Abstract
The nutritional symbiosis between aphids and their obligate symbiont, Buchnera aphidicola, is often characterized as a highly functional partnership in which the symbiont provides the host with essential nutrients. Despite this, some aphid lineages exhibit dietary requirements for nutrients typically synthesized by Buchnera, suggesting that some aspect of the symbiosis is disrupted. To examine this phenomenon in the pea aphid, Acyrthosiphon pisum, populations were assayed using defined artificial diet to determine dietary requirements for essential amino acids (EAAs). Six clones exhibiting dependence on EAAs in their diet were investigated further. In one aphid clone, a mutation in a Buchnera amino acid biosynthesis gene could account for the clone's requirement for dietary arginine. Analysis of aphid F1 hybrids allowed separation of effects of the host and symbiont genomes, and revealed that both affect the requirement for dietary EAAs in the clones tested. Amino acid requirements were minimally affected by secondary symbiont infection. Our results indicate that variation among pea aphids in dependence on dietary amino acids can result from Buchnera mutation as well as variation in the host genotype.
Keywords: amino acid metabolism, nutritional symbiosis, Acyrthosiphon pisum, artificial diet, secondary symbiont, Buchnera aphidicola
1. Introduction
Nutritional symbiotic associations with bacteria are common in insects and have allowed many groups to exploit otherwise unsuitable food sources by synthesizing nutrients lacking in their host's diet [1]. Such relationships appear remarkably persistent over evolutionary time, with associations lasting hundreds of millions of years [2,3]. Over this duration, genetic drift and relaxed selection have led to extensive genome erosion, resulting in the loss of many genes in the genomes of bacterial symbionts [4]. Despite having some of the smallest known cellular genomes, these symbionts retain the biosynthetic capacity to produce the nutrients necessary to supplement the hosts' diet [5–8].
Buchnera aphidicola, the obligate bacterial symbiont of aphids, is one of the most extensively investigated nutritional symbionts. Experimental studies and genome sequences of Buchnera from several different aphid species have revealed that the symbiont synthesizes essential amino acids (EAAs) for the aphid that are limiting in the aphid's phloem sap diet [9,10]. Buchnera of distantly related aphid species retain most EAA biosynthesis genes [4,7,10,11]. Some losses may be associated with a shift in the aphid's diet, as is the case in Buchnera of Schizaphis graminum, which no longer reduces sulphur as part of cysteine biosynthesis, possibly owing to the availability of reduced sulphur in the phloem of the grasses the aphid feeds on [4]. Loss of functional amino acid biosynthesis genes in Buchnera might also be compensated for by the presence of secondary symbionts; in Buchnera of Cinara cedri, tryptophan appears to be synthesized cooperatively by Buchnera and the secondary symbiont Serratia symbiotica [12].
Intraspecific variation in amino acid requirements has been described in several species of aphid [13–15], suggesting that symbiont-produced nutrients are insufficient to sustain the host in some populations. One potential source of this variation is disruption of Buchnera's ability to produce EAAs. This explanation has been proposed previously [14] but has not been tested. Differences among aphid genomes could also be the cause of this variation in dietary requirements, as host genes involved in amino acid synthesis, catabolism and transport, as well as genes that support the symbiont, may vary among aphid clones.
The source of variation in amino acid requirements has ramifications for the maintenance of this variation, aphid population dynamics, as well as its long-term effects on the evolution of this symbiosis. Loss of amino acid provisioning is irreversible in Buchnera, which does not recombine or acquire genes through lateral gene transfer [4]. Such loss may restrict the host range of the aphid, leading to population subdivision and divergence [16,17] or potentially to extinction of the lineage.
This study uses fitness assays to identify clones of the pea aphid, Acyrthosiphon pisum, which exhibit dietary requirements for EAAs. The contribution of Buchnera to this variation was determined by the examination of symbiont genes. The role of secondary symbionts was assessed both in a large panel of aphids with their naturally occurring symbionts as well as in aphid hosts with identical genetic backgrounds and different secondary symbionts. Finally, the fitness of F1 hybrids was used to measure the contributions of host and Buchnera to the variation in aphid amino acid requirements.
