Abstract
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infections result in chronic virus replication and progressive depletion of CD4+ T cells, leading to immunodeficiency and death. In contrast, ‘natural hosts’ of SIV experience persistent infection with high virus replication but no severe CD4+ T cell depletion, and remain AIDS-free. One important difference between pathogenic and non-pathogenic infections is the level of activation and proliferation of CD4+ T cells. We analysed the relationship between CD4+ T cell number and proliferation in HIV, pathogenic SIV in macaques, and non-pathogenic SIV in sooty mangabeys (SMs) and mandrills. We found that CD4+ T cell proliferation was negatively correlated with CD4+ T cell number, suggesting that animals respond to the loss of CD4+ T cells by increasing the proliferation of remaining cells. However, the level of proliferation seen in pathogenic infections (SIV in rhesus macaques and HIV) was much greater than in non-pathogenic infections (SMs and mandrills). We then used a modelling approach to understand how the host proliferative response to CD4+ T cell depletion may impact the outcome of infection. This modelling demonstrates that the rapid proliferation of CD4+ T cells in humans and macaques associated with low CD4+ T cell levels can act to ‘fuel the fire’ of infection by providing more proliferating cells for infection. Natural host species, on the other hand, have limited proliferation of CD4+ T cells at low CD4+ T cell levels, which allows them to restrict the number of proliferating cells susceptible to infection.
Keywords: lymphocyte preservation, lymphocyte proliferation/depletion, CD4-positive T-lymphocytes, Ki-67 antigen/analysis, human/simian immunodeficiency virus, mathematical model
1. Introduction
Human immunodeficiency virus (HIV) types 1 and 2 have entered the human population in the last century as a result of cross-species transmission of the closely related simian immunodeficiency viruses—SIVcpz and SIVsmm—that infect chimpanzees (Pan troglodytes) and sooty mangabeys (Cercocebus atys; SM), respectively (Hahn et al. 2000). SIVsmm is also the precursor of SIVmac, which induces an AIDS-like disease in Asian macaques and represents the most commonly used animal model for studies of AIDS pathogenesis and vaccines (Hirsch et al. 1989).
Pathogenic HIV infection of humans and SIV infection of macaques are characterized by an acute phase lasting for four to six weeks, with high viral loads, profound depletion of CD4+ T cells in mucosal sites and moderate depletion of CD4+ T cells in blood (Smit-McBride et al. 1998; Guadalupe et al. 2003; Brenchley et al. 2004; Mattapallil et al. 2005). This is followed by a chronic phase, which corresponds to a prolonged period of intermediate viral loads and slowly progressive CD4+ T cell depletion occurring in the context of a highly dynamic process of CD4+ T cell turnover in the blood and lymphoid tissues (Mohri et al. 2001; Ribeiro et al. 2002). The final phase of disease is characterized by increasing viral loads, severe CD4+ T cell depletion and development of opportunistic infections. While HIV infection of humans and SIV infection of macaques generally follow a similar course, the rates of disease progression are usually faster in SIV-infected macaques.
Studies of SIV infection in ‘natural host’ non-human primate species such as SMs and mandrills (Mandrillus sphinx, MND) revealed that these animals also develop chronic infection with high viral loads (Onanga et al. 2002; Silvestri et al. 2003). However, in stark contrast to pathogenic HIV/SIV infections, natural SIV hosts maintain healthy CD4+ T cell levels and avoid immunodeficiency (Broussard et al. 2001; Silvestri et al. 2003). This peculiarly benign course of infection is thought to reflect a non-pathogenic virus–host equilibrium that has been established over a prolonged period of coevolution. However, it is unclear whether changes in the virus, in the host response or in both may be responsible for this adaptation. One viral feature that would explain the non-pathogenic infection of natural SIV hosts would be that the virus has evolved a mode of intracellular replication that is not cytopathic for the infected cell. However, recent studies have shown that the in vivo longevity of infected cells in these natural hosts is comparable to that observed in pathogenic infection (Silvestri 2005; Gordon et al. 2008). Several important differences have also been observed between pathogenic and non-pathogenic infections (Gordon et al. 2008; Pandrea et al. 2008a; Sodora et al. 2009): natural hosts show lower levels of immune cell activation during chronic infection (Chakrabarti et al. 2000; Sumpter et al. 2007; Paiardini et al. 2009a), and lower expression of CCR5 on CD4+ T cells (Veazey et al. 2003).
Increased CD4+ T cell proliferation is observed during pathogenic infection with SIV and HIV (Hazenberg et al. 2000; Sousa et al. 2002). In contrast, infected SMs show only a minor increase in CD4+ T cell proliferation, and infected MNDs show very little change in CD4+ T cell proliferation. While the mechanisms underlying this phenotype are still poorly understood, it has been proposed that reduced immune activation plays a key role in protecting natural SIV hosts from progression to AIDS (Silvestri et al. 2005). Since CD4+ T cell proliferation and CCR5 expression on CD4+ T cells are required for optimal virus replication, natural SIV infections should be associated with a more limited availability of susceptible (proliferating) cells for infection. Our modelling suggests that a strong proliferation of CD4+ T cells in response to CD4+ T cell depletion can paradoxically result in lower overall T cell numbers. However, maintaining low levels of CD4+ T cell proliferation despite low CD4+ T cell numbers may be a mechanism by which natural host species such as SMs and mandrills control infection.
