Abstract
Current interest in behavioural syndromes, or ‘animal personalities’, reinforces a need for behavioural ecologists to adopt a multivariate view of phenotypes. Fortunately, many of the methodological and theoretical issues currently being dealt with by behavioural ecologists within the context of behavioural syndromes have previously been investigated by researchers in other areas of evolutionary ecology. As a result of these previous efforts, behavioural syndrome researchers have considerable theory and a wide range of tools already available to them. Here, we discuss aspects of quantitative genetics useful for understanding the multivariate phenotype as well as the relevance of quantitative genetics to behavioural syndrome research. These methods not only allow the proper characterization of the multivariate behavioural phenotype and genotype—including behaviours within, among and independent of behavioural syndrome structures—but also allow predictions as to how populations may respond to selection on behaviours within syndromes. An application of a quantitative genetics framework to behavioural syndrome research also clarifies and refines the questions that should be asked.
Keywords: quantitative genetics, personality, trade-offs, G matrix
1. Introduction
Research into cross-contextual behavioural correlations, also termed ‘behavioural syndromes’ and ‘animal personalities’, has been of increasing interest to behavioural ecologists over the last several years (Sih et al. 2004). This interest has led to the identification of behavioural syndromes in many animal taxa (Réale et al. 2007), which are of interest, in part, because syndromes suggest that behavioural plasticity or flexibility might be constrained (Neff & Sherman 2004). For example, behavioural correlations within the structure of personalities suggest the potential for trade-offs in the evolution of the behavioural phenotype (Sih et al. 2004). Unfortunately, most behavioural syndrome research has focused only on phenotypic correlations, which do not, by themselves, indicate the presence or shape of trade-offs. Instead, the course of evolution is largely shaped by genetic correlations; genetic correlations indicate that evolutionary trajectories are not free to move in any direction, even if the final endpoint is not itself constrained (Blows & Hoffmann 2005; Roff & Fairbairn 2007). Genetic correlations among behaviours can strongly affect both the direction and the magnitude of responses to selection (Lande & Arnold 1983; Phillips & Arnold 1989). For example, strong selection on a particular behaviour may not have a large effect if there is also strong selection but with opposite effects on a genetically correlated behaviour. Importantly, the sign of a phenotypic correlation does not necessarily indicate the sign of the genetic correlation, and hence the course of evolutionary change might not be correctly understood solely by an examination of the phenotypic correlations (Roff 1996, 1997; Hadfield et al. 2007).
While there are now sufficient estimates of the heritability of behavioural traits to make general statements about the rapidity with which individual traits might respond to selection (Stirling et al. 2002), there are too few estimates of genetic correlations to make general statements about how the correlational structure of behaviours might influence evolutionary trajectories (genetic correlations between behaviours are reviewed in Roff (1996) and van Oers et al. (2005)). This necessarily limits the scope of inferences that can be made regarding behavioural syndromes.
In the case of behavioural syndromes, selection responses might be affected because both the distribution of a behaviour under direct selection and the distribution of any behaviour with which it is genetically correlated will change in response to selection (Roff 1997; Lynch & Walsh 1998). This creates the possibility for both additive and counteracting effects of selection and suggests that predicting responses to selection based only on individual behaviours can lead to improper inferences. Researchers must instead consider evolution within a multivariate framework. Adaptive landscapes associated with a multivariate view of the phenotype, and thus behavioural syndromes, are also far more complicated than those for single traits (Gavrilets 2004), making an understanding of selection and optimality more difficult.
While many of these issues are new to behavioural ecologists, the study of phenotypes and selection from a multivariate perspective has a long history in other areas of evolutionary ecology. The issues introduced to behavioural ecology via research on syndromes have been topics of research for several decades in the context of quantitative genetics. Here, we briefly discuss some of the methods and conceptual approaches available to behavioural syndrome researchers for addressing questions that have been developed within the framework of quantitative genetics. We focus on approaches for characterizing and evaluating suites of phenotypic traits and introduce five primary topics: (i) how the multivariate behavioural phenotype and underlying genotype can be properly described; (ii) how the potential plasticity of behavioural traits and the potential for indirect effects complicates the study of behavioural syndromes and multivariate evolution; (iii) how selection on the multivariate phenotype can be modelled; (iv) how behavioural syndromes can be compared among populations; and (v) how modern molecular approaches connect to quantitative genetic approaches. We also introduce research questions important to our understanding of behavioural syndromes associated with many of these topics.
2. The multivariate behavioural phenotype and genotype
(a). Describing the behavioural phenotype
For questions regarding both multivariate evolution and the general discussion of behavioural syndromes, a syndrome's structure (i.e. which behaviours covary and how) is often of more interest than the mean for a particular behaviour in a population. Syndrome structure can therefore be summarized based on phenotypic variances and covariances. However, to properly understand the evolutionary consequences of syndrome structures, researchers should decompose the phenotypic variances and covariances into their constituent genetic and environmental components:
| 2.1 |
where P is a matrix containing the phenotypic variances and covariances, G contains the additive genotypic variances and covariances and E summarizes the environmental effects. P represents the structure of the behavioural syndrome, that is, which behaviours covary and whether these covariances are positive or negative.
Behavioural ecologists regularly report P matrices in standardized form as correlations. These correlations can be used to assess which behaviours covary within syndromes and the direction of the relationships between behaviours (e.g. Bell 2005; Dingemanse et al. 2007; Brodin 2008), quantitatively assess multiple hypotheses of syndrome structure (Dochtermann & Jenkins 2007; Dingemanse et al. 2010a,b) or compare syndrome structures among populations (Bell 2005, 2007; Dingemanse et al. 2007, 2010a,b).