2. Material and methods
(a). Aphid cultures
Acyrthosiphon pisum clones were collected on various host plant species (referred to hereafter as ‘host plant’) from locations around the United States between 1999 and 2008 (electronic supplementary material, table S1). Each of the 35 clonal lineages was derived from single parthenogenetic females then divided into two to four sublines for at least three generations prior to experiments. These were reared on Vicia faba seedlings under 16 L : 8 D at 20°C in an environmental chamber.
(b). Artificial diet and feeding assays
Aphids were fed on an artificial diet to reveal dietary requirements for EAAs. The all-essential-amino acids (AEAA) diet was formulated after the AP3 diet of Febvay et al. [13], which contains sucrose, amino acids, vitamins and trace elements. The no-essential-amino acid (NEAA) diet was created by omitting all EAAs and cysteine from the AP3 diet and increasing the amounts of non-EAAs proportionally until the total nitrogen concentration was equivalent to that of the AEAA diet. Diets lacking individual EAAs were created by omitting a particular EAA and balancing total nitrogen by the addition of equal molar concentrations of glutamate. Adult A. pisum were placed on fresh V. faba seedlings 24 h prior to experimental set-up and allowed to deposit nymphs. Artificial diet feeding was a microtitre plate system with one to two nymphs and 300 µl of artificial diet per well [18]. Plates were kept at conditions described for the aphid cultures. After 7 days, individual aphids were weighed on a microbalance and their mass used as a proxy for fitness (electronic supplementary material, figure S1). Mass at birth was not measured, and variation in initial mass probably contributed to the variation in adult mass. All clones were tested on both the AEAA diet and the NEAA diet. A subset of clones was tested on diet lacking individual EAAs. For all dietary assays, clones were subject to feeding treatments twice independently, with a total of at least 30 aphids measured for each clone × treatment pair.
(c). Sequencing of Buchnera genes
Genomic DNA was isolated from clones exhibiting requirements for EAAs in the individual amino acid deletion assay; specifically these were clones 8-10-1, G17, G6 and G19. PCR was performed to amplify Buchnera genes underlying the biosynthesis of the required amino acid genes; PCR products were directly sequenced (electronic supplementary material, methods and table S2).
(d). Secondary symbiont screening
Presence of secondary symbiont was determined by PCR screen using symbiont-specific diagnostic primers that produce amplicons spanning the intergenic region between the 16S and 23S ribosomal RNA subunits. Forward primers (1279F_Ss, 1279F_Ri and 1279F_Hd) specific for each secondary symbiont were used along with a universal primer (35R) [19].
(e). Establishment of F1 clones
To determine whether Buchnera mutations were responsible for fitness differences observed on the NEAA diet, clones 5A and 8-10-1 were sexually induced using the method of Moran & Dunbar [20] (electronic supplementary material, methods). Fundatrices were collected and initially reared on V. faba and Medicago sativa shoots. Single fundatrices were used to establish clones corresponding to independent genotypes for each direction of the sexual cross and were allowed to reproduce asexually for three generations prior to use in feeding experiments.
(f). Statistical analyses
For all experiments, aphid mass was normalized by natural log transformation and then subjected to a least-squares means analysis in JMP7 to determine significant effects within the model. For the AEAA versus NEAA comparisons and the individual amino acid deletion assays, significant differences between and within clones, respectively, were determined by a Tukey's HSD test. Significant differences within clones on the AEAA and NEAA diets were determined by a Student's t-test.
3. Results
(a). Aphid performance on AEAA and NEAA diets
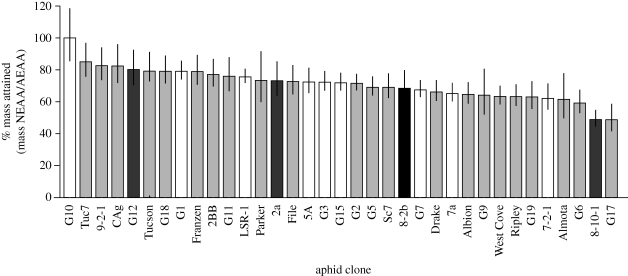
Aphid performance was substantially different between diets with and without EAAs (figure 1 and electronic supplementary material, table S3). Compared with their mass on the AEAA diet, clones lost between 0 and 52 per cent of individual adult mass on the NEAA diet. Between-clone variation in individual adult mass attained on the NEAA diet relative to the AEAA diet was effectively continuous. Clones varied in dependence on EAAs in the diet, as indicated by a significant clone × diet interaction (electronic supplementary material, table S3; F34,4064 = 14.1, p = 6.3 1.0 × 10−16). The effect of the host plant by treatment was significant but minor, and probably reflects differences in the aphid genotype rather than an effect of the plant, as all clones were reared on V. faba prior to feeding assays (electronic supplementary material, methods and table S3).