2. Material and Methods
(a). Experimental methods
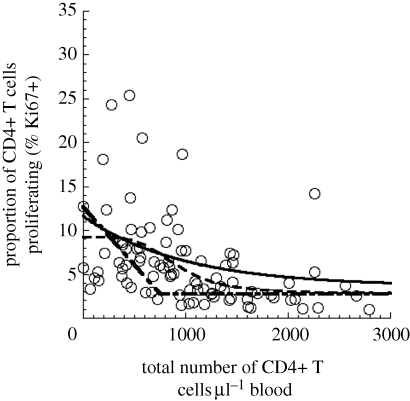
We investigated the relationship between CD4+ T cell counts and level of CD4+ T cell proliferation (measured in terms of Ki67 expression) in natural and non-natural hosts using the experimental data for rhesus macaques (RMs), humans and SMs obtained from previously published studies (figure 1a–c), and unpublished results in mandrills obtained using the same protocols (figure 1d). Four RMs were inoculated with SIVmac239 (Engram et al. 2009), while 31 of them were uninfected (Paiardini et al. 2009b). Of all the 35 RMs, we had single data points from 30 RMs and longitudinal data for five RMs. One hundred and seventeen SMs were naturally infected with SIV, while 85 were uninfected (Sumpter et al. 2007; Brenchley et al. 2008). Of all the 202 SMs, we had single data points from 92 of them and two or more points from the other 110. Twenty-three mandrills were naturally infected with SIV, while 24 of them were uninfected, and each animal was represented by only one data point. All animals were housed at the Yerkes National Primate Research Center and maintained in accordance with National Institutes of Health (NIH) guidelines. The data for HIV patients were pooled from two studies (Paiardini et al. 2005; Dunham et al. 2008), and represent 26 HIV-1-infected therapy-naive adults and nine healthy HIV-uninfected controls, all of whom were represented as single data points.
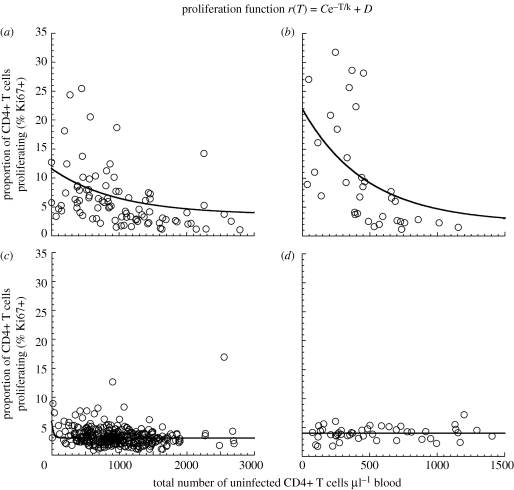
Figure 1.
Relationship between CD4+ T cell number and proliferation level (by Ki67 expression): experimental data are shown from pathogenic infection of (a) rhesus macaques (RMs) or (b) HIV infection, as well as natural host infection of (c) sooty mangabeys (SMs) or (d) mandrills. The experimental data for RMs, humans and SMs are obtained from previously published studies (Paiardini et al. 2005, 2009b; Sumpter et al. 2007; Brenchley et al. 2008; Dunham et al. 2008; Engram et al. 2009) and for mandrills from unpublished results. All of the species except mandrills show a significant negative correlation between CD4+ T cell count and the proportion of cells Ki67+ (Spearman correlation). The relationship between total CD4+ T cell number and proliferation [r(T), solid line] is fitted with an exponential function (top). Parameters for best fit of exponential function to the data for each species, as well as the p-values, are shown in table 1. The baseline proliferation rate (D)—the average proliferation rate when CD4+ T cell counts are high (greater than or equal to 1000 cells per microlitre of blood for SM and RM, and greater than or equal to 500 cells per microlitre of blood for humans)—is very similar between species. However, the maximal proliferation (C + D) is higher in the more pathogenic hosts (RMs and humans) compared with SMs.
We also analysed the relationship between CD4+ T cell counts and level of CD4+ T cell proliferation in uninfected RMs and SMs whose CD4+ T cells were depleted using anti-CD4 antibodies (figure 2). Three uninfected RMs and three uninfected SMs were treated with repeated infusion of the humanized anti-CD4 monoclonal antibody Cdr-OKT4A-huIgG1clone 12F11, and the fractions of CD3 + CD4+ T cells were monitored over time by flow cytometry, as previously described (Klatt et al. 2008). There were 13 measurements taken over a period of 200 days for each of these animals.
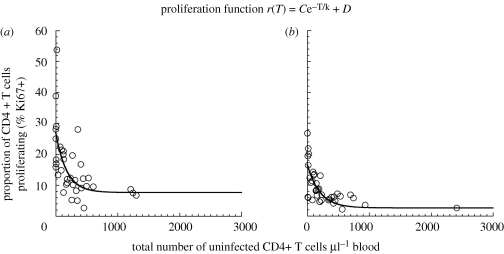
Figure 2.
Relationship between CD4+ T cell number and proliferation level (by Ki67 expression) after depletion of CD4+ T cells with antibody: (a) three uninfected RMs and (b) three uninfected SMs were treated with repeated infusion of the humanized anti-CD4 monoclonal antibody Cdr-OKT4A-huIgG1clone 12F11, and the fractions of CD3+,CD4+ T cells monitored over time by flow cytometry (as previously described; Klatt et al. 2008). Both Ab-depleted species show a significant negative correlation between CD4+ T cell count and the proportion of cells Ki67+ (Spearman correlation), even in the absence of virus. The relationship between total CD4+ T cell number and level of proliferation [r(T), solid line] is likewise fitted with an exponential function and the parameters, as well as the p-values, are shown in table 2.
