Abstract
When previously isolated populations meet and mix, the resulting admixed population can benefit from several genetic advantages, including increased genetic variation, the creation of novel genotypes and the masking of deleterious mutations. These admixture benefits are thought to play an important role in biological invasions. In contrast, populations in their native range often remain differentiated and frequently suffer from inbreeding depression owing to isolation. While the advantages of admixture are evident for introduced populations that experienced recent bottlenecks or that face novel selection pressures, it is less obvious why native range populations do not similarly benefit from admixture. Here we argue that a temporary loss of local adaptation in recent invaders fundamentally alters the fitness consequences of admixture. In native populations, selection against dilution of the locally adapted gene pool inhibits unconstrained admixture and reinforces population isolation, with some level of inbreeding depression as an expected consequence. We show that admixture is selected against despite significant inbreeding depression because the benefits of local adaptation are greater than the cost of inbreeding. In contrast, introduced populations that have not yet established a pattern of local adaptation can freely reap the benefits of admixture. There can be strong selection for admixture because it instantly lifts the inbreeding depression that had built up in isolated parental populations. Recent work in Silene suggests that reduced inbreeding depression associated with post-introduction admixture may contribute to enhanced fitness of invasive populations. We hypothesize that in locally adapted populations, the benefits of local adaptation are balanced against an inbreeding cost that could develop in part owing to the isolating effect of local adaptation itself. The inbreeding cost can be revealed in admixing populations during recent invasions.
Keywords: population admixture, local adaptation, inbreeding depression, biological invasions
1. Introduction
Biological invasions are an ecological and economic concern but they also present interesting natural experiments that may tell us something about how basic ecological and evolutionary processes shape patterns of diversity within species and communities. Single species are placed outside their recent evolutionary context, and some species apparently benefit and become noxious invaders in their new range while others remain unsuccessful. In search of the factors that determine success or failure, the role of post-introduction evolution is receiving increased attention [1–4]. Because population bottlenecks during early introduction can sharply reduce their genetic variation, it may be predicted that introduced species have limited potential to adapt to their novel environments. However, the adaptive potential of some invaders turns out to be considerable. For instance, invasive Hypericum canariense populations show evidence of adaptive differentiation within 50 years after introduction despite introduction bottlenecks [5]. And invasive populations can even show increased genetic variation compared with native range populations [6].
One factor that contributes to the adaptive potential of introduced species is intraspecific hybridization (admixture) in the introduced range between populations that were introduced from different source locations from the native range. Genetic marker studies show that multiple introductions from different source populations are common (reviewed in [5]), and these different populations can meet and establish hybrid populations in the introduced range [6–10]. Such admixture offers many benefits to invaders [11]. First, it increases standing levels of population genetic variation that natural selection can act upon. Second, recombination between genotypes from different source populations can create novel genotypes with new combinations of traits. This may be especially important in the face of novel selection pressures that invaders may encounter in their new range. Third, it allows for masking of deleterious mutations (inbreeding load) that had built up within isolated populations.
These admixture benefits can be important contributors to population fitness. But if population admixture plays an important role in the evolutionary potential and invasive success of introduced populations, then the following question presents itself: why do we not see admixture and subsequent fitness boosts happening all the time in populations within the original native range? Native range populations are often well differentiated (e.g. [12]) and frequently show some level of inbreeding depression [13], suggesting that admixture is not ubiquitous. There are several reasons why admixture might play a more important role in invasive than in native populations. First, obviously, vectors of transcontinental transportation might promote contact between source populations that do not readily come in contact in the native range, for instance by ships arriving at the same port. Second, the benefits of admixture may be larger for populations that experienced a recent bottleneck or that face novel selection pressures. But, surely, native populations could benefit too from increased variation, novel genotypes or heterosis. For instance, admixture could instantly lift the inbreeding depression that is often observed in native populations [13]. So, what prevents native populations from reaping the benefits of admixture?