Figure 1.
EAA-dependence of aphid clones presented as relative adult mass on diet without and with essential amino acids (AEAA versus NEAA). Aphid clones showed 0–52% average decrease in mass on the NEAA diet. Error bars indicate 95% CI of the mean. Shade of bars indicates secondary symbiont type: open bars, uninfected; black bars, infected with Hamiltonella defensa; light grey bars, infected with Serratia symbiotica; dark grey bars, infected with Regiella insecticola.
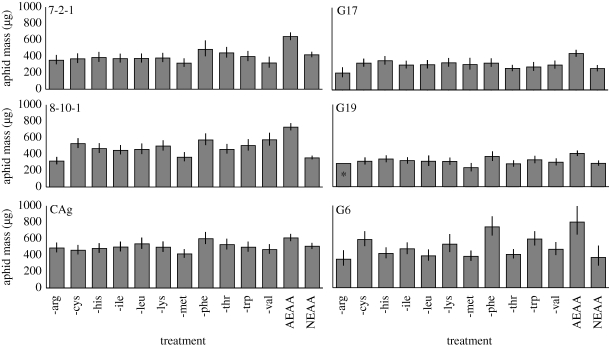
Six clones exhibiting major loss of mass (greater than 37%) on the NEAA relative to the AEAA diet were G17, 8-10-1, G6, Almota, 7-2-1 and G19 (referred hereafter as EAA-dependent). These EAA-dependent clones were subjected to feeding assays using diets lacking individual EAAs to determine if specific amino acids were responsible for EAA-dependence (figure 2 and electronic supplementary material, figure S2). Clone CAg was also tested as controls for performance on these diets. When tested on a diet lacking individual EAAs, a significant effect of clone × treatment was observed (electronic supplementary material, table S3; F60,1774 = 3.3, p = 3.5 × 10−16) indicating variation among clones in the particular EAAs required in the diet. Elimination of individual amino acids from the diet affected all aphids tested, and most such eliminations resulted in significant decreases in aphid mass (p < 0.01, Tukey's HSD). We chose to focus on amino acids whose elimination had a major effect on aphid mass (see §4 for more information), particularly cases in which mass on individual amino acid deletion diets was no more than 102 per cent of mass on the NEAA diet. Using this criterion, several clones were dependent on arginine, while methionine, threonine, leucine, valine and histidine were each needed by at least one clone. Most clones required one to two EAAs. Clone G6 exhibited much greater variation in performance on individual amino acid deletion diets. Six of the diets lacking individual EAAs reduced G6's mass by more than 55 per cent.
Figure 2.
Adult mass for selected clones on diets lacking individual EAAs. Clone names appear in the upper left of each graph. X-axis shows the amino acid omitted from the diet. Error bars represent 95% CI of the mean. The asterisk denotes that only a single aphid from G19 survived the duration of the experiment.
(b). Examining polymorphisms in Buchnera to identify causes of EAA-dependence
To determine if biosynthetic capabilities of Buchnera caused the variation in dependence on particular EAAs, we sequenced the corresponding Buchnera biosynthetic genes and itemized all polymorphisms (table 1 and electronic supplementary material, table S4). Of 89 polymorphic sites, two were single base insertions/deletions, one in argC of 8-10-1 and another in the upstream region of thrB in both clones examined (G19 and G6).
Table 1.