(b). Mathematical model
Mathematical models of HIV infection have been used for over a decade to shed light on a number of important features of infection. We and others have recently investigated the role of CD4+ T cell proliferation and preferential infection of activated and proliferating cells in driving the observed immune dynamics (Ribeiro et al. 2006; Yates et al. 2007). Thus, we adapted the standard model of HIV infection (Perelson et al. 1996; Nowak & May 2000; Lloyd 2001; Perelson 2002) to (i) allow only infection of proliferating CD4+ T cells, where (ii) the proportion of proliferating CD4+ T cells increases as CD4+ T cell count decreases. The model can be expressed as a set of ordinary differential equations:
| 2.1 |
| 2.2 |
and
| 2.3 |
In our model, T is the number of all uninfected CD4+ T cells (both proliferating and non-proliferating); I and V are the levels of infected CD4+ T cells and virus, respectively; λ is the production rate of uninfected CD4+ T cells from the thymus; δT and δI are the death rates of uninfected and infected CD4+ T cells, respectively; p and c are the production and clearance rates of virus, respectively; and β is the infectivity of virus. The important change from the standard model of infection is the inclusion of a proliferation function r(T), which represents the proportion of cells that are proliferating at a given total number of uninfected CD4+ T cells in the pool (T), and ρ is the division rate of these cells. The absolute number P of cells that are proliferating at a given total CD4+ T cell number is therefore the product P = r(T)T. Our model assumes that only proliferating CD4+ T cells can be infected, so that at any time the number of uninfected cells susceptible to infection (or ‘targets for the virus’) is equal to the number of proliferating cells P.
Initially, as in figure 1, we have chosen the proliferation function r(T) to have an exponential form, but have also investigated other functional forms such as piecewise linear or Hill function.
3. Results and discussion
(a). Mechanisms of T cell activation and proliferation
HIV infection is characterized by a chronically high level of CD4+ T cell proliferation. This level of T cell proliferation has been shown to predict the risk of disease progression (Carbone et al. 2000). CD4+ T cell proliferation during HIV infection can arise from a number of mechanisms: (i) antigenic stimulation, (ii) direct activation by the virus or its products, (iii) bystander activation as a result of inflammatory cytokines or other factors, (iv) microbial translocation from the intestinal lumen to the systemic circulation and (v) homeostatic T cell proliferation as a response to CD4+ T cell depletion (Hazenberg et al. 2000; Douek et al. 2001; Vlahakis et al. 2001; Brenchley et al. 2006; Lee et al. 2009).
We investigated the relationship between CD4+ T cell number and CD4+ T cell proliferation in non-pathogenic and pathogenic infections. Previous studies have shown that there is a negative correlation between CD4+ T cell number and proliferation (Hazenberg et al. 2000). To further investigate this phenomenon, we compared the relationship between CD4+ T cell counts and level of CD4+ T cell proliferation in SIV-infected natural host species (SMs and MNDs), SIV-infected RMs (Macaca mulatta; RM), as well as HIV-infected humans (figure 1). We observed a significant negative correlation between CD4+ T cell number and CD4+ T cell proliferation in humans (Spearman r = −0.7434, p < 0.0001), RM (r = −0.5585, p < 0.0001) and for SM (r = −0.1902, p = 0.0004; table 1). However, in MND, there was no significant correlation.
Table 1.
Summary of statistics and parameters of the exponential fit to figure 1. n.s., not significant; n.a., not applicable.
| rhesus macaques (CI) | humans (CI) | sooty mangabeys (CI) | mandrills (CI) | |
|---|---|---|---|---|
| Spearman r | −0.5585 | −0.7434 | −0.1902 | 0.1240 |
| p | <0.0001 | <0.0001 | 0.0004 | 0.4065 (n.s.) |
| intercept C + D (%) | 11 (7.8–14.0) | 22 (12.4–31.5) | 5.8 (4.1–7.4) | n.a. |
| sensitivity k | 977.5 (612.7–2319) | 480.3 (280.4–1673) | 277.4 (184.4–559.9) | n.a. |
| baseline D (%) | 2.8 | 2.1 | 2.9 | 4.0 |
Next, we attempted to fit different relationships to the experimental data in order to compare the infections. Using a simple exponential relationship, we show rising CD4+ T cell proliferation with lower CD4+ T cell counts in SM, RM and human infection. A Mann–Whitney test showed that there was no significant difference (p = 0.5361) in Ki67 levels at high T cell counts (above 1000 cells µl−1 of blood) between SM and RM, hence we constrained the baseline proliferation levels of each species to be the average of those Ki67 values. Importantly, the maximal proliferation level of SM (i.e. intercept of 5.8%; confidence intervals 4.1–7.4%) was much lower than that for RM (11%; confidence intervals 7.8–14%) and HIV-infected humans (22%; confidence intervals 12.4–31.5%). The confidence intervals of the intercepts of the proliferation function for SM and pathogenic hosts (HIV, RM) did not overlap. We used generalized nonlinear regression in order to demonstrate that the intercept C + D is indeed significantly different between SM and RM, and between SM and HIV. We compared the goodness-of-fit when intercept and sensitivity are shared between the datasets to the goodness-of-fit when they are determined independently for each dataset, and in both cases the fit with different intercepts was significantly better (in both cases p < 0.0001).