Here, we propose that local adaptation acts as an isolating force that selects against admixture in the native range. In contrast, a temporary absence of an established pattern of local adaptation in recently introduced populations permits those populations to admix freely in their novel range. In a locally adapted population, admixture can be selected against because it can make the population less well adapted to the environment. Because of this, local adaptation contributes to population isolation and promotes inbreeding among relatives. Some level of inbreeding depression may develop as a consequence, but if this cost is smaller than the fitness benefits of local adaptation, then admixed individuals will continue to be selected against. The balance is changed in the absence of local adaptation in the introduced range, where admixture may more often provide an instant fitness boost when genetically distinct introduced populations come into contact.
2. Costs and benefits of population admixture
In order to explore the interaction between local adaptation and population admixture, we first need to distinguish the various genetic costs and benefits that are associated with admixture (see [14] for a more in-depth discussion). The costs of admixture include an environment-dependent cost (dilution of locally adapted genomes) and an intrinsic genetic cost (hybrid breakdown). The latter is often attributed to the disruption of co-adapted gene complexes that build up in isolated populations and that result in genetic incompatibilities in recombinant individuals, but other types of incompatibilities can be involved too [15,16]. One way to think of this is as adaptation of genes to their genetic background, not necessarily to the environment. The cost of diluting locally adapted genomes is expressed in the native field environment to which populations have adapted. In contrast, the intrinsic genetic cost of genetic incompatibilities is expressed also in other environments.
The benefits of admixture include increased genetic variation and, through segregation and recombination, the formation of individuals with novel trait combinations. These factors are potentially advantageous in both novel and native ranges, but their impact may be modest in the native range if local adaptation is well established (approaching an optimal sorting of available genotypes over the adaptive landscape). In contrast, both factors can be a considerable benefit to recently introduced populations that are genetically impoverished owing to introduction bottlenecks and that may face environmental conditions to which none of the genotypes from the native range are (pre-) adapted.
The heterosis (hybrid vigour) advantage of admixture concerns the masking of the genetic load that has built up in isolated populations owing to inbreeding effects. Mildly deleterious mutations that are expressed in the homozygous state are masked in hybrid heterozygotes [14]. Overdominance, or the fitness advantage of a heterozygous locus over either of the two parental homozygous loci, also contributes to heterosis, but this is generally considered less important than the masking of deleterious recessive alleles [17,18]. Heterosis is relevant in both native and novel environments, but is probably more important in novel environments because of the increased level of inbreeding in initial small populations during early invasion. Heterosis is often considered a transient phenomenon because heterozygous hybrids become increasingly homozygous in subsequent generations (e.g. [4]). But even short-lived heterosis can have a dramatic effect on the chance that introduced populations establish successfully [19].
3. Does local adaptation prevent population admixture in the native range?
A large body of evidence from reciprocal transplant experiments shows that local adaptation, or the superior performance of genotypes grown in their home environment compared with genotypes that were introduced from elsewhere, is a common phenomenon [20,21]. Although not ubiquitous, it is considered an important factor that structures patterns of genetic diversity within species. Selection against non-local genotypes hampers their establishment in non-native environments and contributes to reproductive isolation of populations [22]. This reduces the opportunities for successful population admixture.