While differences in the expressed behavioural syndrome (P) are of intrinsic interest, G may reveal more about behavioural syndrome evolution. Indeed, it is the G underlying a behavioural syndrome that determines whether behavioural correlations inherent in syndromes influence evolutionary trajectories and G also allows the prediction of responses to selection (Lande & Arnold 1983). It is also differences among populations in a behavioural syndrome's G that may provide key insights into the evolution of behavioural syndromes. Although G can be estimated under certain situations (discussed subsequently), P is typically much easier to estimate. Thus the degree to which P approximates G is of key importance when studying trait relationships (Cheverud 1988). If P and G are strongly related (i.e. the effects of E do not drastically alter the covariances in P relative to G), then P can also be used to draw evolutionary inferences.
Cheverud (1988) proposed that in many cases phenotypic correlations closely correspond to genotypic correlations; this argument has been termed the ‘phenotypic gambit’ in a more general context within the behavioural literature (Grafen 1984). Cheverud's proposition has been supported within the general evolutionary ecological literature for morphological traits (Roff 1995; Norry et al. 1997; Waitt & Levin 1998), but not for life history traits (Roff 1995, 1996). More recent reviews also generally support Cheverud's conjecture, although considerable differences between phenotypic and genotypic correlations may remain (Hadfield et al. 2007; Kruuk et al. 2008). Unfortunately, there has been little research into the degree to which behavioural correlations correspond between the phenotypic and genotypic levels, and the applicability of Cheverud's conjecture to behaviour remains unclear (Willis et al. 1991; Roff 1996). If applicable, Cheverud's conjecture would broaden the inferences that researchers can draw from phenotypic estimates of behavioural syndromes. Thus an important topic for behavioural syndrome research is assessing the degree to which behavioural correlations correspond between the phenotypic and genotypic levels.
(b). Describing the behavioural genotype
Unlike P, which can be directly estimated based on observed behavioural responses, the estimation of G is complicated by the effects of multiple factors on the phenotype. For example, the contribution of additive genetic effects to P, which we seek to estimate with G, may be conflated with epistatic effects, dominance, shared environment and maternal effects. Quantitative genetic research provides various tools that allow the disentanglement of these effects, which can be used in behavioural syndrome research to estimate G as it relates to behaviours of interest.
Under experimental conditions, a variety of structured breeding designs can be used to disentangle these effects. For complex pedigrees as typically found in natural populations or designs to extract multiple genetic components (e.g. sex linkage, Fairbairn & Roff 2006), the mixed model statistical approach known as the ‘animal model’ allows estimation of G. Arnold (1994) presented a general discussion of classical approaches for estimating G for behavioural questions. Programs written in SPlus to estimate genetic components for standard designs and to compare components among populations are also available on the website of D.A.R. (http://www.biology.ucr.edu/people/faculty/Roff.html; see Roff 2002 for discussion). Wilson et al. (2010) presented an introduction to the animal model statistical approach, along with tutorial material that is available online. These quantitative genetic approaches directly deal with the questions being asked in behavioural syndrome, so researchers interested in behavioural syndromes should become familiar with these methods. Unfortunately, these tools have rarely been applied to multiple behaviours and the estimation of genetic correlations (but see van Oers et al. 2004; Dingemanse et al. 2009; Réale et al. 2009). We encourage researchers of behavioural syndromes to become familiar with these approaches both from the suggested introductory treatments and from more in-depth texts (e.g. Lynch & Walsh 1998).
(c). Limitations to estimating genotypic variances and covariances
While the approaches we have discussed are relevant to the study of behavioural syndromes, like any approach, they do have limitations that users should be aware of. Two particularly acute issues are the ability to properly estimate genetic covariances and required sample sizes.
For behavioural ecologists, it is important to note that approaches for partitioning phenotypic variation into its genotypic and environmental components assume that genotypic effects and the environment are not correlated, which will not always be the case (Arnold 1994). For example, philopatry may introduce a G by E correlation, which would lead to an overestimation of genotypic variances and covariances and heritabilities. This is potentially problematic as many species in which syndrome structure has been demonstrated can exhibit philopatry or differential rates of dispersal (e.g. stickleback, Cano et al. 2008; marmots, Svendsen 1974 and great tits, Verhulst et al. 1997). These effects may be most pronounced with the use of animal models in natural populations where manipulations to test for these correlations are not possible. In laboratory settings, the possibility that the changed environmental setting affects behavioural expression must also be addressed.
Genetic parameters estimated from the phenotypes of individuals from known breeding designs also often include large error estimates (Roff 1997). As a result, sample sizes generally in the hundreds or more may be necessary for robust estimates and comparisons among groups (Lynch & Walsh 1998). Required sample sizes will also increase with the number of behaviours studied (Wilson et al. 2010). This may often be a problem for behavioural syndrome research because measuring just the phenotypic components of behavioural syndromes is often laborious, limiting practical sample sizes even when not attempting to also estimate the genetic as well as phenotypic covariances.
For standard pedigree designs such as half-sib or offspring on parent prior power analyses can be conducted by analytical methods (Roff 1997), but more complex designs will require a simulation approach. Such models using an individual-based variance components approach are readily programmed (see Roff (2010) and Roff & Fairbairn (2009) for description and examples; coding is also available on the website of D.A.R.). Numerical methods of power analysis of pedigrees analysed using the animal model are also provided by Morrissey et al. (2007).
Additional concerns that have been raised in the study of heritabilities and genetic covariances, which behavioural syndrome researchers should also be aware of, include potential biases in estimation and the validity of laboratory versus field estimates (e.g. Blum 1988; Astles et al. 2006; Hadfield et al. 2010).