In addition, we noted that the fitting of the curves of SM and RM included both cross-sectional and longitudinal data, which raises the potential issue of autocorrelation within the data. In order to make sure that this did not bias the results, we repeated our analysis using only one data point per animal (choosing the point with lowest CD4+ T cell number, where there was more than one point). This did not appreciably affect the estimated relationship between CD4+ T cell count and Ki67 expression.
The correlation between CD4+ T cell numbers and proliferation has also been attributed to direct viral effects or antigenic stimulation (since virus level and CD4+ T cell number are also correlated; Cohen Stuart et al. 2000; Hazenberg et al. 2000; Biancotto et al. 2008). Therefore, we also analysed the relationship between viral load and CD4+ T cell proliferation in non-pathogenic (SM) and pathogenic (RM) infections. We found that the level of proliferation of CD4+ T cells was not significantly correlated to viral load in SM (Spearman r = 0.2260, p = 0.0749), and was surprisingly significantly negatively correlated to viral load in RM (Spearman r = −0.5397, p = 0.0001). The negative correlation seems to be driven mainly by the acute phase of infection around the peak viral load, when the viral levels are very high without a corresponding high level of CD4+ T cell activation. When we took into account only the chronic phase (56 days post-infection or later), there was no significant correlation between viral load and the level of proliferation of CD4+ T cells in either SM or RM (p = 0.132 and p = 0.0896, respectively).
Conversely, if CD4+ T cell proliferation is driven by the decrease in CD4+ T cell numbers and not viral load, this suggests that a higher proportion of CD4+ T cells will proliferate at low CD4+ T cell numbers even without infection. We therefore analysed the relationship between CD4+ T cell proliferation and CD4+ T cell number in uninfected RM and SM whose CD4+ T cells were depleted using anti-CD4 antibodies (figure 2). We observed a strong negative correlation between CD4+ T cell number and proliferation in the complete absence of virus (Spearman r = −0.6974 and −0.6998 for antibody-depleted uninfected SM and RM, respectively, and p < 0.0001 for both), supporting the interpretation that the decrease in CD4+ T cell numbers can be an independent driver of T cell proliferation. Interestingly, the level of proliferation of CD4+ T cells as a function of CD4+ T cell number appeared to be even higher in uninfected animals treated with antibodies when compared with the SIV-infected animals (table 2). Generalized nonlinear regression analysis shows that the intercept C + D is significantly different between antibody-depleted and infected SM (p < 0.0001), as well as between antibody-depleted and infected RM (p < 0.0001). A limitation of our analysis of antibody-depleted animals is that it relies on frequent measurements of CD4+ T cell number and proliferation from only three animals in each group. As the longitudinal data for these animals were pooled together, temporal autocorrelation may exist in the analysis. Nevertheless, the increase in CD4+ T cell proliferation in response to antibody depletion of CD4+ T cells suggests that this can be driven by a homeostatic response to the loss of CD4+ T cells, and occurs independent of SIV infection.
Table 2.
Summary of statistics and parameters of the exponential fit to figure 2.
| Ab-depleted SIV-rhesus macaques (CI) | Ab-depleted SIV-sooty mangabeys (CI) | |
|---|---|---|
| Spearman r | −0.6998 | −0.6974 |
| p | <0.0001 | <0.0001 |
| intercept C + D (%) | 27.2 (21.5–32.8) | 16.6 (13.5–19.7) |
| sensitivity k | 175.1 (107.0–481.8) | 202.8 (139.0–374.7) |
| baseline D (%) | 7.6 | 2.6 |
(b). Modelling the impact of T cell proliferation on infection
Since pathogenic and non-pathogenic infections differ significantly in the way proliferation increases at low CD4+ T cell levels, we used modelling to understand how the relationship between CD4+ T cell number and proliferation alone may impact the outcome of infection (figure 3a), even when the virus–host interactions that are influenced by immune response (contained in the ‘infection parameters’ β, δI, p and c) are the same in both hosts.
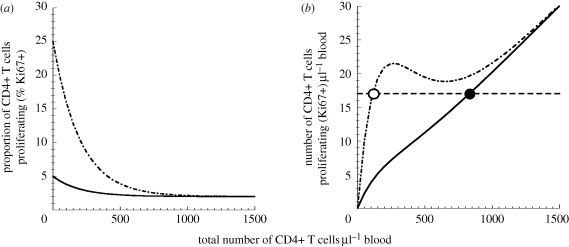
Figure 3.
Higher proliferation leads to fewer remaining uninfected CD4+ T cells. (a) The relationship between the total number of uninfected CD4+ T cells and the proportion of these cells that are proliferating. Curves for pathogenic infection (peak proliferation 25%) are shown as dashed-dotted lines, and those for non-pathogenic infection (peak proliferation 5%) are shown as solid lines. (b) The relationship between the total number of uninfected CD4+ T cells and the number of proliferating CD4+ T cells (i.e. obtained from the curve in (a) by multiplying the proportion of proliferating cells by the number of uninfected cells). The equilibrium number of proliferating (i.e. susceptible) uninfected cells is 17 cells µl−1 of blood. This level is reached when the total pool (proliferating and non-proliferating) of uninfected CD4+ T cells is 847 cells µl−1 of blood for non-pathogenic infection, as shown by the solid line (with 2.0% proliferating), and 117 cells µl−1 of blood for pathogenic infections, as shown by the dashed-dotted lines (with 14.8% proliferating). Equilibrium is shown as dashed line.