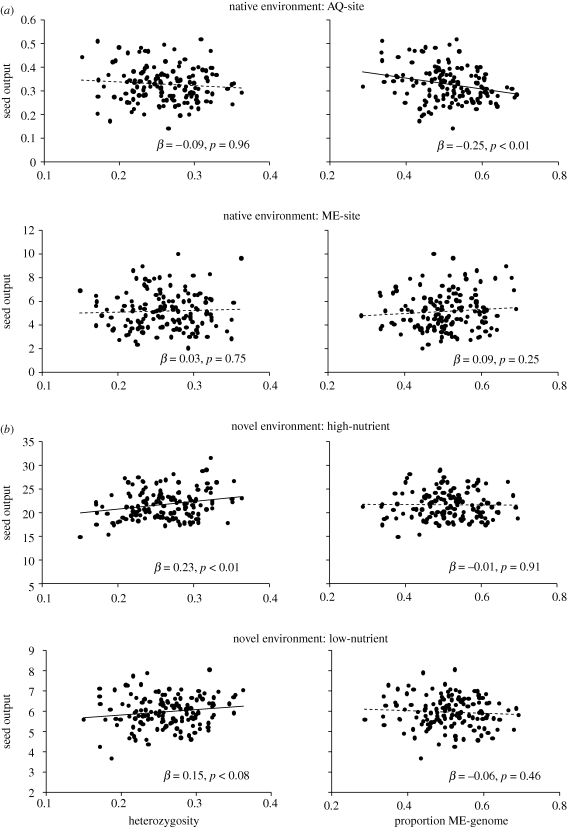
Despite selection against non-local genotypes, successful matings between local and non-local genotypes do occur. What about their hybrid offspring? Increased fitness of hybrid progeny could promote population admixture even when there is selection against the non-local parents. This question can be explored in reciprocal transplant experiments that include recombinant progeny derived from between-population crosses. One such study was performed in wild barley [23]. F3 offspring families from a cross between two locally adapted populations were grown in the native field environments of both parental populations, and it was shown that recombinant performance tended to decrease with increasing amounts of non-local DNA in their genomes (figure 1a). While some individual recombinant genotypes can still enjoy high fitness, this indicates that there is overall selection against dilution of the locally adapted genome.
Figure 1.
Fitness effects of heterozygosity and genome dilution of locally adapted wild barley in native and novel environments. A cross was made between plants from two locally adapted wild barley populations (AQ and ME) and performance of recombinant F3 families (derived by selfing from the F1) was evaluated at the natural field sites of the AQ and ME populations (a, native environments) and in an experimental garden at different nutrient levels (b, novel environments). Using genome-wide markers, the proportion of heterozygous loci (one allele derived from each parent) and the proportion of genome content derived from either parent were estimated for each of 140 F3 families (proportion ME = 1 − (proportion AQ)). Seed output (total biomass [g] of all seeds produced) was positively associated with heterozygosity only in the novel environments, while genome dilution had an effect only under native field conditions where a high proportion of AQ genome was positively associated with seed output at the AQ site but not at the ME site. Regression coefficients and p-values are from simple linear regressions (data from [23], see that paper for details).
Thus, even in the face of genetic exchange between populations, local adaptation can select against population admixture, and hampers the free dispersal and mixing of genotypes throughout the species' distribution range. Of course, the isolating effect of local adaptation is not absolute. Mixing and successful introgressions do occur, as is for instance suggested by generally stronger population differentiation for genome regions that are under local selection than for genome regions that are not involved in local adaptation [24]. But, importantly, local adaptation can act as a general barrier that reduces gene flow. Its isolating effect has been demonstrated to affect also neutral regions of the genome that diverge owing to genetic drift as a consequence (isolation by adaptation; [25,26]).
4. Does local adaptation mask an inbreeding depression cost?
Local adaptation evolves because it promotes population fitness. But if local adaptation leads to population isolation, then some level of inbreeding depression may be an unavoidable and undesired consequence. Isolation promotes matings between local relatives, leading to increased levels of homozygosity and inbreeding depression via the expression of deleterious recessive alleles [16]. Thus, isolation by local adaptation could be a contributing factor to inbreeding depression, but the costs of inbreeding can be masked by the fitness benefits of local adaptation. In this view, local adaptation might be considered as a type of inbreeding where the beneficial effects of fixing local adaptation genes are offset to some degree by the negative effects of inbreeding depression caused by other loci.
Is there empirical evidence that the benefits of local adaptation mask a hidden cost of inbreeding, which develops in part as an indirect consequence of local adaptation? Natural populations often suffer from inbreeding depression owing to isolation, as demonstrated in many species in common garden experiments that show increased fitness of offspring from between-population crosses compared with within-population crosses [13,27–29]. The key question is to what extent population isolation is maintained by local adaptation relative to other isolating factors, such as geographical distance. This question can be tackled empirically by comparing patterns of genetic, geographical and ecological trait divergence across populations. To date, work in this area suggests that isolation by adaptation can contribute significantly to genetic isolation [26,30].