Summary of mutations in selected regions of EAA-dependent A. pisum clones.
| total mutations | 89 |
| SNPsa | 87 |
| indels | 2 |
| coding region | 83 |
| upstream | 6 |
| synonymous | 62 |
| non-synonymous | 21 |
| uniqueb | 19 |
| all linesc | 16 |
| segregatingd | 43 |
aAll mutations are called relative to the genome of Buchnera aphidicola str. APS.
bFound only in a single clone examined, does not include mutations from pLeu of G6 as only this clone was sequenced for this region.
cFound in all four EAA-dependent clones.
dFound in more than one clone but not all clones examined.
Clone 8-10-1 had an insertion of a thymidine at the 610th base pair of argC, causing a frameshift and a premature stop codon at amino acid 206. argC encodes N-acetyl-gamma-glutamyl phosphate reductase, which catalyzes the third step in ornithine biosynthesis [21]. Inactivation of this gene would inhibit biosynthesis of ornithine, an essential precursor of arginine. All other gene sequences for all other clones lacked any obvious mutations (frameshift or nonsense), which would inhibit translation of the gene product.
Of the 87 single nucleotide changes, six were located in the region immediately upstream of a coding region, although none was found in a −10/−35 promoter region as identified by BPROM (http://www.softberry.com), suggesting that these mutations have no effect on gene expression. Of mutations in coding regions for which more than one clone was sequenced, 19 were unique to a single clone, and 16 were shared by all four EAA-dependent clones relative to the reference genome of Buchnera APS (NC_002528).
Of the 87 observed changes, 21 were non-synonymous mutations, but most resulted in a conservative amino acid change. Only a single mutation created a major change in the coded amino acid, defined as a penalty of −3 or more in the amino acid matrix of Muller et al. [22]. An A → T transversion in argE of the related matrilines 8-10-1 and 5A [23] changed an asparagine to isoleucine, but 5A is not an EAA-dependent clone, suggesting that this mutation has little effect. The other non-synonymous mutations involved less drastic changes in the amino acid; five had penalties of −1 to −2, and 15 had 0 or positive values in the matrix.
(c). Effect of secondary symbiont infection on host EAA-dependence
Among tested clones, one (8-2b) was infected with Hamiltonella defensa, four clones were infected with Regiella insecticola and 21 clones were infected with S. symbiotica (figure 1). Serratia symbiotica or R. insecticola did not differentially impact aphid mass on the NEAA and AEAA diet (electronic supplementary material, table S3; F2,31 = 0.895, p = 0.42). The impact of H. defensa could not be assessed as only one clone was infected.
To directly assess the effects of secondary symbionts on aphid EAA-dependence, sublines of a single clone (5A) artificially infected with each of the three secondary symbionts were tested on AEAA and NEAA diets (electronic supplementary material, figure S3). The three symbionts had different effects on aphid mass. Regardless of the treatment (AEAA or NEAA diet), sublines infected with H. defensa or R. insecticola were significantly smaller than S. symbiotica-infected or uninfected sublines (p < 0.05, Tukey's HSD). There was a significant interaction between diet and treatment (electronic supplementary material, table S3; F3,1069 = 3.2, p = 0.022), although the difference in per cent mass attained between clones was small (12%; electronic supplementary material, figure S2). Taken together, these data suggest that secondary symbionts exert a minimal effect on their host's EAA-dependence.
(d). Effect of Buchnera genotype on EAA-dependence
Since Buchnera is exclusively maternally transmitted, sexual crosses of A. pisum clones can be used to change the host genetic background independently of Buchnera genotype. In such crosses, F1 progeny will be 50 per cent related on average at aphid loci and will contain the same Buchnera of the maternal clone. Clone 5A was reciprocally mated with clone 8-10-1 to assess whether the observed mutation in Buchnera of 8-10-1 was responsible for the clone's decrease in fitness on the NEAA diet.