We first modelled the disease course in a pathogenic ‘human-like’ infection, where the maximal proliferation of CD4+ T cells was 25 per cent. This resulted in an acute phase of infection leading to an equilibrium level of 17 uninfected proliferating CD4+ T cells per microlitre of blood (figure 3b). We then modelled the same infection in a non-pathogenic ‘SM-like’ host, where the maximal CD4+ T cell proliferation was only 5 per cent. This decrease in the proliferative response to CD4+ T cell loss in the SM-like host led to a large increase in the total number of uninfected CD4+ T cells present in chronic infection (847 versus 117 cells per microlitre of blood; solid and dashed-dotted lines in figure 3b). Upon analysis, the mechanisms for this are easily understood. In our model, only the proliferating CD4+ T cells are susceptible to infection, whereas the non-proliferating cells are not. The number of these proliferating (i.e. susceptible) cells at the viral set point depends only on infection parameters (Peqm = δIc/pβ), and is independent of the host proliferation level. Thus, the same equilibrium number of uninfected proliferating CD4+ T cells (17 cells per microlitre of blood) is seen in the two hosts (figure 3b). The number of uninfected proliferating cells is maintained at a steady state because any increase in proliferating cells would increase the number of infected cells as well as the viral load. This will in turn infect more proliferating cells, and bring the system back to equilibrium. However, the different rates of CD4+ T cell proliferation in HIV and SM lead to this equilibrium level being achieved in two different ways: in the low proliferation (SM-like) situation, the 17 proliferating cells represent 2 per cent of a total pool (proliferating and non-proliferating) of 847 uninfected CD4+ T cells per microlitre of blood. However, in the high proliferation (HIV in humans or SIV in RMs) situation, this same number of proliferating cells is reached when the total pool is 117 uninfected CD4+ T cells per microlitre of blood (of which 14.8% are proliferating). For identical infection parameters, changing only the maximal level of proliferation reproduces the observed CD4+ T cell behaviours of SM and HIV infection. Thus, our model shows that, paradoxically, a higher level of CD4+ T cell proliferation ultimately leads to fewer remaining uninfected CD4+ T cells (figure 4a).
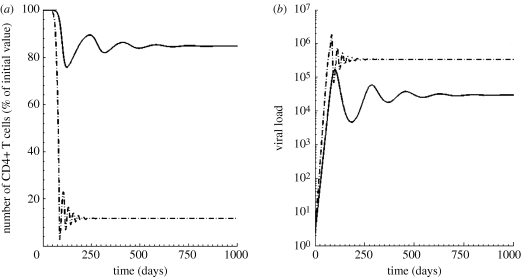
Figure 4.
Course of infection with different levels of proliferation: using identical infection parameters but varying the maximal proliferation, we found that a higher proliferation of CD4+ T cells at low CD4+ T cell counts (dashed-dotted line, pathogenic infection) led to fewer remaining uninfected CD4+ T cells and higher viral loads in chronic infection. By contrast, low proliferation (solid line, non-pathogenic infection) led to preservation of uninfected CD4+ T cells. The viral load in the non-pathogenic model was lower than the pathogenic, consistent with the experimentally observed difference in viral loads between RMs and SMs. Parameters: λ = 10 cells µl−1 d−1, ρ = 0.7 d−1, k = 200 cells µl−1, β = 3.3 × 10−6 ml per RNA copy per day, δI = 0.8 d−1, p = 2.8 × 105 RNA copies per cell per day, c = 20 d−1.
In the model, the massive shrinkage of the pool of all uninfected CD4+ T cells in the pathogenic host was accompanied by an increase in viral load (3.4 × 105 compared with 2.9 × 104 in the non-pathogenic host), when starting with identical infection parameters (figure 4b). This difference in set-point viral loads is similar to the difference in viral loads between RMs and SMs seen in our experimental datasets (average viral loads between 50 and 150 days post-infection are 2.9 × 105 and 3.0 × 104 for RMs and SMs, respectively). Thus, the modelled viral loads closely fit the experimental data from infected SMs and RMs when identical infection parameters are used.
It has been suggested that the maintenance of CD4+ T cells in SMs, compared with RMs and HIV-infected individuals, may be attributed to the longer lifespan of their infected cells or a more effective immune control of the virus. However, the lifespans of infected cells of both SM and humans were found to be in the same order of about 1 day, as deduced from the decline in viral replication following anti-retroviral therapy in both groups (Ho et al. 1995; Wei et al. 1995; Perelson et al. 1996; Silvestri 2005; Gordon et al. 2008). At the same time, previous studies have found no conclusive evidence that the adaptive immune responses of SMs are different from those mounted against pathogenic infections in RMs and humans (Sodora et al. 2009). The high viraemia in naturally infected SMs shows that immune control is incomplete and only exerts a partial immune pressure on the virus (Silvestri 2005; Silvestri et al. 2007; Pandrea et al. 2008b; Paiardini et al. 2009c). Other studies have also shown that the suppression of viral replication by CD8+ T cells in SMs is less or comparable to RMs (Dunham et al. 2006; Sodora et al. 2009). Furthermore, the extent of clonal expansion of SIV-specific CD8+ T cells in SMs, as well as the range of epitopes of SIV that they can recognize, are similar or smaller than those in pathogenic infections in RMs and humans (Dunham et al. 2006; Sodora et al. 2009).
