Abstract
For many years, extracellular matrix (ECM) was considered to function as a tissue support and filler. However, we now know that ECM proteins control many cellular events through their interaction with cell-surface receptors and cytoplasmic signaling pathways. For example, they regulate cell proliferation, cell division, cell adhesion, cell migration, and apoptosis. We focus in this review on a laminin isoform, laminin-332 (formerly termed laminin-5), a major component of the basement membrane (BM) of skin and other epithelial tissues. It is composed of 3 subunits (α3, β3, and γ2) and interacts with at least two integrin receptors expressed by epithelial cells (α3β1 and α6β4 integrin). Mutations in either laminin-332 or integrin α6β4 result in junctional epidermolysis bullosa, a blistering skin disease, while targeting of laminin-332 by autoantibodies in cicatricial pemphigoid leads to dysadhesion of epithelial cells from their underlying connective tissue. Abnormal expression of laminin-332 and its integrin receptors is also a hallmark of certain tumor types and is believed to promote invasion of colon, breast and skin cancer cells. Moreover, there is emerging evidence that laminin-332 and its protease degradation products are not only found at the leading front of several tumors but also likely induce and/or promote tumor cell migration. Thus, in this review, we focus specifically on the role of laminin-332 and its integrin receptors in adhesion, proliferation, and migration/invasion of cancer cells. Finally, we discuss strategies for the development of laminin-332-based antagonists for the treatment of malignant tumors.
Keywords: Integrin, laminin, cancer, basement membrane, proteolysis, cell signaling, cell adhesion, gene expression
INTRODUCTION
Cancer is one of the leading causes of death in developed countries [1]. Therefore, considerable research effort has focused on developing new therapeutic regimens for cancer patients and on improving methods to diagnose cancer so that tumors can be treated as early as possible. One obvious target is the extracellular matrix (ECM).
In normal tissue, cell attachment to the extracellular matrix maintains tissue integrity. However, in tumor tissue, cancer cells infiltrate into the connective tissue (infiltration/invasion), migrate, and enter the blood or lymphatic circulation. Subsequently, they attach to endothelial cells located distant from the original tissue, penetrate through the endothelial cell layer, and then infiltrate new sites: at these new sites, they proliferate and form secondary tumors [2]. In addition, angiogenesis occurs around primary and secondary tumors to nourish the tumors [2]. The ECM is not merely a bystander in the above processes but is an active participant. Both full length matrix proteins and their proteolytic fragments promote survival, adhesion, migration and proliferation of tumor and endothelial cells [3–6]. Thus, understanding tumor cell-ECM interactions and the mechanisms via which ECM and ECM receptors regulate cancer progression is critical to the development of therapies that target cancer progression.
The ECM of normal and diseased tissues is a complex of various molecules. These include various collagens, elastin, fibronectin, laminin, fibulin, perlecan, entactin, and nidogen [7]. Among these, laminin is a glycoprotein produced mainly by epithelial cells and endothelial cells. Laminins are composed of 3 subunits, namely α,β, and γ with each laminin isoform being named based on its component subunits [6]. For instance, laminin-111 (formerly, laminin-1) is composed of α1, β1, and γ1 subunits and laminin-332 (formerly, laminin-5), the subject of this review, consists of α3, β3, and γ2 subunits [8]. Laminin-332 has also been termed kalinin, epiligrin, ladsin, BM600 and nicein [9–12].
The aim of this article is to provide an overview of what we currently know about epithelial-ECM interaction mediated by laminin-332 and its receptors in normal and diseased tissues, with an emphasis on the functional role laminin-332 plays in the dissemination of tumor cells. We conclude with a discussion of therapeutic strategies for cancer treatment using various laminin-332 antagonists. We emphasize that it is not our goal to provide a comprehensive review of the expression and regulation of laminin-332 in various cancers. We direct you to several recent reviews that provide a detailed account of the association of laminin-332 with cancer [13,14].
SECTION 1: STRUCTURE OF LAMININ-332, INTERACTIONS OF ITS SUBUNITS AND ASSEMBLY OF MATRIX ADHESIVE COMPLEXES
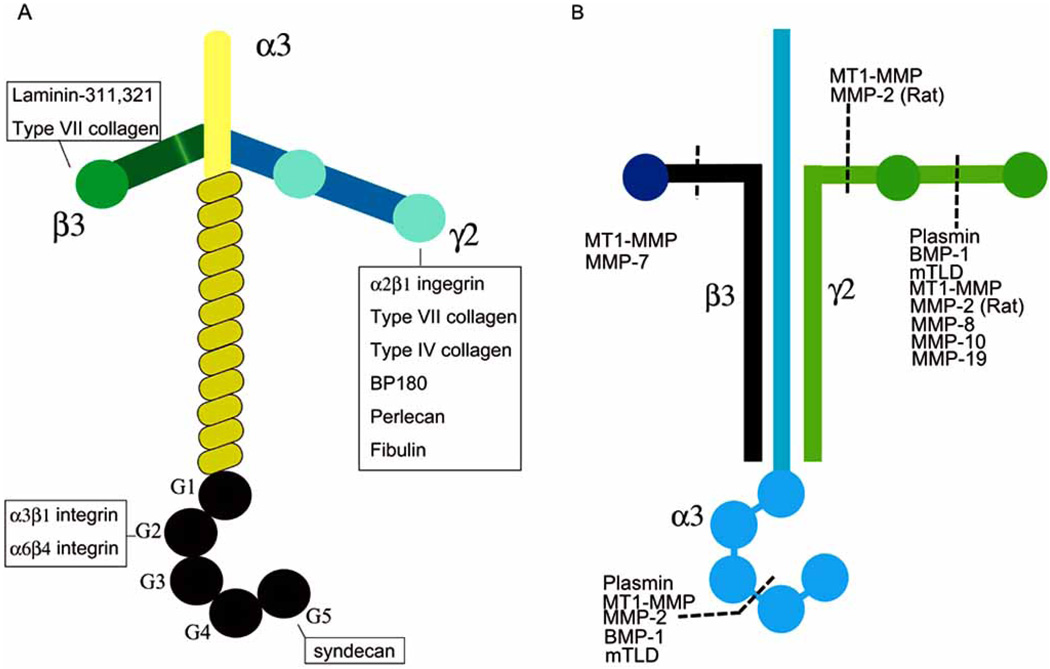
All laminin isoforms are high-molecular-weight trimeric glycoproteins that assemble into cross-like structures (Fig. 1A) [2,6]. Thus far, there are 5 α subunits, 3 β subunits and 3 γ subunits that associate in different combination into at least 15 laminin isoforms [6,15]. These laminin isoforms show distinct tissue distributions and their expression often is precisely regulated during development.