One source of bias is introduced by the method of statistical estimation. The previously discussed animal model estimates heritabilities and genetic covariances using mixed models and restricted error maximum likelihood (REML; Wilson et al. 2010). Unfortunately, REML estimates of genetic correlations are biased to be different from either 0 or |1| (Astles et al. 2006). As a result, behavioural correlations might be biased upward or downward by REML estimation depending on their actual values. Animal model estimates of individual behavioural responses at the genetic level are also complicated by a leptokurtic distribution (Hadfield et al. 2010). Where distributions are very far from normal and no transformation is available that produces a suitable continuous distribution, one can use a threshold transformation that divides the data at the median into a 0,1 distribution (Roff 2001) or use alternative Markov chain Monte Carlo approaches (Hadfield 2010).
Estimates of heritabilities and genetic correlations may also be biased depending on whether estimated under laboratory or field conditions. Heritabilities estimated in the laboratory are often presumed to be overestimated because environmental variation in the laboratory is expected to be lower than in the field (Blum 1988). Because environmental variation is a component of the denominator for the calculation of heritability, greater environmental variation in the field would be expected to lead to lower heritability estimates in the field and an associated overestimation in the laboratory. However, contrary to this presumption, Weigensberg & Roff (1996) found no difference between laboratory estimates of behaviours and field estimates, the latter actually being higher, though not significantly so. In a broader review, Bell et al. (2009) found that field estimates of the repeatability of behaviours—which typically set an upper limit to heritability—were higher than laboratory estimates. These reviews suggest that laboratory estimates may be appropriate for questions regarding behavioural syndrome structure.
3. Reaction norms, indirect genetic effects and behavioural syndromes
Two areas of quantitative genetics that are of particular conceptual importance to researchers in behavioural syndromes are reaction norms and indirect genetic effects.
A reaction norm, the quantitative formulation of phenotypic plasticity, represents the modelling of a genotype's phenotype as a function of a continuous environmental gradient (Nussey et al. 2007). Reaction norms are particularly relevant to the study of behaviours as they are often labile, and thus responses often vary greatly with environmental and social conditions (Smiseth et al. 2008). Dingemanse et al. (2010a,b) suggested that the general framework for describing reaction norms provided by quantitative genetics could be extended to those behaviours observed as components of behavioural syndromes. In the context of behavioural syndromes, this suggests that not only average behavioural responses but also how behaviour changes with environmental variation covary within a syndrome structure. While researchers should refer to Nussey et al. (2007) for an in-depth technical discussion of how to statistically model reaction norms—phrased informally, when the behaviour of an individual is measured over a continuous range of variations, regressions can be performed at the individual level with an estimation of an intercept and a slope for each individual. The presence of statistical differences among individuals within a mixed model can then be determined.
Unfortunately, the characterization of reaction norms at the genotypic level may not be a tractable goal for behavioural syndrome researchers. The methods for characterizing reaction norms for single traits require very large sample sizes (Nussey et al. 2007), and characterizing these for a suite of traits is likely to be logistically prohibitive. However, differences between individuals in how behavioural reactions interact with the environment at the phenotypic level have been demonstrated in some cases to be components of overarching syndromes (Koolhaas et al. 1999; Dochtermann & Jenkins 2007), and studies of these interactions were reviewed by Dingemanse et al. (2010b). Thus, the conceptual basis of reaction norms within quantitative genetics can be used at the phenotypic level and incorporated into syndrome research. Indeed, the degree to which individuals are able to modify their responses is likely to be a key aspect of the evolution and evolutionary implications of behavioural syndromes (McElreath & Strimling 2006; Wolf et al. 2008).
Indirect genetic effects are likely to be similarly important to behavioural syndrome research from a conceptual standpoint. These effects can be defined as the effects of one individual's phenotype on the phenotype of another, usually related, individual (Moore et al. 1997; Wolf et al. 1998). In a practical sense, this represents the influence of an individual's phenotype on the environment of another (see fig. 1 in Moore et al. 1997). The degree to which the phenotype affecting the environment is heritable also determines the degree to which indirect genetic effects are heritable and the most commonly considered forms of indirect genetic effects are maternal effects on offspring phenotypes.
Indirect genetic effects via maternal phenotypes have been found to be important in behaviours ranging from song rates in birds (e.g. Forstmeier et al. 2004), female choosiness (Forstmeier et al. 2004), ‘boldness’ (Tobler & Sandell 2007) and social status and aggression in mammals (Dloniak et al. 2006; Onyango et al. 2008, but see East et al. 2009). Because all of these behaviours have been found to be components of syndromes in other organisms, the role of indirect genetic effects on behavioural syndrome research requires further study. Proper understanding of the role of indirect genetic effects will also allow a greater understanding on the degree to which behaviours within and independent of syndrome structures can evolve in response to changing environments.
4. Modelling the multivariate response to selection
Genotypic variances and covariances for behaviours can be used to predict how populations will or can respond to selection. Most behavioural ecologists are probably familiar with the breeder's equation that predicts how individual behaviours change in response to selection:
| 4.1 |
where  is the change in the population's mean expression of the behaviour of interest, h2 is the narrow-sense heritability of the behaviour and s is the selection differential. The narrow-sense heritability represents the additive genetic variance of a behaviour relative to the total phenotypic variance.
is the change in the population's mean expression of the behaviour of interest, h2 is the narrow-sense heritability of the behaviour and s is the selection differential. The narrow-sense heritability represents the additive genetic variance of a behaviour relative to the total phenotypic variance.