Direct evidence that local adaptation masks a cost of inbreeding comes from the wild barley experiment described above. Recombinant offspring genotypes that were hybrid (heterozygous) at more loci enjoyed higher fitness in novel environments (figure 1b). This heterosis effect suggests that parental (homozygous) genomes suffered from inbreeding depression. In wild barley, inbreeding depression could be a consequence of high natural rates of self-fertilization [31]. But, strikingly, the positive effect of heterozygosity was visible only in novel environments but not in the native field environments. Here, only dilution of the local genome, not heterozygosity, was associated with fitness (figure 1a). Thus, parental genotypes are both locally adapted and suffer from inbreeding depression, but owing to the overriding effect of local adaptation, it is more important to have undiluted local genomes than to release the inbreeding depression burden. The advantage of being locally adapted outweighs the disadvantage of being inbred, but there is a hidden cost of inbreeding.
5. Biological invasions: loss of local adaptation shifts the cost–benefit ratio of admixture
When populations are introduced to an entirely novel range, a pattern of local adaptation will not instantly be present but introduced populations will initially be distributed rather randomly. For instance, recently introduced Verbascum thapsus populations seem to show a pattern of local mal-adaptation in their novel range [5]. The absence of local adaptation removes a barrier to admixture because there is no selection against dilution of local genomes. Compared with the native range, the benefits of admixture are probably higher in the new range (§1) and now the costs are also much smaller. This shifted balance can permit introduced populations, unlike native populations, to freely benefit from admixture.
If local adaptation masks the negative effects of inbreeding in the native range, then admixture in the novel range can instantly lift this inbreeding cost via heterosis and the sheltering of the genetic load. The shifted balance in the costs and benefits of admixture therefore predicts that the population genetic forces that are involved in attaining high fitness are fundamentally altered during the early invasion process. In native range populations, high fitness may be achieved by maintaining local adaptation and balancing its advantage against the costs of inbreeding, and as long as the net fitness effect is positive it does not pay to admix much. In recent invaders, in contrast, high fitness may be achieved most easily by unconstrained admixture and lifting the inbreeding depression cost. This heterosis benefit may not necessarily be sufficient to explain invasiveness, but it could provide an important fitness boost compared with non-admixed populations and may play a significant role during the establishment phase of recent invaders [19].
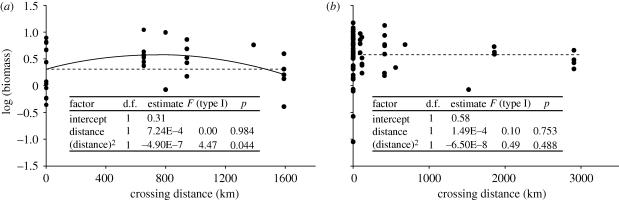
Testing the effects of local adaptation and inbreeding depression on the cost–benefit balance of admixture, and how this differs between native and introduced populations, requires common garden and reciprocal transplant experiments that assess performance of parental populations and inter-population crosses in both the native and introduced ranges. To our knowledge, such a comprehensive analysis has not yet been done for any species. But recent work in native and introduced Silene latifolia (the white campion) reveals some interesting differences in the effects of admixture and the levels of inbreeding depression in native versus introduced populations. Silene latifolia occurs in differentiated populations in its native European range [32,33] and was introduced to North America during the late eighteenth or early nineteenth century [10]. Genetic evidence shows that multiple introductions from different source populations took place and that at least some population admixture occurred in the introduced range [10]. Introduced populations tend to have higher scores for fitness and performance-related traits than populations from the native range, as consistently demonstrated in common garden experiments that were carried out in both the native range [34] and the introduced range [35]. Wolfe et al. [36] extended these common garden experiments and evaluated offspring from within- and between-population crosses, both from the native and the novel range. A re-analysis of their data reveals that (i) native populations show evidence of inbreeding depression but introduced populations do not, and (ii) introduced populations have higher biomass than natural native populations but their biomass is similar to that of hybrids between native populations from intermediate crossing distances (figure 2). In these common garden experiments, inbreeding depression in native populations is expressed as higher biomass of between-population crosses than within-population crosses. There is a penalty of outbreeding when parental populations are too far apart, which has been documented also in other S. latifolia studies from the native range [37]. Intermediate-distance crosses therefore have the highest biomass, which is comparable in magnitude to the biomass levels observed in introduced populations (figure 2).