Fig. (1).
(A) The structure of laminin-332. Laminin-332 is composed of three subunits, α3, β3, and γ2 The domains of laminin-332 that bind other molecules are indicated. (B) Cleavage sites in laminin-332. The proteases which degrade laminin-332 are marked.
Unlike some other laminins, laminin-332 is specifically and extensively degraded by proteases either following secretion or as a consequence of tissue remodeling (Fig. 1B). Specifically, the 200-kD laminin α3 chain is cleaved to a 165-kD subunit via the action of bone morphogenic protein (BMP)-1 or serine proteases, while the 155-kD precursor of the γ2 chain is cleaved to a 105-kD subunit via the action of membrane type 1-matrix metalloproteinase (MT1-MMP), MMP-2 or BMP-1 [16,17]. During tissue remodeling, the γ2 subunit is further proteolyzed by MMP-2 to an 80 kD species [18]. The β3 laminin chain has also recently been found to be processed by the action of matrix metalloproteinase-7 (MMP-7, matrilysin-1) or membrane type1 (MT1)-MMP in several cell lines [19,20]. The proteolytic cleavage of the subunits of laminin-332 is biologically important and likely alters its interaction not only with cell-surface receptors but also with other ECM molecules [21].
Each laminin subunit has a number of distinct functional domains via which they assemble into the trimeric molecule, bind to other ECM molecules and/or interact with cell surface receptors (Fig. 1A). For example, the globular (G) domain at the carboxy-terminal of the laminin α chain interacts with cell-surface receptors such as integrin, syndecan, and α-dystroglycan [13,22–24]. In regard to the α3 laminin subunit, its G domain interacts with two integrins (α3β1 and α64 integrin) and syndecan [25,26].
In the BM of normal skin it is believed that the β3 laminin subunit interacts with other laminins [9]. Moreover, the β3 laminin subunit binds to the amino-terminal noncollagenous (NC) 1 domain of type VII collagen [27]. Both of these interactions likely strengthen attachment of keratinocytes to the connective tissue. This notion is supported by the finding that mutations in the gene encoding the β3 laminin subunit cause generalized atrophic benign epidermolysis bullosa [28].
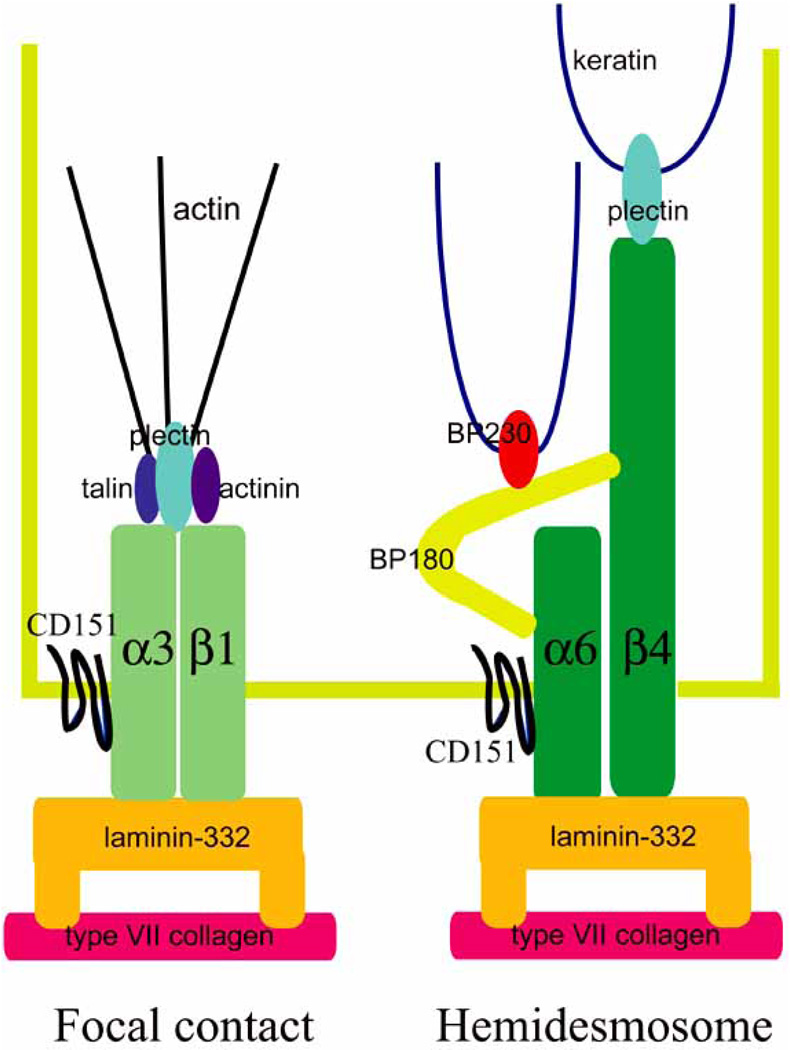
Complexes composed of laminin-332 and α3β1 integrin are assembled by actively migrating cells and likely exist in focal contact adhesive structures, which are not only dynamic attachments but also mediate cell movement (Fig. 2) [29]. In contrast, laminin-332/α6β4 integrin complexes nucleate the assembly of hemidesmosomes in stratified squamous epithelial tissues and in some glands whereas in simple epithelial cells such as those that line the gut the same complex assembles into a variant of the hemidesmosome (a “type II hemidesmosome”) (Fig. 2) [30,31]. Hemidesmosomes are considered to mediate more stable anchorage of cells to their ECM than focal contacts [32,33]. They also differ structurally in that the hemidesmosome tethers the keratin intermediate filament cytoskeleton to the cell surface while in focal contacts α3β1 and other integrins anchor the actin cytoskeleton [32,33].