(c). Generality of the model
The model above used an exponential function to characterize the relationship between CD4+ T cell number and the proportion of proliferating cells, and demonstrated how differences in the peak level of CD4+ T cell proliferation can determine disease outcome. In addition to the peak proliferation levels, natural SIV hosts also differ in the shape of the curve (determined by the sensitivity ‘k’), which effectively determines how steeply the CD4+ T cell proliferation level rises to its peak as CD4+ T cell number declines. SMs show little increase in CD4+ T cell proliferation until low CD4+ T cell levels are reached (k = 277.4), compared with RMs (k = 977.5), while MNDs appear not to respond to depletion at all (figure 1 and table 1). We modelled how the shape of the curve affected the outcome, and found that a lower sensitivity led to higher set-point CD4+ T cell levels, even if the peak level of proliferation were the same. This suggests that the sensitivity of the proliferative response to CD4+ T cell depletion (independent of the maximum proliferation level) can also determine the difference between pathogenic and non-pathogenic infection.
The exponential function used to compare CD4+ T cell proliferation in pathogenic and non-pathogenic infection is one of many possible relationships. However, the outcome was not dependent on the use of an exponential function, as different functions (such as the Hill function, or piecewise linear relationship; Hazenberg et al. 2000) for the relationship between CD4+ T cell levels and level of proliferation also produced the same divergent behaviour between pathogenic and non-pathogenic infection (figure 5).
Figure 5.
Functions used to describe the relationship between CD4+ T cell number and proliferation level: Using the data points for RMs as an example, the simple exponential (solid line), Hill function (dashed line) and piecewise linear (dashed-dotted line) functions were fitted to describe the relationship between CD4+ T cell number and proliferation level. However, the same divergent behaviour between pathogenic and non-pathogenic infection is observed regardless of the form of the functions used in the model.
(d). T cell proliferation and disease course in HIV infection
Our experimental data and modelling suggest that the level of CD4+ T cell proliferation is negatively correlated with CD4+ T cell number, and it plays a key role in determining the differences in the outcome between pathogenic HIV/SIV infection in RMs and humans, and non-pathogenic SIV infection in SMs and MNDs. Even with similar viral cytopathicity, a low level of CD4+ T cell proliferation leads to higher set-point total CD4+ T cell levels in natural hosts.
CD4+ T cell proliferation may play a dual role during infection. On the one hand, increased proliferation of CD4+ T cells in response to low CD4+ T cell numbers acts to replace CD4+ T cells and restore homeostasis. On the other hand, increasing the proliferation of CD4+ T cells also provides more cells that are susceptible to infection. The high level of CD4+ T cell proliferation in RMs essentially ‘fuels the fire’ of infection by producing a large proportion of proliferating cells that are susceptible to SIV infection. Conversely, although the low homeostatic proliferation in SM slows the replacement of CD4+ T cells, it leads to a higher equilibrium level of uninfected cells during infection.
Our studies of the relationship between CD4+ T cell number and proliferation in HIV-infected individuals involved pooling data from a number of subjects to determine a ‘population average response’ (figure 1d). The significant deviation observed in the data suggests that individual variation in the proliferation level of CD4+ T cells in the human population may be a contributor to the observed different rates of disease progression in HIV. This is consistent with the observation that CD4+ T cell activation and proliferation is an independent predictor of CD4+ T cell depletion in HIV infection (i.e. those with higher activation tend to end up with low set-point levels of CD4+ T cells; Carbone et al. 2000). Studies of natural hosts of SIV infection can therefore provide important insights into the pathogenesis of HIV infection, and suggest that variation in the proliferation level of CD4+ T cells may be an important determinant of outcome. Future work is required to determine whether manipulation of the level of CD4+ T cell proliferation may provide a potential mechanism to improve outcomes in HIV.
Acknowledgements
Financial support was provided by National Health and Medical Research Council (Australia), the Australian Research Council and NIH (USA) grants (AI-66998 and HL-75766, to G.S.).