However, when examining behaviours that may be part of a behavioural syndrome, there is the potential for correlated responses to selection. These correlated responses can accentuate or dampen evolutionary responses depending on how traits covary with each other and with fitness. Lande & Arnold (1983) addressed this problem for correlated traits in general by modelling responses to selection based not only on the additive genetic variances of traits but also on their covariances, producing a multivariate equivalent of the breeder's equation:
| 4.2 |
where P is the phenotypic covariance matrix (the observed behavioural syndrome structure), G is the genotypic covariance matrix discussed earlier, S is a vector of selection differentials and 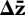 is a vector of the change in a population's mean behavioural response. If genetic variances and covariances (G) are known along with the relationships between traits (S), then a population's response to selection can be estimated. Behavioural variation will often consist of discrete sets of decisions for which the above continuous distributions may appear inapplicable: the solution is to use the threshold model of quantitative genetics in which there is assumed to be a continuously distributed underlying trait called the liability and a threshold (or thresholds). Individuals with values lying above the threshold take one action, whereas individuals below take the alternative: for an example of its use in this context, see King & Roff (2010).
is a vector of the change in a population's mean behavioural response. If genetic variances and covariances (G) are known along with the relationships between traits (S), then a population's response to selection can be estimated. Behavioural variation will often consist of discrete sets of decisions for which the above continuous distributions may appear inapplicable: the solution is to use the threshold model of quantitative genetics in which there is assumed to be a continuously distributed underlying trait called the liability and a threshold (or thresholds). Individuals with values lying above the threshold take one action, whereas individuals below take the alternative: for an example of its use in this context, see King & Roff (2010).
5. Among population comparisons
Another question in behavioural syndrome research which can be addressed using quantitative genetic methods is how syndromes differ among populations. Structural differences in P and G among populations can suggest differences in current or historical selective regimes and may be a key to understanding behavioural syndromes. However, attempts at comparing behavioural syndrome structures among populations have focused on the comparison of bivariate phenotypic correlations rather than difference in the overall syndrome structure (e.g. Bell & Stamps 2004; Bell 2005; Dingemanse et al. 2007).
Various statistical tools applicable to the study of behavioural syndromes have been developed as alternatives to the comparison of bivariate correlations. While many of these approaches were developed explicitly for comparing G matrices among populations, they can be easily implemented with phenotypic data. These statistical tools include a comparison of principal components (CPC) to determine the similarity of P and G matrices among populations (Phillips & Arnold 1999). The CPC method evaluates whether matrices are statistically unrelated, whether they share principal components, whether they exhibit similar principal components but not variances or whether they are identical (Phillips & Arnold 1999). This approach has been used extensively within evolutionary ecology as a whole, but has not been used to compare behavioural syndromes (behavioural P matrices) among populations.
To evaluate multivariate trait differences and their association with environmental conditions among populations, multivariate analysis of variance (MANOVA) approaches can be used (Roff 2002). MANOVA approaches have been used to estimate and compare G matrices for morphological traits and to determine the environmental causes underlying differences (Bégin et al. 2004), and could similarly be used in behavioural syndrome research to statistically test proposed causes of syndrome differences. To our knowledge, despite their potential, MANOVA analyses have not been employed in the study of behavioural syndromes.
Other methods for comparing matrices among populations include the random and selection skewers methods (Cheverud & Marroig 2007; Calsbeek & Goodnight 2009), which would allow researchers to estimate the evolutionary consequences of behavioural syndromes. These methods complement recent applications of structural equation modelling and confirmatory factor analysis, which may allow a greater range of inferences to be drawn than possible with strictly exploratory methods (Dochtermann & Jenkins 2007; Dingemanse et al. 2010a,b).
6. Classical quantitative genetics and genomics
Molecular approaches such as identifying quantitative trait loci (QTL) or microarray analysis are becoming more generally available to assess genetic architecture at the level of DNA. These approaches complement quantitative genetics and promise to provide a platform on which the functional and statistical aspects of the genome can be combined. The statistical tools developed in quantitative genetics, such as mixed model ANOVAs, are ideal for the analysis of genomic data (Kerr & Churchill 2001; Wolfinger et al. 2001). Genomic analyses can also address some of the fundamental assumptions of quantitative genetics. For example, the field of quantitative genetics is based on a statistical model that assumes the action of many loci of small effect, although the possibility of some genes having large effect can be subsumed into the general framework. Thus it came as a surprise when early QTL analyses suggested that variation was due primarily to a few genes of large effect. However, later work showed that this finding was a consequence of a statistical artefact (Beavis 1994; Roff 1997), and more recent work has indicated that while some genes may have relatively large effects, many genes are likely to be involved in trait determination (Roff 1997; Mackay & Lyman 2005). This is particularly true of behavioural syndromes, which are likely to involve the interaction of numerous different types of pathways (see also Coppens et al. 2010). Quantitative genetics provides a framework for the synthesis of the emergent properties, expressed as means, variances and covariances, of these different pathways. Genomic analysis can provide us with insight into underlying mechanisms as illustrated by work on Drosophila (Anholt & Mackay 2001), African cichlids (Hofmann et al. 1999) and Atlantic salmon (Aubin-Horth et al. 2005a,b) and in regards to questions specific to behavioural syndrome research (Bell & Aubin-Horth 2010; van Oers & Mueller 2010).
7. Conclusions
The emergence of behavioural syndromes as a prominent aspect of research in behavioural ecology represents an encouraging tendency to shift from a univariate to a multivariate view of the behavioural phenotype. This view allows a more realistic understanding of behavioural evolution and a more complete, albeit complicated, understanding of how proximate and ultimate factors shape how organisms interact with their environments. However, for the study of behavioural syndromes to properly advance, it is important that researchers be cognizant of the research done in other areas of evolutionary ecology. Many of the complexities of multivariate evolution have had potential explanations proposed in other areas of research. Indeed, the study of how and why behaviours vary and covary is essentially a study of behaviour from a quantitative genetics framework.