Fig. (2).
The molecular components of the hemidesmosome, and focal contacts.
SECTION 2: INVOLVEMENT OF LAMININ-332 IN CANCER DEVELOPMENT
Cancer is a malignant transformation of epithelial cells. The ECM of the BM acts as a barrier for cancer cell infiltration but ECM molecules are also co-opted by cancer cells as they invade and migrate from an initial tumor [2,13].
Cancer development can be divided into the following 7 stages: mutation of normal cells, cancer cell proliferation, infiltration/invasion, migration, metastasis, angiogenesis, and additional cell proliferation in the metastatic lesion [2]. Laminin-332 is associated with each of these stages. We have divided the subsequent sections into 6 parts: (1) changes in laminin-332 expression in cancer, (2) proteolytic cleavage of laminin-332 and its relation to cancer, (3) laminin-332 signal transduction in cancer, (4) interaction of laminin-332 and its receptors with other molecules in cancer development, (5) growth factor and tumor promoter regulation of laminin-332 expression and potential role in cancer development, and (6) role of laminin-332 in cancer angiogenesis.
SECTION 2, PART 1: CHANGES IN LAMININ-332 EXPRESSION IN CANCER
The expression of laminin-332 has been reported to be altered at the transcriptional, translational, and posttranslational levels in cancer [34,35]. Methylation of the laminin α3 chain mRNA (LAMA3) promoter or silencing of the genes encoding laminin-332 subunits (α3,β3,γ2) correlates well with tumor stage or breast cancer size [34]. In addition, genetic inactivation through the methylation of the genes encoding laminin-332 subunits has been reported in lung and bladder cancer [35,36]. Moreover, in colon cancer, the laminin γ2 subunit mRNA (LAMC2) promoter has been observed to be activated by transforming growth factor β1 or hepatocyte growth factor (HGF), leading to overexpression of the laminin γ2 subunit protein [37]. Upregulation of expression of laminin-332 in cancer is also controled by a number of other molecules. In colorectal carcinoma, an upregulation of β-catenin induces not only an increase in the expression of MT1-MMP but also activates laminin γ2 subunit gene expression through T-cell factor binding elements and HGF, resulting in an enhancement of tumor development and cancer cell invasion [38,39].
At the protein level there are data indicating that laminin-332 is both up- and down-regulated in cancer. Elevated expression of laminin-332 in cancer is considered a poor diagnostic factor and has been related to tumor invasiveness in the cervical cancer, pancreatic carcinoma, hypopharyngeal cancer, urinary bladder urothelial cancer, small-sized lung adenocarcinoma, malignant glioma, gastric cancer, squamous cell carcinoma (SCC) of the tongue, colorectal adenoma and hepatocellular carcinoma [40–47].
The precise patterns of localization of subunits of laminin-332 and their proteolytic fragments have also been studied in various cancers. For example, in colon carcinoma, urinary bladder carcinoma, colorectal carcinoma, SCC of the oral cavity, tongue, skin, kidney, prostate, lung, vagina, hand and neck, gastric carcinoma, alveolar carcinoma, breast cancer, splenic carcinoma, cervical adenocarcinoma, esophageal carcinoma, adenoid cystic carcinoma, ductal tumor of the pancreas, glioma, clear cell carcinoma of the ovary, and epidermoid anal cancer, cells or tissues adjacent to the tumor front express the laminin γ2 chain or its fragment [48–65]. The latter fragment is also known as the laminin γ2 short arm (SA) and includes an epidermal growth factor (EGF)-like domain. The laminin γ2 fragment is also found in circulating blood in cancer patients and has been reported to be a tumor marker [66].
It should be noted that, contrary to the above, laminin-332 expression has been reported by a number of groups to be down regulated in colorectal carcinoma, breast cancer, prostate carcinoma, and oral SCC [61,67–72]. There are a number of explanations for this apparent discrepancy. The most obvious is that investigators have used different laminin subunit antibodies with distinct antigenic determinants which may or may not be masked in cancer tissue specimens. Another is that laminin-332 antigens may show differential stability depending on how tissue is preserved. Nonetheless, the general consensus in the field is that in many cancers laminin-332 is upregulated and is often found at the migrating edge of tumor cells.
SECTION 2, PART 2: PROTEOLYTIC CLEAVAGE OF LAMININ-332 AND ITS RELATION TO CANCER
Cancer cells and the surrounding mesenchymal cells produce and secrete proteases, which degrade ECM molecules [73,74]. Degradation of the ECM is a necessary prerequisite for the dissemination of cancer cells. However, certain of the proteolytic fragments of ECM proteins also have functions, independent of those of the intact matrix molecule. This is the case with both fragments of the α3 and γ2 subunits of laminin-332 as we will discuss next.
Laminin-332 is subject to proteolysis, mediated by a number of distinct proteases (Fig. 1B). For example, proteins of the astacin family, BMP-1, and its related enzyme, mammalian tolloid (mTLD) have been reported to cleave both the laminin α3 and γ2 subunits of laminin-332 [75]. Thus far, the precise biological role of the latter protease in cancer is unclear although, interestingly, mTLD is the predominant astacin expressed in skin and squamous cell carcinoma [75]. The serine protease plasmin, which is a product of plasminogen degradation by tissue-type plasminogen activator (t-PA), not only binds the G domain of the laminin α3 subunit but cleaves the molecule within the same domain, resulting in a decrease in molecular weight from 190-kD to 160-kD [21,76]. The processing site is a spacer region between G3 and G4 at the amino acids Gln(1337)-Asp(1338) [77]. The G4/G5 fragment that is released is believed to stimulate cell migration [25].