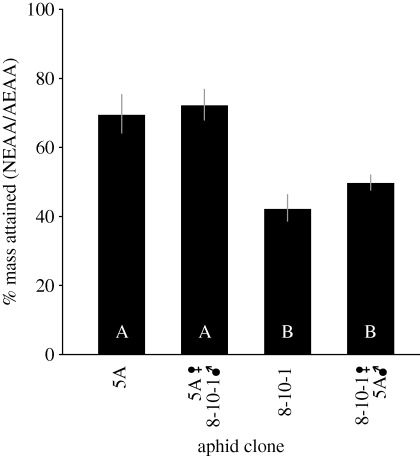
Clones from both sexual crosses (8-10-1f × 5Am or 5Af × 8-10-1m) showed a significant decrease in mass on the NEAA diet when compared with the AEAA diet (8-10-1f × 5Am: t-ratio1144 = −26.91, p = 1 × 10−15; 5Af × 8-10-1m: t-ratio718 = −10.48, p < 1 × 10−15, Student's t-test), as did both parental clones (figure 1). The offspring of the 5Af × 8-10-1m cross had an average 28 per cent decrease in mass (figure 3), which is not significantly different from that observed for the parental 5A clone when tested simultaneously, but significantly different from parental 8-10-1, which lost an average of 56 per cent mass on the NEAA diet (p < 0.05, Tukey's HSD). The clones from the 8-10-1f × 5Am cross exhibited a 46 per cent decrease on average when reared on the NEAA diet, which was similar to the decrease observed for the 8-10-1 parental clone but significantly greater than the performance of the 5A parental clone (p < 0.05, Tukey's HSD). Thus, the argC mutation in Buchnera of 8-10-1 was a major factor in the EAA-dependence of the F1 hybrids from the 8-10-1 matriline. The offspring of both crosses and 8-10-1 parental clones attained similar mass on the AEAA diet (data not shown).
Figure 3.
EAA-dependence of aphid clones representing full siblings from a sexual cross. Aphids from 8-10-1f × 5Am contain Buchnera with inactivated argC; aphids from 5Af × 8-10-1m contain Buchnera with intact argC. Error bars represent 95% CI. Different letters within bars indicate significant differences (p < 0.05, Tukey's HSD).
(e). Effect of host genotype on EAA-dependence
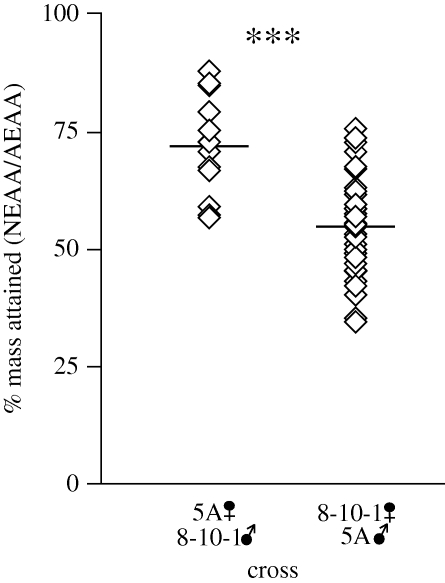
Clones from the same cross showed highly significant differences in EAA-dependence (figure 4 and electronic supplementary material, table S3; F47,2153 = 12.5, p = 5.9 × 10−79, standard least squares). Clones from the 8-10-1f × 5Am cross lost between 25 and 67 per cent of adult mass on the NEAA diet, while clones from 5Af × 8101m cross lost 12–43% of adult mass on the NEAA diet. These data imply that the Buchnera argC mutation does cause EAA-dependence, but that host-encoded factors also play a substantial role.
Figure 4.
Effect of aphid genotype on EAA-dependence of F1 hybrids. Diamonds indicate the mean value for each F1 genotype within each direction of the cross. The horizontal lines denote the mean per cent mass attained for all genotypes from each direction of the cross. ***p = 1.1 × 10−15, standard least squares.
4. Discussion
Aphid clones used in this study varied in requirements for dietary EAAs. In one instance, increased EAA-dependence reflected an inactivating mutation in an amino acid biosynthesis gene of Buchnera, but such inactivating mutations were absent in three of four clones exhibiting major EAA-dependence. The one inactivating mutation observed was confirmed as a factor in EAA-dependence through crossing experiments in which the Buchnera with the mutation were established in novel host-genetic backgrounds. However, different F1 genotypes with identical Buchnera varied substantially in performance, implying that host genotype also contributes to EAA-dependence. Host genotypes are probably the source of variation among other aphid clones examined in these experiments, as no other mutations were found in Buchnera genes that could explain their requirement for dietary amino acids.