While we have stressed the importance of estimating genotypic covariances between behaviours, it is important to again emphasize that many of the tools as well as conceptual aspects of quantitative genetics are applicable even if heritabilities and genotypic covariances cannot be estimated due to logistic constraints. For example, the reaction norm perspective is important at the individual level. While individual variation in consistency has been observed as part of syndromes (e.g. Koolhaas et al. 1999; Dochtermann & Jenkins 2007), formally placing this variation in the context of reaction norms provides additional predictions from available theory. Likewise, tools such as common principal components analysis and MANOVA can be readily implemented with phenotypic data in the absence of genetic information. If already aware of the tools and theory available in quantitative genetics, syndrome researchers can dedicate energy that would otherwise be applied to reinventing the wheel to asking novel evolutionary questions about how and why behavioural syndromes evolve.
Acknowledgements
We thank Denis Réale, Niels Dingemanse, Anahita Kazem and Jonathon Wright for insightful discussions of the topics covered in this paper and for their invitation to participate in this issue of Philosophical Transactions. We also thank Mathias Kölliker and an anonymous reviewer for insightful comments on an earlier version of this paper.
Footnotes
One contribution of 16 to a Theme Issue ‘Evolutionary and ecological approaches to the study of personality’.
References
- Anholt R. R. H., Mackay T. F. C.2001The genetic architecture of odor-guided behavior in Drosophila melanogaster. Behav. Genet. 31, 17–27 10.1023/A:1010201723966 (doi:10.1023/A:1010201723966) [DOI] [PubMed] [Google Scholar]
- Arnold S. J.1994Multivariate inheritance and evolution: a review of concepts. In Quantitative genetic studies of behavioral evolution (ed. Boake C. R. B.), pp. 17–48 Chicago, IL: The University of Chicago Press [Google Scholar]
- Astles P. A., Moore A. J., Preziosi R. F.2006A comparison of methods to estimate cross-environment genetic correlations. J. Evol. Biol. 19, 114–122 10.1111/j.1420-9101.2005.00997.x (doi:10.1111/j.1420-9101.2005.00997.x) [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N., Landry C. R., Letcher B. H., Hofmann H. A.2005aAlternative life histories shape brain gene expression profiles in males of the same population. Proc. R. Soc. B 272, 1655–1662 10.1098/rspb.2005.3125 (doi:10.1098/rspb.2005.3125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N., Letcher B. H., Hofmann H. A.2005bInteraction of rearing environment and reproductive tactic on gene expression profiles in Atlantic salmon. J Hered. 96, 261–278 10.1093/jhered/esi030 (doi:10.1093/jhered/esi030) [DOI] [PubMed] [Google Scholar]
- Beavis W. D.1994The power and deceit of QTL experiments: lessons from comparative QTL studies. In Proceedings of the Forty-ninth Annual Corn and Sorghum Industry Research Conference 1994, pp. 250–266 Washington, DC: American Seed Trade Association [Google Scholar]
- Bégin M., Roff D. A., Debat V.2004The effect of temperature and wing morphology on quantitative genetic variation in the cricket Gryllus firmus, with an appendix examining the statistical properties of the Jackknife-manova method of matrix comparison. J. Evol. Biol. 17, 1255–1267 10.1111/j.1420-9101.2004.00772.x (doi:10.1111/j.1420-9101.2004.00772.x) [DOI] [PubMed] [Google Scholar]
- Bell A. M.2005Behavioural differences between individuals and two populations of stickleback (Gasterosteus aculeatus). J. Evol. Biol. 18, 464–473 10.1111/j.1420-9101.2004.00817.x (doi:10.1111/j.1420-9101.2004.00817.x) [DOI] [PubMed] [Google Scholar]
- Bell A. M.2007Future directions in behavioural syndromes research. Proc. R. Soc. B 274, 755–761 10.1098/rspb.2006.0199 (doi:10.1098/rspb.2006.0199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. M., Aubin-Horth N.2010What can whole genome expression data tell us about the ecology and evolution of personality? Phil. Trans. R. Soc. B 365, 4001–4012 10.1098/rstb.2010.0185 (doi:10.1098/rstb.2010.0185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A. M., Stamps J. A.2004Development of behavioural differences between individuals and populations of sticklebacks, Gasterosteus aculeatus. Anim. Behav. 68, 1339–1348 10.1016/j.anbehav.2004.05.007 (doi:10.1016/j.anbehav.2004.05.007) [DOI] [Google Scholar]
- Bell A. M., Hankison S. J., Laskowski K. L.2009The repeatability of behaviour: a meta-analysis. Anim. Behav. 77, 771–783 10.1016/j.anbehav.2008.12.022 (doi:10.1016/j.anbehav.2008.12.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blows M. W., Hoffmann A. A.2005A reassessment of genetic limits to evolutionary change. Ecology 86, 1371–1384 10.1890/04-1209 (doi:10.1890/04-1209) [DOI] [Google Scholar]
- Blum A. 1988Plant breeding for stress environments. Boca Raton, FL: CRC Press [Google Scholar]