Proteases belonging to the MMP family, zinc-dependent endoproteases, in particular MMP-2 or MT1-MMP cleave the γ2 subunit of laminin-332 [17,18,78]. MT1-MMP directly, or indirectly through MMP-2, is reported to cleave the laminin γ2 subunit into 100-, 85-, 27-, and 25-kD subunits. Among these products, the 27-kD fragment (γ2SA), mentioned above, has been reported to stimulate the EGF receptor and thus can induce cancer cell migration [79]. MMP-3, MMP-12, MMP-13, MMP-19, and MMP-20 have also been reported to cleave the γ2 laminin subunit, and enhance epithelial cell migration [80,81]. With regard to the laminin β3 chain, MMP-7 (matrilysin) cleaves the laminin β3 chain into a 90-kD subunit at Ala(515)-Ile(516) [19,82]. This 90-kD product has been reported to stimulate the migration of colon carcinoma cells [19]. Protease cleavage sites within the subunits of laminin-332 are summarized in Table 1 [16–19,21,77,78,81,82].
Table 1.
The Reported Amino Acid Proteolytic Cleavage Sites within Laminin-332
| Chain | A.A. Position | Protease |
|---|---|---|
| α3 | Lys(191)-Asp(192) | BMP-1 |
| Gln(1337)-Asp(1338) | serine protease | |
| β3 | Ala(515)-Ile(516) | MMP-7 |
| γ2 | Gly(434)-Asp(435) | BMP-1, MT1-MMP |
| Gly(559)-Asp(560) | MT1-MMP | |
| Gly(579)-Ser(580) | MT1-MMP | |
| γ2rat | Gly(413)-Asp(414) | MT1-MMP |
| Ala(586)-Leu(587) | MT1-MMP, MMP-2, 3, 12,13, 20 | |
| Leu(587)-Thr(588) | MMP-8 | |
| Gly(413)-Asp(414) | MMP-14 |
SECTION 2, PART 3: LAMININ-332 SIGNAL TRANSDUCTION IN CANCER
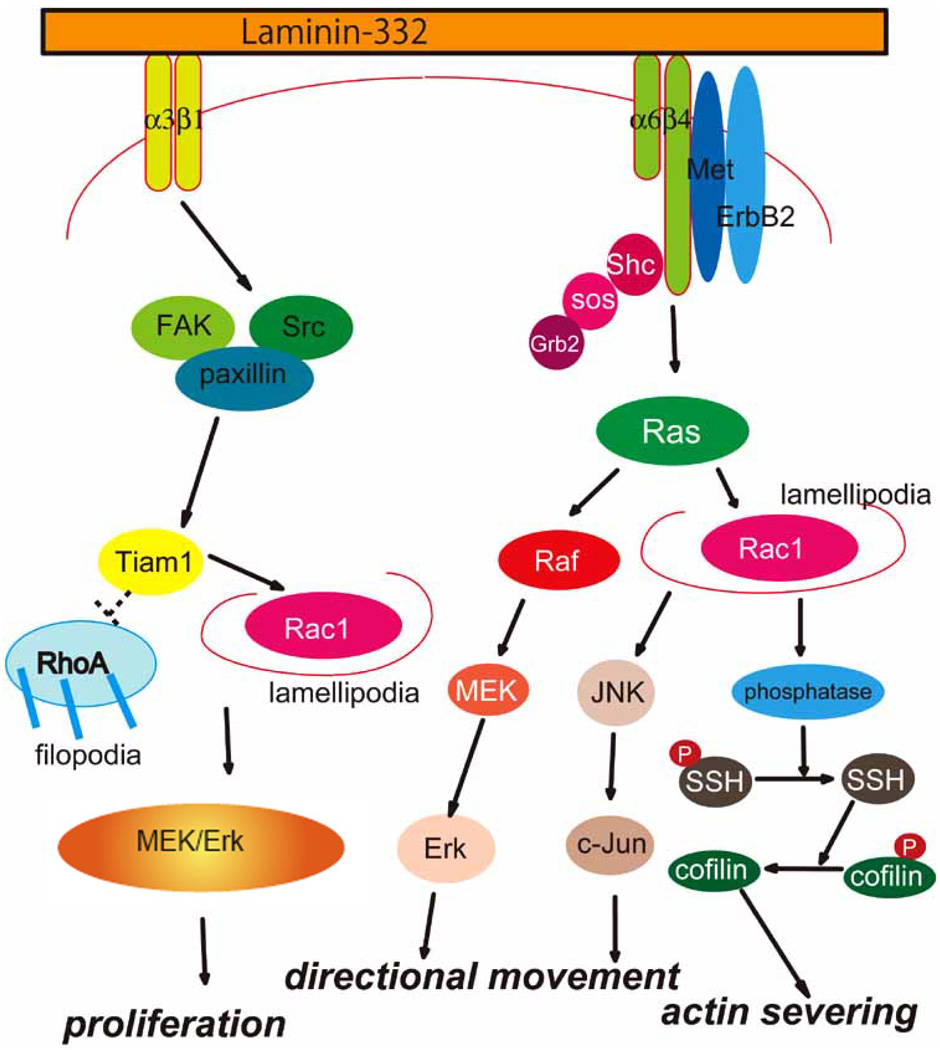
Although the proteolytic fragments of the subunits of laminin-332 support tumor dissemination, it is also clear that laminin-332 enhances cancer development by its impact on various cell surface receptors and their associated signaling pathways (Fig. 3). In this regard, the role of laminin-332-α3β1 integrin signaling in cancer cell migration has been the most extensively investigated. However, there is also evidence that laminin-332-α6β4 integrin-mediated signaling also plays a role in cancer cell motility.
Fig. (3).
A summary of signal transduction pathways associated with laminin-332 in normal and cancer cells.
The enhanced invasiveness of pancreatic carcinoma, colorectal adenocarcinoma, head and neck SCC, and melanoma cells is considered to be a result of enhanced laminin-332-α3β1 integrin signaling [83]. For example, laminin-332-α3β1 integrin binding leads to the activation of the mitogen-activated protein (MAP) kinase pathway and may therefore promote cancer cell proliferation [84]. In oral cancer cells, the activity of RhoA is suppressed when cells adhere to laminin-332 via α3β1 integrin [85]. In addition, the engagement of laminin-332 by α3β1 integrin interaction activates cdc42 and its effector the serine threonine kinase PAK1 leading to an enhancement of cell motility [85]. Laminin-332-α3β1 integrin signaling may also activate the FAK/Src/Rac1 pathway and the formation of lamellipodia [86]. α3β1 integrin signaling is also involved in laminin-332 matrix assembly by normal and tumor cells [87,88]. This appears to involve Rho-GTPases, stress fiber and focal contact formation and is dependent on the protein T-lymphoma invasion and metastasis 1 (Tiam1) [87,88].