Elimination of individual EAAs from the diet significantly reduced aphid mass in almost all cases, suggesting that such elimination imposes an energetic or metabolic cost to the aphid–symbiont pair, even when Buchnera has the capacity to make the missing amino acid. Consistent with this hypothesis, the complete Buchnera genome of clone 8-10-1 exhibits no sequence differences in genes underlying any amino acid biosynthesis pathway except for the frameshift in argC found in this study [23], yet 8-10-1 showed small decreases in mass on diets lacking individual amino acids other than arginine.
Previous studies have failed to reveal an obvious connection between infection with secondary symbionts and aphid performance on artificial diets [24], though secondary symbiont infection appears to exacerbate deleterious effects of sub-optimal host plants on aphids [25]. Our experiments, carried out for three common secondary symbiont types infecting the same A. pisum clone, revealed no major impact of secondary symbionts on aphid amino acid requirements, though secondary symbionts did significantly reduce aphid mass on the AEAA diet. These results suggest that the impact of secondary symbionts on aphid fitness may not be related to amino acid nutrition. Population cage studies have shown that, in the absence of parasitoid pressure, the frequencies of H. defensa and S. symbiotica decrease, suggesting that infection with these symbionts imparts a cost to the aphid [26]. The recently completed genome sequences of H. defensa and R. insecticola revealed the absence of most EAA biosynthesis pathways, implying that these symbionts obtain these nutrients from the aphid's free amino acid pool [27,28].
In clone 8-10-1, the need for dietary arginine along with the frameshift in argC suggests that Buchnera cannot produce this EAA. While some eukaryotes have evolved the ability to synthesize arginine from aspartate and citrulline as part of the urea cycle, the A. pisum genome lacks genes encoding arginosuccinate synthase and arginosuccinate lyase, the two enzymes necessary to synthesize arginine in this manner [29]. Analyses of arginine levels in the phloem sap of several host plants of A. pisum reveal it to be one of the most abundant EAAs in the phloem sap of all host plants tested [30]. Phloem arginine levels in V. faba are unlikely to completely compensate for disrupted arginine biosynthesis by Buchnera, but the relatively higher level of this amino acid may reduce selection against the loss of arginine biosynthesis enough to allow fixation of this inactivating mutation in laboratory cultures. In wild populations, selection against such mutations is probably stronger and effective population sizes larger, resulting in more efficient purging of mutations in amino acid biosynthesis genes in Buchnera.
Recent re-sequencing of Buchnera genomes from A. pisum lineages has revealed that the symbiont's mutation rate is an order of magnitude higher than that of free-living relatives such as Escherichia coli [23]. This high mutation rate is counteracted by host-level selection that acts to maintain Buchnera's amino acid production capabilities [31,32]. This study has shown that differences among A. pisum clones in dietary amino acid requirements can result from differences in the amino acid biosynthesis genes of Buchnera, but often reflect genetic variation in the aphid hosts. The underlying host genes could encode products that interact with the symbiont [33] or that affect chemosensory abilities, as certain amino acids, notably methionine, are known phagostimulants of A. pisum [34].
The variation in the other A. pisum clones' dietary requirements may affect their ability to use different plants, and thereby their population structure. Host plants of A. pisum vary in their phloem amino acid concentrations, and aphid performance is partly associated with plant phloem amino acid levels [30]. Adaptation by either A. pisum or Buchnera to the phloem profile of specific host plants could help to affect the establishment of host races of A. pisum [35].
Acknowledgements
The authors would like to thank J. Stavrinides for assistance in developing the feeding assay, N. Gerardo and S. Eigenbrode for collection of aphid clones, K. Hammond, K. MacDow, K. Z. Sunitsch and E. Hartley for help in data collection and aphid maintenance, and three anonymous reviewers for their help in improving the manuscript. This work was funded by NSF grant 0723472 to N.A.M. K.J.V. was supported on an NSF IGERT fellowship and an NSF G-K12 fellowship during this research.