Figure 2.
Effect of population crossing distance on offspring biomass in native and introduced ranges of S. latifolia. Crosses were made within and between populations from the species' native European range (a, six populations) and the introduced North American range (b, 15 populations). F1 progeny individuals were all raised in one common garden experiment and their biomass is plotted against the geographical distance between their parental populations (within-population crosses: distance = 0 km; between-population crosses: distance > 0 km). Dashed lines indicate the mean value of within-population crosses. Inset tables show results of second-order polynomial regression analysis; p-values indicate significance of sequential addition of linear and quadratic distance terms to the model (type I analysis, proc GLM in SAS, SAS Institute, Cary, NC, USA). Model fit is generally low (native range: r2 = 0.14; introduced range: r2 = 0.01), but in the native range, model fit was significantly improved by adding the quadratic term. Data are from [36]; aboveground biomass at the end of the 2004 growing season, MLBS transplant site.
These results are predicted by the hypothesis that introduced populations, after multiple introductions from different source populations, admix freely in their novel range and benefit from heterosis, whereas native range populations remain differentiated and consequently suffer from inbreeding depression. Any of the admixture benefits could be involved (including increased genetic variation and novel genotypes), but the data suggest that heterosis alone could in principle account for the observed superior fitness of invasive populations. However, the results remain open to alternative interpretations, including invasive spread of incidental high-fitness, non-inbred populations from the native range or post-introduction evolution in the absence of admixture [36]. The data also do not tell us whether isolation-by-adaptation contributed to the observed inbreeding depression of native populations. In fact, this particular species shows good evidence for isolation by distance in its native range [33,38]. Also, some adaptation to novel conditions may have evolved already in the introduced populations as well (for instance, in herbivore defences [34]). Transplantation studies are currently being conducted in the native and introduced ranges that will be able to explicitly examine the extent of local adaptation of Silene populations in both ranges. Additional detailed studies, such as the wild barley experiment (figure 1), are required to unravel the effects of population-level local adaptation on admixture and inbreeding depression in both the introduced and native ranges. The Silene system may provide a good opportunity for such an extended analysis.
6. Conclusion and perspectives
Patterns of local adaptation are established and reinforced over long time scales and are therefore more pronounced in native ranges than in recently introduced ranges. When populations meet, we expect that this readily leads to admixture in introduced ranges, whereas admixture may be restricted in native ranges because there is selection against immigrants and against dilution of locally adapted genomes. Native populations may therefore attain high fitness by maintaining local adaptation and limiting admixture, even when this comes at an inbreeding depression cost. Recently introduced populations, in contrast, may attain high fitness most easily by admixture, which lifts the inbreeding depression cost that had built up in the native range. Additional advantages of admixture, such as the generation of novel genotypes, can subsequently provide further fitness enhancements.
Interestingly, and in agreement with expectations if local adaptation selects against admixture, successful admixture between populations within the native range may occur preferentially during periods of range expansion, when novel territory is colonized that was not occupied already by differentiated populations. In Silene vulgaris, genetic evidence of admixture within the species' native range traces back to post-glacial migration periods from southern refugia to northern latitudes. The genetic footprint of this old admixture is visible in present-day populations, but it is no longer associated with increased fitness (consistent with a non-permanent effect of heterosis). In contrast, more recent admixture in the species' introduced range provides a dramatic fitness boost to invasive populations [39].
(a). What can this tell us about local adaptation?