As we have already mentioned, in normal cells laminin-332-α6β4 integrin interaction is at the site of hemidesmosomes. However, in cancer cells, it is now believed that laminin-332-α6β4 integrin interaction triggers a number of signaling cascades that promote both cell migration and cancer cell survival. Laminin-332-α6β4 integrin association causes clustering of receptor tyrosine kinases (RTKs), such as ErbB2, EGF receptor, and Met (a hepatocyte growth factor receptor), thereby resulting in the phosphorylation of the cytoplasmic domain of β4 integrin [89]. These events lead to a recruitment of the adaptor protein, Shc to the β4cytoplasmic tail; this protein then undergoes phosphorylation and recruits the Grb2/Sos complex, leading to activation of the Ras/Raf/MEK/Erk pathway or the Ras/Rac/JNK/c-Jun pathway [90,91]. Moreover, Akt/PkB kinase and lipid kinase pathways have also been reported to be activated following α6β4 integrin clustering by laminin-332 ligand [92,93]. In keratinocytes, Rac1 signaling leads to MAP kinase and NF-κB activation resulting in cell proliferation, differentiation, apoptosis, cytoskeletal reorganization and migration [94].
Recently, signaling mediated by α6β4 integrin has been shown to be involved in the assembly of laminin-332 tracks which determine cell migration behavior. Sehgal et al. showed that β4 integrin-deficient (JEB) keratinocytes display aberrant migration; they move in circles, a behavior that mirrors the circular arrays of laminin-332 in their matrix [95]. In contrast, wild-type keratinocytes, and JEB keratinocytes induced to express β4 integrin, assemble laminin-332 in linear tracks over which they migrate. Moreover, laminin-332-dependent migration of JEB keratinocytes along linear tracks is restored when cells are plated on wild-type keratinocyte matrix. The activities of Rac1 and the actin cytoskeleton-severing protein cofilin are low in JEB keratinocytes compared with wild-type cells but are rescued following expression of wild-type β4 integrin protein in JEB cells. Moreover, Rac1 or cofilin inactivation results in wild-type keratinocytes moving in circles over rings of laminin-332 in their matrix. Recently, Kligys et al. have identified key components that regulate the ability of β4 integrin to determine cofilin activation [96]. They analyzed how cofilin phosphorylation is regulated by certain phosphatases, termed slingshots (SSH1-3), downstream of signaling by α6β4 integrin/Rac1 in human keratinocytes. Moreover, expression of phosphatase-dead versions of all three SSH proteins results in phosphorylation/inactivation of cofilin, changes in actin cytoskeleton organization, loss of cell polarity and assembly of aberrant arrays of laminin-332 in human keratinocytes. SSH activity is regulated by 14-3-3 protein binding. Taken together these findings suggest novel mechanisms in which α6β4̣ integrin signaling via Rac1, 14-3-3 proteins and SSH family members regulates cofilin activation, cell polarity and matrix assembly, leading to specific epidermal cell migration behavior.
SECTION 2, PART 4: INTERACTION OF LAMININ-332 AND ITS RECEPTORS WITH OTHER MOLECULES IN CANCER DEVELOPMENT
In cancer cells and tissues, laminin-332 is reported to interact with a number of other matrix molecules. In oral SCC invasion, laminin-332 and tenascin-C co-deposition has been detected [97,98]. Moreover, in poorly differentiated esophageal adenocarcinoma, the co-expression of laminin-332 and tenascin-C has been reported [99]. Moreover, the down-regulation of tenascin-C along with laminin-332 is greater in scirrhous hepatocellular carcinoma (HCC) than in nonscirrhous HCC [100].
The coexpression of laminin-332 with type VII collagen indicates poor prognosis in esophageal SCC [101]. Moreover, Ortiz-Urda et al. reported that a specific collagen VII fragment including the NC1 domain promotes tumor cell invasion in a laminin-332-dependent fashion in recessive dystrophic epidermolysis bullosa (RDEB) keratinocytes [102]. This may partly explain the association of epidermal cancer with RDEB.
The activity of laminin-332 binding integrins is regulated by molecules called tetraspanins in both normal and tumor cells. As their name suggests, tetraspanin molecules are characterized by four transmembrane domains [103–106]. The tetraspanin CD151 interacts with both integrins α3β1 and α6β4 and is believed to strengthen the cell attachment to ECM molecules [107,108]. Moreover, CD151 modulates integrin-dependent signals, such as Ras, or CDC42 [109,110]. Recently, Zijlstra et al. have reported that CD151 regulates the dissemination of tumor cells in vivo [111]. This is consistent with a proposed role for CD151 in regulating cell migration [112].
SECTION 2, PART 5: GROWTH FACTOR AND TUMOR PROMOTER REGULATION OF LAMININ-332 EXPRESSION AND POTENTIAL ROLE IN CANCER DEVELOPMENT
The role of growth factors and tumor promoters in regulating laminin-332 synthesis remains uncertain. Some workers have reported that epidermal growth factor, insulin-like growth factor-1, interferon-γ and keratinocyte growth factor, transforming growth factor (TGF)-α tumor necrosis factor (TNF)-α, TGF-β1 and TPA trigger an increase in laminin-332 production by epithelial cells in vitro [52,113,114]. In contrast, others have presented evidence that at least one of the above growth factors (TGF-β) induces a decrease in the expression of laminin-332 [115]. Moreover, TGF-β1 causes dramatic changes in cellular interactions of keratinocytes with laminin-332. It inhibits the interaction between laminin-332 and α3β1 and α6β4 integrin by downregulating the surface expression of these integrins [116]. In contrast, it promotes interaction of unprocessed laminin-332 in the matrix of the TGF-β treated cells with heparin sulfate proteoglycan, thereby enhancing cell migration [116], but the upregulation of the binding between unprocessed form of laminin-332 (including G4/5) and heparin sulfate proteoglycan increases [116]. As regards EGF, EGF receptor and the α6β4 integrin are often overexpressed in highly invasive SCCs [117–119]. EGF treatment induces tyrosine phosphorylation of the cytoplasmic domain of β4 integrin and disruption of hemidesmosomes [120]. Interestingly, recent data have suggested that laminin-332 receptors work synergistically with growth factors in regulating cancer cells. For example, the production of HGF by tumor cells coincides with an increase in expression of Met [121]. This increase results in phosphorylation of the cytoplasmic tail of β4 integrin, leading to an increase in migration and invasion of tumor cells [122]. Indeed, at the surface α6β4 integrin interacts with a number of receptor tyrosine kinases including EGF-R, ErbB2, Ron, as well as Met. When these kinases are stimulated, the β4 integrin tail undergoes phosphorylation, thereby inducing not only disassembly of hemidesmosomes but also an enhancement of cell motility [122–125].