References
- Biancotto A., Iglehart S. J., Vanpouille C., Condack C. E., Lisco A., Ruecker E., Hirsch I., Margolis L. B., Grivel J. C.2008HIV-1 induced activation of CD4+ T cells creates new targets for HIV-1 infection in human lymphoid tissue ex vivo. Blood 111, 699–704 10.1182/blood-2007-05-088435 (doi:10.1182/blood-2007-05-088435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J. M., et al. 2004CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200, 749–759 10.1084/jem.20040874 (doi:10.1084/jem.20040874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J. M., et al. 2006Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12, 1365–1371 10.1038/nm1511 (doi:10.1038/nm1511) [DOI] [PubMed] [Google Scholar]
- Brenchley J. M., et al. 2008Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112, 2826–2835 10.1182/blood-2008-05-159301 (doi:10.1182/blood-2008-05-159301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard S. R., Staprans S. I., White R., Whitehead E. M., Feinberg M. B., Allan J. S.2001Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75, 2262–2275 10.1128/JVI.75.5.2262-2275.2001 (doi:10.1128/JVI.75.5.2262-2275.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone J., Gil J., Benito J. M., Navarro J., Munoz-Fernandez A., Bartolome J., Zabay J. M., Lopez F., Fernandez-Cruz E.2000Increased levels of activated subsets of CD4 T cells add to the prognostic value of low CD4 T cell counts in a cohort of HIV-infected drug users. AIDS 14, 2823–2829 10.1097/00002030-200012220-00003 (doi:10.1097/00002030-200012220-00003) [DOI] [PubMed] [Google Scholar]
- Chakrabarti L. A., et al. 2000Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74, 1209–1223 10.1128/JVI.74.3.1209-1223.2000 (doi:10.1128/JVI.74.3.1209-1223.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Stuart J. W., Hazebergh M. D., Hamann D., Otto S. A., Borleffs J. C., Miedema F., Boucher C. A., de Boer R. J.2000The dominant source of CD4+ and CD8+ T-cell activation in HIV infection is antigenic stimulation. J. Acquir. Immune Defic. Syndr. 25, 203–211 [DOI] [PubMed] [Google Scholar]
- Douek D. C., et al. 2001Evidence for increased T cell turnover and decreased thymic output in HIV infection. J. Immunol. 167, 6663–6668 [DOI] [PubMed] [Google Scholar]
- Dunham R., et al. 2006The AIDS resistance of naturally SIV-infected sooty mangabeys is independent of cellular immunity to the virus. Blood 108, 209–217 10.1182/blood-2005-12-4897 (doi:10.1182/blood-2005-12-4897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham R. M., et al. 2008CD127 and CD25 expression defines CD4+ T cell subsets that are differentially depleted during HIV infection. J. Immunol. 180, 5582–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engram J. C., et al. 2009Vaccine-induced, simian immunodeficiency virus-specific CD8+ T cells reduce virus replication but do not protect from simian immunodeficiency virus disease progression. J. Immunol. 183, 706–717 10.4049/jimmunol.0803746 (doi:10.4049/jimmunol.0803746) [DOI] [PubMed] [Google Scholar]
- Gordon S. N., et al. 2008Short-lived infected cells support virus replication in sooty mangabeys naturally infected with simian immunodeficiency virus: implications for AIDS pathogenesis. J. Virol. 82, 3725–3735 10.1128/JVI.02408-07 (doi:10.1128/JVI.02408-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe M., Reay E., Sankaran S., Prindiville T., Flamm J., McNeil A., Dandekar S.2003Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77, 11708–11717 10.1128/JVI.77.21.11708-11717.2003 (doi:10.1128/JVI.77.21.11708-11717.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn B. H., Shaw G. M., De Cock K. M., Sharp P. M.2000AIDS as a zoonosis: scientific and public health implications. Science 287, 607–614 10.1126/science.287.5453.607 (doi:10.1126/science.287.5453.607) [DOI] [PubMed] [Google Scholar]
- Hazenberg M. D., Stuart J. W., Otto S. A., Borleffs J. C., Boucher C. A., de Boer R. J., Miedema F., Hamann D.2000T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active antiretroviral therapy (HAART). Blood 95, 249–255 [PubMed] [Google Scholar]
- Hirsch V. M., Olmsted R. A., Murphey-Corb M., Purcell R. H., Johnson P. R.1989An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339, 389–392 10.1038/339389a0 (doi:10.1038/339389a0) [DOI] [PubMed] [Google Scholar]
- Ho D. D., Neumann A. U., Perelson A. S., Chen W., Leonard J. M., Markowitz M.1995Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 10.1038/373123a0 (doi:10.1038/373123a0) [DOI] [PubMed] [Google Scholar]
- Klatt N. R., et al. 2008Availability of activated CD4 T cells dictates the level of viremia in naturally SIV-infected sooty mangabeys. J. Clin. Invest. 118, 2039–2049 10.1172/JCI33814 (doi:10.1172/JCI33814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. I., Ciccone E. J., Read S. W., Asher A., Pitts R., Douek D. C., Brenchley J. M., Sereti I.2009Evidence for translocation of microbial products in patients with idiopathic CD4 lymphocytopenia. J. Infect. Dis. 199, 1664–1670 10.1086/598953 (doi:10.1086/598953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A. L.2001The dependence of viral parameter estimates on the assumed viral life cycle: limitations of studies of viral load data. Proc. Biol. Sci. 268, 847–854 10.1098/rspb.2000.1572 (doi:10.1098/rspb.2000.1572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil J. J., Douek D. C., Hill B., Nishimura Y., Martin M., Roederer M.2005Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434, 1093–1097 10.1038/nature03501 (doi:10.1038/nature03501) [DOI] [PubMed] [Google Scholar]
- Mohri H., et al. 2001Increased turnover of T lymphocytes in HIV-1 infection and its reduction by antiretroviral therapy. J. Exp. Med. 194, 1277–1287 10.1084/jem.194.9.1277 (doi:10.1084/jem.194.9.1277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M., May R. M.2000. In Virus dynamics: mathematical principles of immunology and virology. Oxford, UK: Oxford University Press [Google Scholar]