- Brodin T.2008Behavioral syndrome over the boundaries of life—carryovers from larvae to adult damselfly. Behav. Ecol. 20, 30–37 10.1093/beheco/arn111 (doi:10.1093/beheco/arn111) [DOI] [Google Scholar]
- Calsbeek B., Goodnight C. J.2009Empirical comparison of G matrix test statistics: finding biologically relevant change. Evolution 63, 2627–2635 10.1111/j.1558-5646.2009.00735.x (doi:10.1111/j.1558-5646.2009.00735.x) [DOI] [PubMed] [Google Scholar]
- Cano J. M., Makinen H. S., Merila J.2008Genetic evidence for male-biased dispersal in the three-spined stickleback (Gasterosteus aculeatus). Mol. Ecol. 17, 3234–3242 10.1111/j.1365-294X.2008.03837.x (doi:10.1111/j.1365-294X.2008.03837.x) [DOI] [PubMed] [Google Scholar]
- Cheverud J. M.1988A comparison of genetic and phenotypic correlations. Evolution 42, 958–968 10.2307/2408911 (doi:10.2307/2408911) [DOI] [PubMed] [Google Scholar]
- Cheverud J. M., Marroig G.2007Comparing covariance matrices: random skewers method compared to the common principal components model. Genet. Mol. Biol. 30, 461–469 10.1590/S1415-47572007000300027 (doi:10.1590/S1415-47572007000300027) [DOI] [Google Scholar]
- Coppens C. M., de Boer S. F., Koolhaas J. M.2010Coping styles and behavioural flexibility: towards underlying mechanisms. Phil. Trans. R. Soc. B 365, 4021–4028 10.1098/rstb.2010.0217 (doi:10.1098/rstb.2010.0217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemanse N. J., Wright J., Kazem A. J. N., Thomas D. K., Hickling R., Dawnay N.2007Behavioural syndromes differ predictably between 12 populations of three-spined stickleback. J. Anim. Ecol. 76, 1128–1138 10.1111/j.1365-2656.2007.01284.x (doi:10.1111/j.1365-2656.2007.01284.x) [DOI] [PubMed] [Google Scholar]
- Dingemanse N. J., van der Plas F., Wright J., Réale D., Schrama M., Roff D. A., Van der Zee E., Barber I.2009Individual experience and evolutionary history of predation affect expression of heritable variation in fish personality and morphology. Proc. R. Soc. B 276, 1285–1293 10.1098/rspb.2008.1555 (doi:10.1098/rspb.2008.1555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemanse N. J., Dochtermann N., Wright J.2010aA method for exploring the structure of behavioural syndromes to allow formal comparison within and between datasets. Anim. Behav. 79, 439–450 10.1016/j.anbehav.2009.11.024 (doi:10.1016/j.anbehav.2009.11.024) [DOI] [Google Scholar]
- Dingemanse N. J., Kazem A. J. N., Réale D., Wright J.2010bBehavioural reaction norms: animal personality meets individual plasticity. Trends Ecol. Evol. 25, 81–89 10.1016/j.tree.2009.07.013 (doi:10.1016/j.tree.2009.07.013) [DOI] [PubMed] [Google Scholar]
- Dloniak S. M., French J. A., Holekamp K. E.2006Rank-related maternal effects of androgens on behaviour in wild spotted hyaenas. Nature 440, 1190–1193 10.1038/nature04540 (doi:10.1038/nature04540) [DOI] [PubMed] [Google Scholar]
- Dochtermann N. A., Jenkins S. H.2007Behavioural syndromes in Merriam's kangaroo rats (Dipodomys merriami): a test of competing hypotheses. Proc. R. Soc. B 274, 2343–2349 10.1098/rspb.2007.0622 (doi:10.1098/rspb.2007.0622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- East M. L., Honer O. P., Wachter B., Wilhelm K., Burke T., Hofer H.2009Maternal effects on offspring social status in spotted hyenas. Behav. Ecol. 20, 478–483 10.1093/beheco/arp020 (doi:10.1093/beheco/arp020) [DOI] [Google Scholar]
- Fairbairn D. J., Roff D. A.2006The quantitative genetics of sexual dimorphism: assessing the importance of sex-linkage. Heredity 97, 319–328 10.1038/sj.hdy.6800895 (doi:10.1038/sj.hdy.6800895) [DOI] [PubMed] [Google Scholar]
- Forstmeier W., Coltman D. W., Birkhead T. R.2004Maternal effects influence the sexual behavior of sons and daughters in the zebra finch. Evolution 58, 2574–2583 [DOI] [PubMed] [Google Scholar]
- Gavrilets S.2004Fitness landscapes and the origin of species. Monographs in Population Biology; 41. Princeton, NJ: Princeton University Press [Google Scholar]
- Grafen A. 1984Natural selection, kin selection and group selection. In Behavioural ecology: an evolutionary approach (eds Krebs J. R., Davies N. B.), pp. 62–84, 2nd edn Sunderland, MA: Sinauer [Google Scholar]
- Hadfield J. D.2010MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Statist. Softw. 33, 1–22 [Google Scholar]
- Hadfield J. D., Nutall A., Osorio D., Owens I. P. F.2007Testing the phenotypic gambit: phenotypic, genetic and environmental correlations of colour. J. Evol. Biol. 20, 549–557 10.1111/j.1420-9101.2006.01262.x (doi:10.1111/j.1420-9101.2006.01262.x) [DOI] [PubMed] [Google Scholar]
- Hadfield J. D., Wilson A. J., Garant D., Sheldon B. C., Kruuk L. E. B.2010The misuse of BLUP in ecology and evolution. Am. Nat. 175, 116–125 10.1086/648604 (doi:10.1086/648604) [DOI] [PubMed] [Google Scholar]