SECTION 2, PART 6. ROLE OF LAMININ-332 IN CANCER ANGIOGENESIS
Although laminin-332 is best known as an epithelial BM component, it has also been reported in the BM of blood vessels [126]. Its receptor α6β4 integrin has also been localized to endothelial cells [127]. In fact, β4 integrin knockout mice have reduced angiogenesis in skin wounds, suggesting the importance of β4 integrin-laminin-332 interaction in angiogenesis [89]. In human gliomas, the expression of MMP2, MT1-MMP, the laminin γ2 chain, and angiopoietin-2 are associated with tumor angiogenesis [128]. Moroever, there is an upregulation in expression of the γ2 subunit of laminin-332 during vasculogenesis in melanoma [129].
FUTURE PERSPECTIVES: LAMININ-332 AND CANCER TREATMENT
As we have already discussed, in normal tissues laminin-332 acts to tether cells and inhibits their movement by inducing assembly of stable adhesive devices called hemidesmosomes. In a variety of tumors, laminin-332 functions in a completely different fashion by promoting migration. The importance of laminin-332 in the development of a variety of cancers makes it an attractive target for cancer therapeutics. However, laminin-332-based cancer therapies are not without their problems since treatments of necessity must target tumor laminin-332 but not laminin-332 in normal tissues since the latter is essential for maintaining tissue integrity. Based on this important caveat, we propose the following strategies to generate potential laminin-332-based therapeutic agents; (1) develop and apply specific antagonists that inhibit laminin-332 in tumors but not laminin-332 in normal tissue, (2) develop and apply specific inhibitors of proteases that degrade laminin-332 in tumors but not normal tissue, (3) develop signal transduction-oriented modifiers that promote stable interactions of cells with laminin-332 while inhibiting cell migration on laminin-332, (4) develop recombinant laminin-332 isoforms that promote robust cellular adhesion without promoting cell motility, (5) develop molecules that enhance the production of endogeneous highly adhesive laminin-332, and (6) develop molecules that enhance the degradation of laminin-332 isoforms that promote cell migration.
There are already promising results using some of the above approaches. Marinkovich and his coworkers have recently demonstrated the utility of a laminin-332 antibody antagonist in the treatment of SCC of the skin. These workers first showed that the G4/5 domain of laminin-332 is expressed in most human SCCs. As we have already mentioned this domain is absent in normal tissues. More important is the fact that, these same workers treated an animal model of SCC with an antibody against the G4/5 domain. Remarkably, this antibody treatment inhibits SCC tumor proliferation, increases tumor cell apoptosis, and inhibits human SCC tumorigenesis. It does so without impacting normal tissue structure [130].
In addition to antibody antagonists, a number of groups have identified pharmaceutical reagents and synthetic peptides that may be capable of inhibiting expression of laminin-332 or some of the biological functions of laminin-332 in tumors. For example, COL-3 is a chemically modified tetracycline reported to inhibit expression of the laminin γ2 chain gene in melanoma [131]. Moreover, certain peptides have the potential to perturb laminin-332-induced signal transduction and laminin-332-induced cell migration. The synthetic D-amino acid peptide HYD1 (KIKMVISWKG) reversibly inhibits cytoskeleton-dependent tumor cell migration on laminin-332 [132] while peptides containing the tripeptide motif KLP, which is homologous to laminin-332, have been demonstrated to inhibit the growth of peritoneal tumors [133].
Future development of new reagents that inhibit the ability of laminin-332 to drive tumor growth and/or dissemination will be greatly facilitated once we better understand not only the precise regulation of laminin-332 proteolytic processing but also the different signaling cascades regulated by laminin-332 in normal versus tumor cells.
Footnotes
Copyright of Current Medical Chemistry is the property of Bentham Science Publishers Ltd. and its content may not be copied or emailed to multiple sites or posted to a listserv without the copyright holder's express written permission. However, users may print, download, or email articles for individual use.