References
- 1.Baumann P.2005Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Ann. Rev. Microbiol. 59, 155–189 10.1146/annurev.micro.59.030804.121041 (doi:10.1146/annurev.micro.59.030804.121041) [DOI] [PubMed] [Google Scholar]
- 2.Moran N. A., Munson M. A., Baumann P., Ishikawa H.1993A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. B 253, 167–171 10.1098/rspb.1993.0098 (doi:10.1098/rspb.1993.0098) [DOI] [Google Scholar]
- 3.Moran N. A., Tran P., Gerardo N. M.2005Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71, 8802–8810 10.1128/AEM.71.12.8802-8810.2005 (doi:10.1128/AEM.71.12.8802-8810.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamas I., Klasson L., Canback B., Naslund A. K., Eriksson A. S., Wernegreen J. J., Sandström J. P., Moran N. A., Andersson S. G. E.200250 million years of genomic stasis in endosymbiotic bacteria. Science 296, 2376–2379 10.1126/science.1071278 (doi:10.1126/science.1071278) [DOI] [PubMed] [Google Scholar]
- 5.McCutcheon J. P., McDonald B. R., Moran N. A.2009Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl Acad. Sci. USA 106, 15 394–15 399 10.1073/pnas.0906424106 (doi:10.1073/pnas.0906424106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCutcheon J. P., McDonald B. R., Moran N. A.2009Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565. 10.1371/journal.pgen.1000565 (doi:10.1371/journal.pgen.1000565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Brocal V., Gil R., Ramos S., Lamelas A., Postigo M., Michelena J. M., Silva F. J., Moya A., Latorre A.2006A small microbial genome: the end of a long symbiotic relationship? Science 314, 312–313 10.1126/science.1130441 (doi:10.1126/science.1130441) [DOI] [PubMed] [Google Scholar]
- 8.Wu D., et al. 2006Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4, 1079–1092 10.1371/journal.pbio.0040188 (doi:10.1371/journal.pbio.0040188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas A. E., Prosser W. A.1992Synthesis of the essential amino-acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 38, 565–568 10.1016/0022-1910(92)90107-O (doi:10.1016/0022-1910(92)90107-O) [DOI] [Google Scholar]
- 10.Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H.2000Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 10.1038/35024074 (doi:10.1038/35024074) [DOI] [PubMed] [Google Scholar]
- 11.Van Ham R. C. H. J., et al. 2003Reductive genome evolution in Buchnera aphidicola. Proc. Natl Acad. Sci. USA 100, 581–586 10.1073/pnas.0235981100 (doi:10.1073/pnas.0235981100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosalbes M. J., Lamelas A., Moya A., Latorre A.2008The striking case of tryptophan provision in the cedar aphid Cinara cedri. J. Bacteriol. 190, 6026–6029 10.1128/JB.00525-08 (doi:10.1128/JB.00525-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Febvay G., Delobel B., Rahbe Y.1988Influence of the amino-acid balance on the improvement of an artificial diet for a biotype of Acyrthosiphon pisum (Homoptera, Aphididae). Can. J. Zool. 66, 2449–2453 10.1139/z88-362 (doi:10.1139/z88-362) [DOI] [Google Scholar]
- 14.Srivastava P. N., Gao Y., Levesque J., Auclair J. L.1985Differences in amino acid requirements between 2 biotypes of the pea aphid, Acyrthosiphon pisum. Can. J. Zool. 63, 603–606 10.1139/z85-087 (doi:10.1139/z85-087) [DOI] [Google Scholar]
- 15.Wilkinson T. L., Douglas A. E.2003Phloem amino acids and the host plant range of the polyphagous aphid, Aphis fabae. Entomol. Exp. Appl. 106, 103–113 10.1046/j.1570-7458.2003.00014.x (doi:10.1046/j.1570-7458.2003.00014.x) [DOI] [Google Scholar]
- 16.Dres M., Mallet J.2002Host races in plant-feeding insects and their importance in sympatric speciation. Phil. Trans. R. Soc. Lond. B 357, 471–492 10.1098/rstb.2002.1059 (doi:10.1098/rstb.2002.1059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller F. P.1985Biotype formation and sympatric speciation in aphids (Homoptera, Aphidinea). Entomol. Genet. 10, 161–181 [Google Scholar]