- Hofmann H. A., Benson M. E., Fernald R. D.1999Social status regulates growth rate: consequences for life-history strategies. Proc. Natl Acad. Sci. USA 96, 14 171–14 176 10.1073/pnas.96.24.14171 (doi:10.1073/pnas.96.24.14171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr M. K., Churchill G. A.2001Statistical design and the analysis of gene expression microarray data. Genet. Res. 77, 123–128 10.1017/S0016672301005055 (doi:10.1017/S0016672301005055) [DOI] [PubMed] [Google Scholar]
- King E. G., Roff D. A.2010Modeling the evolution of phenotypic plasticity in resource allocation in wing dimorphic insects. Am. Nat. 175, 702–716 10.1086/652434 (doi:10.1086/652434) [DOI] [PubMed] [Google Scholar]
- Koolhaas J. M., Korte S. M., De Boer S. F., Van Der Vegt B. J., Van Reenen C. G., Hopster H., De Jong I. C., Ruis M. A. W., Blokhuis H. J.1999Coping styles in animals: current status in behavior and stress-physiology. Neurosci. Biobehav. Rev. 23, 925–935 10.1016/S0149-7634(99)00026-3 (doi:10.1016/S0149-7634(99)00026-3) [DOI] [PubMed] [Google Scholar]
- Kruuk L. E. B., Slate J., Wilson A. J.2008New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Syst. 39, 525–548 10.1146/annurev.ecolsys.39.110707.173542 (doi:10.1146/annurev.ecolsys.39.110707.173542) [DOI] [Google Scholar]
- Lande R., Arnold S. J.1983The measurement of selection on correlated characters. Evolution 37, 1210–1226 10.2307/2408842 (doi:10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B.1998Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates [Google Scholar]
- Mackay T. F. C., Lyman R. F.2005Drosophila bristles and the nature of quantitative genetic variation. Phil. Trans. R. Soc. B 360, 1513–1527 10.1098/rstb.2005.1672 (doi:10.1098/rstb.2005.1672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElreath R., Strimling P.2006How noisy information and individual asymmetries can make ‘personality’ an adaptation: a simple model. Anim. Behav. 72, 1135–1139 10.1016/j.anbehav.2006.04.001 (doi:10.1016/j.anbehav.2006.04.001) [DOI] [Google Scholar]
- Moore A. J., Brodie E. D., Wolf J. B.1997Interacting phenotypes and the evolutionary process. 1. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362 10.2307/2411187 (doi:10.2307/2411187) [DOI] [PubMed] [Google Scholar]
- Morrissey M. B., Wilson A. J., Pemberton J. M., Ferguson M. M.2007A framework for power and sensitivity analyses for quantitative genetic studies of natural populations, and case studies in Soay sheep (Ovis aries). J. Evol. Biol. 20, 2309–2321 10.1111/j.1420-9101.2007.01412.x (doi:10.1111/j.1420-9101.2007.01412.x) [DOI] [PubMed] [Google Scholar]
- Neff B. D., Sherman P. W.2004Behavioral syndromes versus darwinian algorithms. Trends Ecol. Evol. 19, 621–622 10.1016/j.tree.2004.09.017 (doi:10.1016/j.tree.2004.09.017) [DOI] [Google Scholar]
- Norry F. M., Vilardi J. C., Hasson E.1997Genetic and phenotypic correlations among size-related traits, and heritability variation between body parts in Drosophila buzzatii. Genetica 101, 131–139 10.1023/A:1018360804439 (doi:10.1023/A:1018360804439) [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Wilson A. J., Brommer J. E.2007The evolutionary ecology of individual phenotypic plasticity in wild populations. J. Evol. Biol. 20, 831–844 10.1111/j.1420-9101.2007.01300.x (doi:10.1111/j.1420-9101.2007.01300.x) [DOI] [PubMed] [Google Scholar]
- Onyango P. O., Gesquiere L. R., Wango E. O., Alberts S. C., Altmann J.2008Persistence of maternal effects in baboons: mother's dominance rank at son's conception predicts stress hormone levels in subadult males. Horm. Behav. 54, 319–324 10.1016/j.yhbeh.2008.03.002 (doi:10.1016/j.yhbeh.2008.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips P. C., Arnold S. J.1989Visualizing multivariate selection. Evolution 43, 1209–1222 10.2307/2409357 (doi:10.2307/2409357) [DOI] [PubMed] [Google Scholar]
- Phillips P. C., Arnold S. J.1999Hierarchical comparison of genetic variance–covariance matrices. I. Using the Flury hierarchy. Evolution 53, 1506–1515 10.2307/2640896 (doi:10.2307/2640896) [DOI] [PubMed] [Google Scholar]
- Réale D., Reader S. M., Sol D., McDougall P. T., Dingemanse N. J.2007Integrating animal temperament within ecology and evolution. Biol. Rev. 82, 291–318 10.1111/j.1469-185X.2007.00010.x (doi:10.1111/j.1469-185X.2007.00010.x) [DOI] [PubMed] [Google Scholar]
- Réale D., Martin J., Coltman D. W., Poissant J., Festa-Bianchet M.2009Male personality, life-history strategies and reproductive success in a promiscuous mammal. J. Evol. Biol. 22, 1599–1607 10.1111/j.1420-9101.2009.01781.x (doi:10.1111/j.1420-9101.2009.01781.x) [DOI] [PubMed] [Google Scholar]
- Roff D. A.1995The estimation of genetic correlations from phenotypic correlations—a test of Cheverud's conjecture. Heredity 74, 481–490 10.1038/hdy.1995.68 (doi:10.1038/hdy.1995.68) [DOI] [Google Scholar]
- Roff D. A.1996The evolution of genetic correlations: an analysis of patterns. Evolution 50, 1392–1403 10.2307/2410877 (doi:10.2307/2410877) [DOI] [PubMed] [Google Scholar]