References
- 1.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Lancet. 2006;367:1747. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 2.Engbring JA, Kleinman HK. J. Pathol. 2003;200:465. doi: 10.1002/path.1396. [DOI] [PubMed] [Google Scholar]
- 3.Gumbiner BM. Cell. 1996;84:345. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 4.Aumailley M, Smyth N. J. Anat. 1998;193:1. doi: 10.1046/j.1469-7580.1998.19310001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mooney A, Jackson K, Bacon R, Streuli C, Edwards G, Bassuk J, Savill J. Am. J. Pathol. 1999;155:599. doi: 10.1016/s0002-9440(10)65155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miner JH, Yurchenco PD. Annu. Rev. Cell Dev. Biol. 2004;20:255. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- 7.Charonis A, Sideraki V, Kaltezioti V, Alberti A, Vlahakos D, Wu K, Tsilibary E. Curr. Med. Chem. 2005;12:1495. doi: 10.2174/0929867054039071. [DOI] [PubMed] [Google Scholar]
- 8.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, Engel J, Engvall E, Hohenester E, Jones JC, Kleinman HK, Marinkovich MP, Martin GR, Mayer U, Meneguzzi G, Miner JH, Miyazaki K, Patarroyo M, Paulsson M, Quaranta V, Sanes JR, Sasaki T, Sekiguchi K, Sorokin LM, Talts JF, Tryggvason K, Uitto J, Virtanen I, von der Mark K, Wewer UM, Yamada Y, Yurchenco PD. Matrix Biol. 2005;24:326. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Marinkovich MP, Lunstrum GP, Burgeson RE. J. Biol. Chem. 1992;267:17900. [PubMed] [Google Scholar]
- 10.Carter WG, Ryan MC, Gahr PJ. Cell. 1991;65:599. doi: 10.1016/0092-8674(91)90092-d. [DOI] [PubMed] [Google Scholar]
- 11.Miyazaki K, Kikkawa Y, Nakamura A, Yasumitsu H, Umeda M. Proc. Natl. Acad. Sci. USA. 1993;90:11767. doi: 10.1073/pnas.90.24.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marinkovich MP, Verrando P, Keene DR, Meneguzzi G, Lunstrum GP, Ortonne JP, Burgeson RE. Lab. Invest. 1993;69:295. [PubMed] [Google Scholar]
- 13.Miyazaki K. Cancer Sci. 2006;97:91. doi: 10.1111/j.1349-7006.2006.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marinkovich MP. Nat. Rev. Cancer. 2007;7:370. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen NM, Senior RM. Dev. Biol. 2006;294:271. doi: 10.1016/j.ydbio.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Amano S, Scott IC, Takahara K, Koch M, Champliaud MF, Gerecke DR, Keene DR, Hudson DL, Nishiyama T, Lee S, Greenspan DS, Burgeson RE. J. Biol. Chem. 2000;275:22728. doi: 10.1074/jbc.M002345200. [DOI] [PubMed] [Google Scholar]
- 17.Koshikawa N, Minegishi T, Sharabi A, Quaranta V, Seiki M. J. Biol. Chem. 2005;280:88. doi: 10.1074/jbc.M411824200. [DOI] [PubMed] [Google Scholar]
- 18.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Science. 1997;277:225. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 19.Remy L, Trespeuch C, Bachy S, Scoazec JY, Rousselle P. Cancer Res. 2006;66:11228. doi: 10.1158/0008-5472.CAN-06-1187. [DOI] [PubMed] [Google Scholar]
- 20.Nakashima Y, Kariya Y, Miyazaki K. J. Cell Biochem. 2007;100(3):545–556. doi: 10.1002/jcb.21032. [DOI] [PubMed] [Google Scholar]
- 21.Goldfinger LE, Stack MS, Jones JC. J. Cell Biol. 1998;141:255. doi: 10.1083/jcb.141.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang M, Koshikawa N, Schenk S, Quaranta V. J. Biol. Chem. 2001;276:33045. doi: 10.1074/jbc.M100798200. [DOI] [PubMed] [Google Scholar]
- 23.Sugawara K, Tsuruta D, Ishii M, Jones JC, Kobayashi H. Exp. Dermatol. 2008;17:473. doi: 10.1111/j.1600-0625.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 24.Hozumi K, Suzuki N, Nielsen PK, Nomizu M, Yamada Y. J. Biol. Chem. 2006;281:32929. doi: 10.1074/jbc.M605708200. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto O, Bachy S, Odenthal U, Bernaud J, Rigal D, Lortat-Jacob H, Smyth N, Rousselle P. J. Biol. Chem. 2003;278:44168. doi: 10.1074/jbc.M300726200. [DOI] [PubMed] [Google Scholar]
- 26.Baker SE, Hopkinson SB, Fitchmun M, Andreason GL, Frasier F, Plopper G, Quaranta V, Jones JC. J. Cell Sci. 1996;109:2509. doi: 10.1242/jcs.109.10.2509. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, Marinkovich MP, Jones JC, O'Toole EA, Li YY, Woodley DT. J. Invest. Dermatol. 1999;112:177. doi: 10.1046/j.1523-1747.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- 28.McGrath JA, Pulkkinen L, Christiano AM, Leigh IM, Eady RA, Uitto J. J. Invest. Dermatol. 1995;104:467. doi: 10.1111/1523-1747.ep12605904. [DOI] [PubMed] [Google Scholar]
- 29.Litjens SH, de Pereda JM, Sonnenberg A. Trends. Cell Biol. 2006;16:376. doi: 10.1016/j.tcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Fontao L, Dirrig S, Owaribe K, Kedinger M, Launay JF. Exp. Cell Res. 1997;231:319. doi: 10.1006/excr.1996.3465. [DOI] [PubMed] [Google Scholar]
- 31.Uematsu J, Nishizawa Y, Sonnenberg A, Owaribe K. J. Biochem. 1994;115:469. doi: 10.1093/oxfordjournals.jbchem.a124361. [DOI] [PubMed] [Google Scholar]
- 32.Jones JC, Hopkinson SB, Goldfinger LE. Bioessays. 1998;20:488. doi: 10.1002/(SICI)1521-1878(199806)20:6<488::AID-BIES7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 33.Borradori L, Sonnenberg A. J. Invest. Dermatol. 1999;112:411. doi: 10.1046/j.1523-1747.1999.00546.x. [DOI] [PubMed] [Google Scholar]