- Onanga R., et al. 2002High levels of viral replication contrast with only transient changes in CD4(+) and CD8(+) cell numbers during the early phase of experimental infection with simian immunodeficiency virus SIVmnd-1 in Mandrillus sphinx. J. Virol. 76, 10256–10263 10.1128/JVI.76.20.10256-10263.2002 (doi:10.1128/JVI.76.20.10256-10263.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M., et al. 2005Loss of CD127 expression defines an expansion of effector CD8+ T cells in HIV-infected individuals. J. Immunol. 174, 2900–2909 [DOI] [PubMed] [Google Scholar]
- Paiardini M., et al. 2009aBone marrow-based homeostatic proliferation of mature T cells in nonhuman primates: implications for AIDS pathogenesis. Blood 113, 612–621 10.1182/blood-2008-06-159442 (doi:10.1182/blood-2008-06-159442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M., Hoffman J., Cervasi B., Ortiz A. M., Stroud F., Silvestri G., Wilson M. E.2009bT-cell phenotypic and functional changes associated with social subordination and gene polymorphisms in the serotonin reuptake transporter in female rhesus monkeys. Brain Behav. Immun. 23, 286–293 10.1016/j.bbi.2008.10.006 (doi:10.1016/j.bbi.2008.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M., Pandrea I., Apetrei C., Silvestri G.2009cLessons learned from the natural hosts of HIV-related viruses. Annu. Rev. Med. 60, 485–495 10.1146/annurev.med.60.041807.123753 (doi:10.1146/annurev.med.60.041807.123753) [DOI] [PubMed] [Google Scholar]
- Pandrea I., et al. 2008aSIVagm dynamics in African Green Monkeys. J. Virol. 82, 3713–3724 10.1128/JVI.02402-07 (doi:10.1128/JVI.02402-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandrea I., Sodora D. L., Silvestri G., Apetrei C.2008bInto the wild: simian immunodeficiency virus (SIV) infection in natural hosts. Trends Immunol. 29, 419–428 10.1016/j.it.2008.05.004 (doi:10.1016/j.it.2008.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson A. S.2002Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2, 28–36 10.1038/nri700 (doi:10.1038/nri700) [DOI] [PubMed] [Google Scholar]
- Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D.1996HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586 10.1126/science.271.5255.1582 (doi:10.1126/science.271.5255.1582) [DOI] [PubMed] [Google Scholar]
- Ribeiro R. M., Mohri H., Ho D. D., Perelson A. S.2002In vivo dynamics of T cell activation, proliferation, and death in HIV-1 infection: why are CD4+ but not CD8+ T cells depleted? Proc. Natl Acad. Sci. USA 99, 15572–15577 10.1073/pnas.242358099 (doi:10.1073/pnas.242358099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro R. M., Hazenberg M. D., Perelson A. S., Davenport M. P.2006Naive and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J. Virol. 80, 802–809 10.1128/JVI.80.2.802-809.2006 (doi:10.1128/JVI.80.2.802-809.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G.2005Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS? J. Med. Primatol. 34, 243–252 10.1111/j.1600-0684.2005.00122.x (doi:10.1111/j.1600-0684.2005.00122.x) [DOI] [PubMed] [Google Scholar]
- Silvestri G., Sodora D. L., Koup R. A., Paiardini M., O'Neil S. P., McClure H. M., Staprans S. I., Feinberg M. B.2003Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18, 441–452 [DOI] [PubMed] [Google Scholar]
- Silvestri G., Fedanov A., Germon S., Kozyr N., Kaiser W. J., Garber D. A., McClure H., Feinberg M. B., Staprans S. I.2005Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and non-natural rhesus macaque hosts. J. Virol. 79, 4043–4054 10.1128/JVI.79.7.4043-4054.2005 (doi:10.1128/JVI.79.7.4043-4054.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri G., Paiardini M., Pandrea I., Lederman M. M., Sodora D. L.2007Understanding the benign nature of SIV infection in natural hosts. J. Clin. Invest. 117, 3148–3154 10.1172/JCI33034 (doi:10.1172/JCI33034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit-McBride Z., Mattapallil J. J., McChesney M., Ferrick D., Dandekar S.1998Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 72, 6646–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodora D. L., et al. 2009Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat. Med. 15, 861–865 10.1038/nm.2013 (doi:10.1038/nm.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A. E., Carneiro J., Meier-Schellersheim M., Grossman Z., Victorino R. M.2002CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 169, 3400–3406 [DOI] [PubMed] [Google Scholar]
- Sumpter B., et al. 2007Correlates of preserved CD4(+) T cell homeostasis during natural, nonpathogenic simian immunodeficiency virus infection of sooty mangabeys: implications for AIDS pathogenesis. J. Immunol. 178, 1680–1691 [DOI] [PubMed] [Google Scholar]
- Veazey R., Ling B., Pandrea I., McClure H., Lackner A., Marx P.2003Decreased CCR5 expression on CD4+ T cells of SIV-infected sooty mangabeys. AIDS Res. Hum. Retroviruses 19, 227–233 10.1089/088922203763315731 (doi:10.1089/088922203763315731) [DOI] [PubMed] [Google Scholar]
- Vlahakis S. R., Algeciras-Schimnich A., Bou G., Heppelmann C. J., Villasis-Keever A., Collman R. G., Paya C. V.2001Chemokine-receptor activation by env determines the mechanism of death in HIV-infected and uninfected T lymphocytes. J. Clin. Invest. 107, 207–215 10.1172/JCI11109 (doi:10.1172/JCI11109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., et al. 1995Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122 10.1038/373117a0 (doi:10.1038/373117a0) [DOI] [PubMed] [Google Scholar]
- Yates A., Stark J., Klein N., Antia R., Callard R.2007Understanding the slow depletion of memory CD4+ T cells in HIV infection. PLoS Med. 4, e177. 10.1371/journal.pmed.0040177 (doi:10.1371/journal.pmed.0040177) [DOI] [PMC free article] [PubMed] [Google Scholar]