Native populations often show evidence of inbreeding depression and increased vigour upon outcrossing with other populations [13]. Because adaptation reinforces isolation [26,30], it stands to reason that local adaptation is also a contributing factor to the observed inbreeding depression. This suggests that local adaptation may force populations down a path of compromise, balancing the advantages of fixing local adaptation genes to the disadvantages of local inbreeding. The inbreeding cost will depend on several factors, including the level of local adaptation that is present in the system and the geographical scales at which local adaptation and inbreeding effects are expressed. The genetic architectures of fitness and inbreeding are also relevant: if many genes are involved that are distributed throughout the genome, then it will be difficult to uncouple both factors in early-generation recombinant hybrids. The cost of inbreeding may be hidden in native environments by a larger positive effect of local adaptation. But it will be expressed as a fitness increase of admixed populations in novel environments, such as the introduced range of recent invaders. Thus, we might learn something basic about local adaptation by studying invasive populations that experience a temporary release from local adaptation.
(b). What can this tell us about invasions?
Provided that multiple introductions from different source populations have occurred, the benefits of admixture become freely available to introduced populations that do not yet show a pattern of local adaptation. Because the benefits are potentially large, admixture may play an important role during early invasions. Native populations often show evidence of inbreeding depression [13], and one instant reward of admixture in the introduced range is the release of this genetic burden. Such heterosis effects can contribute significantly to the establishment and early success of invasive species [19]. When tested together in a common garden experiment, invaders can show enhanced fitness-related traits compared with populations from their native range (e.g. [40]). If there is evidence of admixture, the effects of heterosis might be a default explanation for such observations, perhaps providing a null expectation against which other explanations (such as trait evolution) need to be tested.
Acknowledgements
We thank Steve Keller and two anonymous referees for helpful comments on the manuscript. This study was supported by the Netherlands Organisation for Scientific Research (NWO-ALW). This is publication 4829 Netherlands Institute of Ecology (NIOO-KNAW).
References
- 1.Blossey B., Nötzold R.1995Evolution of increased competitive ability in invasive nonindigenous plants—a hypothesis. J. Ecol. 83, 887–889 [Google Scholar]
- 2.Lee C. E.2002Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391 10.1016/S0169-5347(02)02554-5 (doi:10.1016/S0169-5347(02)02554-5) [DOI] [Google Scholar]
- 3.Müller-Schärer H., Schaffner U., Steinger T.2004Evolution in invasive plants: implications for biological control. Trends Ecol. Evol. 19, 417–422 10.1016/j.tree.2004.05.010 (doi:10.1016/j.tree.2004.05.010) [DOI] [PubMed] [Google Scholar]
- 4.Prentis P. J., Wilson J. R. U., Dormontt E. E., Richardson D. M., Lowe A. J.2008Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–294 10.1016/j.tplants.2008.03.004 (doi:10.1016/j.tplants.2008.03.004) [DOI] [PubMed] [Google Scholar]
- 5.Dlugosch K. M., Parker I. M.2008Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449 10.1111/j.1365-294X.2007.03538.x (doi:10.1111/j.1365-294X.2007.03538.x) [DOI] [PubMed] [Google Scholar]
- 6.Lavergne S., Molofsky J.2007Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl Acad. Sci. USA 104, 3883–3888 10.1073/pnas.0607324104 (doi:10.1073/pnas.0607324104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facon B., Pointier J. P., Jarne P., Sarda V., David P.2008High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr. Biol. 18, 363–367 10.1016/j.cub.2008.01.063 (doi:10.1016/j.cub.2008.01.063) [DOI] [PubMed] [Google Scholar]
