from the Crohn’s and Colitis Foundation of America.
Footnotes
Author involvement
Conceived and designed the experiments: HL SVS DM. Performed the experiments: HL ALT YY. Analyzed the data: HL. Contributed reagents/materials/analysis tools: HL ALT GD YY HTTN. Wrote the paper: HL SVS DM.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Dr. Hamed Laroui, Email: hlaroui@emory.edu.
Dr. Arianne L. Theiss, Email: atheiss@emory.edu.
Dr. Yutao Yan, Email: yyan2@emory.edu.
Dr. Guillaume Dalmasso, Email: gdalmas@emory.edu.
Dr. Hang T.T. Nguyen, Email: hnguye9@emory.edu.
Dr. Didier Merlin, Email: dmerlin@emory.edu.
References
- 1.Holtmann MH, Neurath MF. Anti-TNF strategies in stenosing and fistulizing Crohn’s disease. Int J Colorectal Dis. 2005;20(1):1–8. doi: 10.1007/s00384-004-0634-0. [DOI] [PubMed] [Google Scholar]
- 2.de Vries A, Hazlewood L, Fitch PM, Seckl JR, Foster P, Howie SE. High-fat feeding redirects cytokine responses and decreases allergic airway eosinophilia. Clin Exp Allergy. 2009;39(5):731–9. doi: 10.1111/j.1365-2222.2008.03179.x. [DOI] [PubMed] [Google Scholar]
- 3.D’Haens G. Anti-TNF therapy for Crohn’s disease. Curr Pharm Des. 2003;9(4):289–94. doi: 10.2174/1381612033391982. [DOI] [PubMed] [Google Scholar]
- 4.Toub N, Bertrand JR, Tamaddon A, Elhamess H, Hillaireau H, Maksimenko A, et al. Efficacy of siRNA nanocapsules targeted against the EWS-Fli1 oncogene in Ewing sarcoma. Pharm Res. 2006;23(5):892– 900. doi: 10.1007/s11095-006-9901-9. [DOI] [PubMed] [Google Scholar]
- 5.Lieberman J, Song E, Lee SK, Shankar P. Interfering with disease: opportunities and roadblocks to harnessing RNA interference. Trends Mol Med. 2003;9(9):397–403. doi: 10.1016/S1471-4914(03)00143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007;59(2–3):134–40. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raper SE. Gene therapy: the good, the bad, and the ugly. Surgery. 2005;137(5):487–92. doi: 10.1016/j.surg.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64(21):7668–72. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 10.Gunther M, Wagner E, Ogris M. Specific targets in tumor tissue for the delivery of therapeutic genes. Curr Med Chem Anticancer Agents. 2005;5(2):157–71. doi: 10.2174/1568011053174855. [DOI] [PubMed] [Google Scholar]
- 11.Hood JD, Bednarski M, Frausto R, Guccione S, Reisfeld RA, Xiang R, et al. Tumor regression by targeted gene delivery to the neovasculature. Science. 2002;296(5577):2404–7. doi: 10.1126/science.1070200. [DOI] [PubMed] [Google Scholar]
- 12.Kim B, Tang Q, Biswas PS, Xu J, Schiffelers RM, Xie FY, et al. Inhibition of ocular angiogenesis by siRNA targeting vascular endothelial growth factor pathway genes: therapeutic strategy for herpetic stromal keratitis. Am J Pathol. 2004;165(6):2177–85. doi: 10.1016/S0002-9440(10)63267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pardridge WM. Intravenous, non-viral RNAi gene therapy of brain cancer. Expert Opin Biol Ther. 2004;4(7):1103–13. doi: 10.1517/14712598.4.7.1103. [DOI] [PubMed] [Google Scholar]
- 14.Fattal E, Vauthier C, Aynie I, Nakada Y, Lambert G, Malvy C, et al. Biodegradable polyalkylcyanoacrylate nanoparticles for the delivery of oligonucleotides. J Control Release. 1998;53(1– 3):137–43. doi: 10.1016/s0168-3659(97)00246-0. [DOI] [PubMed] [Google Scholar]
- 15.Lamprecht A, Schafer U, Lehr CM. Size-dependent bioadhesion of micro- and nanoparticulate carriers to the inflamed colonic mucosa. Pharm Res. 2001;18(6):788–93. doi: 10.1023/a:1011032328064. [DOI] [PubMed] [Google Scholar]
- 16.Laroui H, Dalmasso G, Nguyen HT, Yan Y, Sitaraman SV, Merlin D. Drug-loaded nanoparticles targeted to the colon with polysaccharide hydrogel reduce colitis in a mouse model. Gastroenterology. 2010;138(3):843–53. e1–2. doi: 10.1053/j.gastro.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Zambaux MF, Bonneaux F, Gref R, Maincent P, Dellacherie E, Alonso MJ, et al. Influence of experimental parameters on the characteristics of poly(lactic acid) nanoparticles prepared by a double emulsion method. J Control Release. 1998;50(1–3):31–40. doi: 10.1016/s0168-3659(97)00106-5. [DOI] [PubMed] [Google Scholar]
- 18.Ngamwongsatit P, Banada PP, Panbangred W, Bhunia AK. WST-1-based cell cytotoxicity assay as a substitute for MTT-based assay for rapid detection of toxigenic Bacillus species using CHO cell line. J Microbiol Methods. 2008;73(3):211–5. doi: 10.1016/j.mimet.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc Natl Acad Sci U S A. 2001;98(23):13249–54. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberman EA, Topaly VP, Tsofina LM, Jasaitis AA, Skulachev VP. Mechanism of coupling of oxidative phosphorylation and the membrane potential of mitochondria. Nature. 1969;222(5198):1076–8. doi: 10.1038/2221076a0. [DOI] [PubMed] [Google Scholar]
- 21.Ketterer B, Neumcke B, Laeuger P. Transport mechanism of hydrophobic ions across through lipid bilayers. J Memb Biol. 1971;5:225–45. doi: 10.1007/BF01870551. [DOI] [PubMed] [Google Scholar]
- 22.Yamanaka YJ, Leong KW. Engineering strategies to enhance nanoparticle-mediated oral delivery. J Biomater Sci Polym Ed. 2008;19(12):1549–70. doi: 10.1163/156856208786440479. [DOI] [PubMed] [Google Scholar]
- 23.Del Cacho E, Gallego M, Lopez-Bernard F, Sanchez-Acedo C, Lillehoj HS. Isolation of chicken follicular dendritic cells. J Immunol Methods. 2008;334(1–2):59–69. doi: 10.1016/j.jim.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Shankar SP, Babensee JE. Comparative characterization of cultures of primary human macrophages or dendritic cells relevant to biomaterial studies. J Biomed Mater Res A. 2010;92(2):791–800. doi: 10.1002/jbm.a.32406. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama K, Iwasaki F, Takizawa T, Yanagie H, Niidome T, Yamada E, et al. Novel receptor-mediated gene delivery system comprising plasmid/protamine/sugar-containing polyanion ternary complex. Biomaterials. 2004;25(16):3267–73. doi: 10.1016/j.biomaterials.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]