- 18.Stavrinides J., McCloskey J. K., Ochman H.2009Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl. Environ. Microbiol. 75, 2230–2235 10.1128/AEM.02860-08 (doi:10.1128/AEM.02860-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunbar H. E., Wilson A. C., Ferguson N. R., Moran N. A.2007Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 5, e96. 10.1371/journal.pbio.0050096 (doi:10.1371/journal.pbio.0050096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran N. A., Dunbar H. E.2006Sexual acquisition of beneficial symbionts in aphids. Proc. Natl Acad. Sci. USA 103, 12 803–12 806 10.1073/pnas.0605772103 (doi:10.1073/pnas.0605772103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keseler I. M., et al. 2009EcoCyc: a comprehensive view of Escherichia coli biology. Nucl. Acids Res. 37, D464–D470 10.1093/nar/gkn751 (doi:10.1093/nar/gkn751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller T., Spang R., Vingron M.2002Estimating amino acid substitution models: a comparison of Dayhoff's estimator, the resolvent approach and a maximum likelihood method. Mol. Biol. Evol. 19, 8–13 [DOI] [PubMed] [Google Scholar]
- 23.Moran N. A., McLaughlin H. J., Sorek R.2009The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323, 379–382 10.1126/science.1167140 (doi:10.1126/science.1167140) [DOI] [PubMed] [Google Scholar]
- 24.Douglas A. E., Francois C. L. M. J., Minto L. B.2006Facultative ‘secondary’ bacterial symbionts and the nutrition of the pea aphid, Acyrthosiphon pisum. Physiol. Entomol. 31, 262–269 10.1111/j.1365-3032.2006.00516.x (doi:10.1111/j.1365-3032.2006.00516.x) [DOI] [Google Scholar]
- 25.Chandler S. M., Wilkinson T. L., Douglas A. E.2008Impact of plant nutrients on the relationship between an herbivorous insect and its symbiotic bacteria. Proc. R. Soc. B 275, 565–570 10.1098/rspb.2007.1478 (doi:10.1098/rspb.2007.1478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver K. M., Campos J., Moran N. A., Hunter M. S.2008Population dynamics of defensive symbionts in aphids. Proc. R. Soc. B 275, 293–299 10.1098/rspb.2007.1192 (doi:10.1098/rspb.2007.1192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degnan P. H., Leonardo T. E., Cass B. N., Hurwitz B., Stern D., Gibbs R. A., Richards S., Moran N. A.2009Dynamics of genome evolution in facultative symbionts of aphids. Environ. Microbiol. 10.1111/j.1462-2920.2009.02085.x (doi:10.1111/j.1462-2920.2009.02085.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Degnan P. H., Yu Y., Sisneros N., Wing R. A., Moran N. A.2009Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl Acad. Sci. USA 106, 9063–9068 10.1073/pnas.0900194106 (doi:10.1073/pnas.0900194106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The International Aphid Genome Consortium 2010Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8, e1000313. 10.1371/journal.pbio.1000313 (doi:10.1371/journal.pbio.1000313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandström J., Pettersson J.1994Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 40, 947–955 10.1016/0022-1910(94)90133-3 (doi:10.1016/0022-1910(94)90133-3) [DOI] [Google Scholar]
- 31.Moran N. A.1996Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl Acad. Sci. USA 93, 2873–2878 10.1073/pnas.93.7.2873 (doi:10.1073/pnas.93.7.2873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rispe C., Moran N. A.2000Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Am. Nat. 156, 425–441 10.1086/303396 (doi:10.1086/303396) [DOI] [PubMed] [Google Scholar]
- 33.Nakabachi A., Shigenobu S., Sakazume N., Shiraki T., Hayashizaki Y., Carninci P., Ishikawa H., Kudo T., Fukatsu T.2005Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl Acad. Sci. USA 102, 5477–5482 10.1073/pnas.0409034102 (doi:10.1073/pnas.0409034102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava P. N., Auclair J. L.1975Role of single amino acids in phagostimulation, growth, and survival of Acyrthosiphon pisum. J. Insect Physiol. 21, 1865–1871 10.1016/0022-1910(75)90255-3 (doi:10.1016/0022-1910(75)90255-3) [DOI] [Google Scholar]
- 35.Peccoud J., Ollivier A., Plantegenest M., Simon J. C.2009A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl Acad. Sci. USA 106, 7495–7500 10.1073/pnas.0811117106 (doi:10.1073/pnas.0811117106) [DOI] [PMC free article] [PubMed] [Google Scholar]