- 8.Kolbe J. J., Larson A., Losos J. B., de Queiroz K.2008Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biol. Lett. 4, 434–437 10.1098/rsbl.2008.0205 (doi:10.1098/rsbl.2008.0205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenthal D. M., Ramakrishnan A. P., Cruzan M. B.2008Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 17, 4657–4669 10.1111/j.1365-294X.2008.03844.x (doi:10.1111/j.1365-294X.2008.03844.x) [DOI] [PubMed] [Google Scholar]
- 10.Taylor D. R., Keller S. R.2007Historical range expansion determines the phylogenetic diversity introduced during contemporary species invasion. Evolution 61, 334–345 10.1111/j.1558-5646.2007.00037.x (doi:10.1111/j.1558-5646.2007.00037.x) [DOI] [PubMed] [Google Scholar]
- 11.Ellstrand N. C., Schierenbeck K. A.2000Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl Acad. Sci. USA 97, 7043–7050 10.1073/pnas.97.13.7043 (doi:10.1073/pnas.97.13.7043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leinonen T., O'Hara R. B., Cano J. M., Merilä J.2008Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J. Evol. Biol. 21, 1–17 10.1111/j.1420-9101.2007.01445.x (doi:10.1111/j.1420-9101.2007.01445.x) [DOI] [PubMed] [Google Scholar]
- 13.Keller L. F., Waller D. M.2002Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 10.1016/S0169-5347(02)02489-8 (doi:10.1016/S0169-5347(02)02489-8) [DOI] [Google Scholar]
- 14.Hufford K. M., Mazer S. J.2003Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol. Evol. 18, 147–155 10.1016/S0169-5347(03)00002-8 (doi:10.1016/S0169-5347(03)00002-8) [DOI] [Google Scholar]
- 15.Galloway L. F., Etterson J. R.2005Population differentiation and hybrid success in Campanula americana: geography and genome size. J. Evol. Biol. 18, 81–89 10.1111/j.1420-9101.2004.00801.x (doi:10.1111/j.1420-9101.2004.00801.x) [DOI] [PubMed] [Google Scholar]
- 16.Lynch M.1991The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45, 622–629 10.2307/2409915 (doi:10.2307/2409915) [DOI] [PubMed] [Google Scholar]
- 17.Charlesworth D., Willis H. H.2009The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 10.1038/nrg2664 (doi:10.1038/nrg2664) [DOI] [PubMed] [Google Scholar]
- 18.Lynch M., Walsh B.1998Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer [Google Scholar]
- 19.Drake J. M.2006Heterosis, the catapult effect and establishment success of a colonizing bird. Biol. Lett. 2, 304–307 10.1098/rsbl.2006.0459 (doi:10.1098/rsbl.2006.0459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi J., et al. 2001Local adaptation enhances performance of common plant species. Ecol. Lett. 4, 536–544 10.1046/j.1461-0248.2001.00262.x (doi:10.1046/j.1461-0248.2001.00262.x) [DOI] [Google Scholar]
- 21.Leimu R., Fisher M.2008A meta-analysis of local adaptation in plants. PLoS ONE, 3, e4010. 10.1371/journal.pone.0004010 (doi:10.1371/journal.pone.0004010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosil P., Vines T. H., Funk D. J.2005Perspective: reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59, 705–719 10.1554/04-428 (doi:10.1554/04-428) [DOI] [PubMed] [Google Scholar]
- 23.Verhoeven K. J. F., Vanhala T. K., Biere A., Nevo E., Van Damme J. M. M.2004The genetic basis of adaptive population differentiation: a quantitative trait locus analysis of fitness traits in two wild barley populations from contrasting habitats. Evolution 58, 270–283 [PubMed] [Google Scholar]
- 24.Merilä J., Crnokrak P.2001Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 14, 892–903 10.1046/j.1420-9101.2001.00348.x (doi:10.1046/j.1420-9101.2001.00348.x) [DOI] [Google Scholar]