- Roff D. A.1997Evolutionary quantitative genetics. New York, NY: Chapman and Hall [Google Scholar]
- Roff D. A.2001The threshold model as a general purpose normalizing transformation. Heredity 86, 404–411 10.1046/j.1365-2540.2001.00844.x (doi:10.1046/j.1365-2540.2001.00844.x) [DOI] [PubMed] [Google Scholar]
- Roff D.2002Comparing G matrices: a MANOVA approach. Evolution 56, 1286–1291 10.1111/j.0014-3820.2002.tb01439.x (doi:10.1111/j.0014-3820.2002.tb01439.x) [DOI] [PubMed] [Google Scholar]
- Roff D. A.2010Modeling evolution: an introduction to numerical methods. Oxford: Oxford University Press [Google Scholar]
- Roff D. A., Fairbairn D. J.2007The evolution of trade-offs: where are we? J. Evol. Biol. 20, 433–447 10.1111/j.1420-9101.2006.01255.x (doi:10.1111/j.1420-9101.2006.01255.x) [DOI] [PubMed] [Google Scholar]
- Roff D. A., Fairbairn D. J.2009Modeling experimental evolution using individual-based variance-components models. In Experimental evolution (eds Garland T., Rose M.), pp. 31–64 Berkeley, CA: University of California Press [Google Scholar]
- Sih A., Bell A. M., Johnson J. C., Ziemba R. E.2004Behavioral syndromes: an integrative overview. Q. Rev. Biol. 79, 241–277 10.1086/422893 (doi:10.1086/422893) [DOI] [PubMed] [Google Scholar]
- Smiseth P. T., Wright J., Kolliker M.2008Parent–offspring conflict and co-adaptation: behavioural ecology meets quantitative genetics. Proc. R. Soc. B 275, 1823–1830 10.1098/rspb.2008.0199 (doi:10.1098/rspb.2008.0199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D. G., Réale D., Roff D. A.2002Selection, structure and the heritability of behaviour. J. Evol. Biol. 15, 277–289 10.1046/j.1420-9101.2002.00389.x (doi:10.1046/j.1420-9101.2002.00389.x) [DOI] [Google Scholar]
- Svendsen G. E.1974Behavioral and environmental factors in the spatial distribution and population dynamics of a yellow-bellied marmot population. Ecology 55, 760–771 10.2307/1934412 (doi:10.2307/1934412) [DOI] [Google Scholar]
- Tobler M., Sandell M. I.2007Yolk testosterone modulates persistence of neophobic responses in adult zebra finches, Taeniopygia guttata. Horm. Behav. 52, 640–645 10.1016/j.yhbeh.2007.07.016 (doi:10.1016/j.yhbeh.2007.07.016) [DOI] [PubMed] [Google Scholar]
- van Oers K., Mueller J. C.2010Evolutionary genomics of animal personality. Phil. Trans. R. Soc. B 365, 3991–4000 10.1098/rstb.2010.0178 (doi:10.1098/rstb.2010.0178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers K., de Jong G., Drent P. J., van Noordwijk A. J.2004A genetic analysis of avian personality traits: correlated, response to artificial selection. Behav. Genet. 34, 611–619 10.1007/s10519-004-5588-z (doi:10.1007/s10519-004-5588-z) [DOI] [PubMed] [Google Scholar]
- van Oers K., de Jong G., van Noordwijk A. J., Kempenaers B., Drent P. J.2005Contribution of genetics to the study of animal personalities: a review of case studies. Behaviour 142, 1185–1206 10.1163/156853905774539364 (doi:10.1163/156853905774539364) [DOI] [Google Scholar]
- Verhulst S., Perrins C. M., Riddington R.1997Natal dispersal of Great Tits in a patchy environment. Ecology 78, 864–872 10.1890/0012-9658(1997)078[0864:NDOGTI]2.0.CO;2 (doi:10.1890/0012-9658(1997)078[0864:NDOGTI]2.0.CO;2) [DOI] [Google Scholar]
- Waitt D. E., Levin D. A.1998Genetic and phenotypic correlations in plants: a botanical test of Cheverud's conjecture. Heredity 80, 310–319 10.1046/j.1365-2540.1998.00298.x (doi:10.1046/j.1365-2540.1998.00298.x) [DOI] [Google Scholar]
- Weigensberg I., Roff D. A.1996Natural heritabilities: can they be reliably estimated in the laboratory? Evolution 50, 2149–2157 10.2307/2410686 (doi:10.2307/2410686) [DOI] [PubMed] [Google Scholar]
- Willis J. H., Coyne J. A., Kirkpatrick M.1991Can one predict the evolution of quantitative characters without genetics. Evolution 45, 441–444 10.2307/2409678 (doi:10.2307/2409678) [DOI] [PubMed] [Google Scholar]
- Wilson A. D. M., Réale D., Clements M., Morrissey M., Postma E., Wailing C., Kruuk L. E. B., Nussey D. H.2010An ecologist's guide to the animal model. J. Anim. Ecol. 79, 13–26 10.1111/j.1365-2656.2009.01639.x (doi:10.1111/j.1365-2656.2009.01639.x) [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Brodie E. D., Cheverud J. M., Moore A. J., Wade M. J.1998Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13, 64–69 10.1016/S0169-5347(97)01233-0 (doi:10.1016/S0169-5347(97)01233-0) [DOI] [PubMed] [Google Scholar]
- Wolf M., van Doorn G. S., Weissing F. J.2008Evolutionary emergence of responsive and unresponsive personalities. Proc. Natl Acad. Sci. USA 105, 15 825–15 830 10.1073/pnas.0805473105 (doi:10.1073/pnas.0805473105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger R. D., Gibson G., Wolfinger E. D., Bennett L., Hamadeh H., Bushel P., Afshari C., Paules R. S.2001Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8, 625–637 10.1089/106652701753307520 (doi:10.1089/106652701753307520) [DOI] [PubMed] [Google Scholar]