- 25.Funk D. J., Nosil P., Etges W. J.2006Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc. Natl Acad. Sci. USA 103, 3209–3213 10.1073/pnas.0508653103 (doi:10.1073/pnas.0508653103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nosil P., Egan S. P., Funk D. J.2008Heterogeneous genomic differentiation between walking-stick ecotypes: ‘Isolation by adaptation’ and multiple roles for divergent selection. Evolution 62, 316–336 10.1111/j.1558-5646.2007.00299.x (doi:10.1111/j.1558-5646.2007.00299.x) [DOI] [PubMed] [Google Scholar]
- 27.Fenster C. B.1991Gene flow in Chamaechrista fasciculata (Leguminosae). 2. Gene establishment. Evolution 45, 410–422 10.2307/2409674 (doi:10.2307/2409674) [DOI] [PubMed] [Google Scholar]
- 28.Fenster C. B., Galloway L. F.2000Population differentiation in an annual legume: genetic architecture. Evolution 54, 1157–1172 10.1554/0014-3820(2000)054[1157:PDIAAL]2.0.CO;2 (doi:10.1554/0014-3820(2000)054[1157:PDIAAL]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 29.Van Treuren R., Bijlsma R., Ouborg N. J., Vandelden W.1993The significance of genetic erosion in the process of extinction. 4. Inbreeding depression and heterosis effects caused by selfing and outcrossing in Scabiosa columbaria. Evolution 47, 1669–1680 10.2307/2410211 (doi:10.2307/2410211) [DOI] [PubMed] [Google Scholar]
- 30.Nosil P., Funk D. J., Ortiz-Barrientos D.2009Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402 10.1111/j.1365-294X.2008.03946.x (doi:10.1111/j.1365-294X.2008.03946.x) [DOI] [PubMed] [Google Scholar]
- 31.Brown A. H. D., Zohary D., Nevo E.1978Outcrossing rates and heterozygosity in natural populations of Hordeum spontaneum Koch in Israel. Heredity 41, 49–62 10.1038/hdy.1978.63 (doi:10.1038/hdy.1978.63) [DOI] [Google Scholar]
- 32.Jolivet C., Bernasconi G.2007Molecular and quantitative genetic differentiation in European populations of Silene latifolia (Caryophyllaceae). Ann. Bot. 100, 119–127 10.1093/aob/mcm088 (doi:10.1093/aob/mcm088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastenbroek O., Prentice H. C., Heringa J., Hogeweg P.1984Corresponding patterns of geographic variation among populations of Silene latifolia (= S. alba = S. pratensis) (Caryophyllaceae). Plant Syst. Evol. 145, 227–242 10.1007/BF00983951 (doi:10.1007/BF00983951) [DOI] [Google Scholar]
- 34.Wolfe L. M., Elzinga J. A., Biere A.2004Increased susceptibility to enemies following introduction in the invasive plant Silene latifolia. Ecol. Lett. 7, 813–820 10.1111/j.1461-0248.2004.00649.x (doi:10.1111/j.1461-0248.2004.00649.x) [DOI] [Google Scholar]
- 35.Blair A. C., Wolfe L. M.2004The evolution of an invasive plant: an experimental study with Silene latifolia. Ecology 85, 3035–3042 10.1890/04-0341 (doi:10.1890/04-0341) [DOI] [Google Scholar]
- 36.Wolfe L. M., Blair A. C., Penna B. M.2007Does intraspecific hybridization contribute to the evolution of invasiveness?: an experimental test. Biol. Invas. 9, 515–521 10.1007/s10530-006-9046-0 (doi:10.1007/s10530-006-9046-0) [DOI] [Google Scholar]
- 37.Hathaway L., Andersson S., Prentice H. C.2009Experimental crosses within European Silene latifolia (Caryophyllaceae): intraspecific differentiation, distance effects, and sex ratio. Bot. Bot. 87, 231–240 10.1139/B08-137 (doi:10.1139/B08-137) [DOI] [Google Scholar]
- 38.Keller S. R., Sowell D. R., Neiman M., Wolfe L. M., Taylor D. R.2009Adaptation and colonization history affect the evolution of clines in two introduced species. New Phytol. 183, 678–690 10.1111/j.1469-8137.2009.02892.x (doi:10.1111/j.1469-8137.2009.02892.x) [DOI] [PubMed] [Google Scholar]
- 39.Keller S. R., Taylor D. R.2010Genomic admixture increases fitness during a biological invasion. J. Evol. Biol. 23, 1720–1731 10.1111/j.1420-9101.2010.02037.x (doi:10.1111/j.1420-9101.2010.02037.x) [DOI] [PubMed] [Google Scholar]
- 40.Blumenthal D. M., Hufbauer R. A.2007Increased plant size in exotic populations: a common-garden test with 14 invasive species. Ecology 88, 2758–2765 10.1890/06-2115.1 (doi:10.1890/06-2115.1) [DOI] [PubMed] [Google Scholar]