- 34.Sathyanarayana UG, Padar A, Huang CX, Suzuki M, Shigematsu H, Bekele BN, Gazdar AF. Clin. Cancer Res. 2003;9:6389. [PubMed] [Google Scholar]
- 35.Sathyanarayana UG, Toyooka S, Padar A, Takahashi T, Brambilla E, Minna JD, Gazdar AF. Clin. Cancer Res. 2003;9:2665. [PubMed] [Google Scholar]
- 36.Sathyanarayana UG, Maruyama R, Padar A, Suzuki M, Bondaruk J, Sagalowsky A, Minna JD, Frenkel EP, Grossman HB, Czerniak B, Gazdar AF. Cancer Res. 2004;64:1425. doi: 10.1158/0008-5472.can-03-0701. [DOI] [PubMed] [Google Scholar]
- 37.Olsen J, Kirkeby LT, Brorsson MM, Dabelsteen S, Troelsen JT, Bordoy R, Fenger K, Larsson LI, Simon-Assmann P. Biochem. J. 2003;371:211. doi: 10.1042/BJ20021454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hlubek F, Jung A, Kotzor N, Kirchner T, Brabletz T. Cancer Res. 2001;61:8089. [PubMed] [Google Scholar]
- 39.Hlubek F, Spaderna S, Jung A, Kirchner T, Brabletz T. Int. J. Cancer. 2004;108:321. doi: 10.1002/ijc.11522. [DOI] [PubMed] [Google Scholar]
- 40.Lyons AJ, Jones J. Int. J. Oral Maxillofac. Surg. 2007;36:671. doi: 10.1016/j.ijom.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama M, Sato Y, Okamoto M, Hirohashi S. Laryngoscope. 2004;114:1259. doi: 10.1097/00005537-200407000-00022. [DOI] [PubMed] [Google Scholar]
- 42.Moriya Y, Niki T, Yamada T, Matsuno Y, Kondo H, Hirohashi S. Cancer. 2001;91:1129. doi: 10.1002/1097-0142(20010315)91:6<1129::aid-cncr1109>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 43.Lohi J, Oivula J, Kivilaakso E, Kiviluoto T, Frojdman K, Yamada Y, Burgeson RE, Leivo I, Virtanen I. A.P.M.I.S. 2000;108:161. doi: 10.1034/j.1600-0463.2000.d01-40.x. [DOI] [PubMed] [Google Scholar]
- 44.Fukushima Y, Ohnishi T, Arita N, Hayakawa T, Sekiguchi K. Int. J. Cancer. 1998;76:63. doi: 10.1002/(sici)1097-0215(19980330)76:1<63::aid-ijc11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 45.Tani T, Lumme A, Linnala A, Kivilaakso E, Kiviluoto T, Burgeson RE, Kangas L, Leivo I, Virtanen I. Am. J. Pathol. 1997;151:1289. [PMC free article] [PubMed] [Google Scholar]
- 46.Rabinovitz I, Mercurio AM. Biochem. Cell Biol. 1996;74:811. doi: 10.1139/o96-087. [DOI] [PubMed] [Google Scholar]
- 47.Giannelli G, Bergamini C, Fransvea E, Marinosci F, Quaranta V, Antonaci S. Lab. Invest. 2001;81:613. doi: 10.1038/labinvest.3780270. [DOI] [PubMed] [Google Scholar]
- 48.Habermann J, Lenander C, Roblick UJ, Kruger S, Ludwig D, Alaiya A, Freitag S, Dumbgen L, Bruch HP, Stange E, Salo S, Tryggvason K, Auer G, Schimmelpenning H. Scand. J. Gastroenterol. 2001;36:751. doi: 10.1080/003655201300192021. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson PJ, Rubio C, Lenander C, Auer G, Glimelius B. Ann. Oncol. 2005;16:893. doi: 10.1093/annonc/mdi179. [DOI] [PubMed] [Google Scholar]
- 50.Hellman K, Hellstrom AC, Silfversward C, Salo S, Aspenblad U, Nilsson B, Frankendal B, Tryggvasson K, Auer G. Int. J. Gynecol. Cancer. 2000;10:391. doi: 10.1046/j.1525-1438.2000.010005391.x. [DOI] [PubMed] [Google Scholar]
- 51.Lenander C, Habermann JK, Ost A, Nilsson B, Schimmelpenning H, Tryggvason K, Auer G. Anal. Cell Pathol. 2001;22:201. doi: 10.1155/2001/137404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizushima H, Miyagi Y, Kikkawa Y, Yamanaka N, Yasumitsu H, Misugi K, Miyazaki K. J. Biochem. (Tokyo) 1996;120:1196. doi: 10.1093/oxfordjournals.jbchem.a021541. [DOI] [PubMed] [Google Scholar]
- 53.Skyldberg B, Salo S, Eriksson E, Aspenblad U, Moberger B, Tryggvason K, Auer G. J. Natl. Cancer Inst. 1999;91:1882. doi: 10.1093/jnci/91.21.1882. [DOI] [PubMed] [Google Scholar]
- 54.Ono Y, Nakanishi Y, Ino Y, Niki T, Yamada T, Yoshimura K, Saikawa M, Nakajima T, Hirohashi S. Cancer. 1999;85:2315. [PubMed] [Google Scholar]
- 55.Noel JC, Fernandez-Aguilar S, Fayt I, Buxant F, Ansion MH, Simon P, Anaf V. Acta Obstet. Gynecol. Scand. 2005;84:1119. doi: 10.1111/j.0001-6349.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- 56.Kato N, Sasou S, Teshima S, Motoyama T. Virchows Arch. 2007;450:273. doi: 10.1007/s00428-006-0354-7. [DOI] [PubMed] [Google Scholar]
- 57.Kumamoto M, Kuratomi Y, Yasumatsu R, Nakashima T, Masuda M, Inokuchi A. Auris. Nasus. Larynx. 2006;33:167. doi: 10.1016/j.anl.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Stoltzfus P, Salo S, Eriksson E, Aspenblad U, Tryggvason K, Auer G, Avall-Lundqvist E. Int. J. Gynecol. Pathol. 2004;23:215. doi: 10.1097/01.pgp.0000130107.95607.f6. [DOI] [PubMed] [Google Scholar]
- 59.Masaki T, Matsuoka H, Sugiyama M, Abe N, Izumisato Y, Goto A, Sakamoto A, Atomi Y. Anticancer Res. 2003;23:4113. [PubMed] [Google Scholar]
- 60.Hindermann W, Berndt A, Haas KM, Wunderlich H, Katenkamp D, Kosmehl H. Cancer Detect. Prev. 2003;27:109. doi: 10.1016/s0361-090x(03)00025-4. [DOI] [PubMed] [Google Scholar]
- 61.Aoki S, Nakanishi Y, Akimoto S, Moriya Y, Yoshimura K, Kitajima M, Sakamoto M, Hirohashi S. Dis. Colon Rectum. 2002;45:1520. doi: 10.1007/s10350-004-6460-1. [DOI] [PubMed] [Google Scholar]
- 62.Katoh K, Nakanishi Y, Akimoto S, Yoshimura K, Takagi M, Sakamoto M, Hirohashi S. Oncology. 2002;62:318. doi: 10.1159/000065063. [DOI] [PubMed] [Google Scholar]
- 63.Patel V, Aldridge K, Ensley JF, Odell E, Boyd A, Jones J, Gutkind JS, Yeudall WA. Int. J. Cancer. 2002;99:583. doi: 10.1002/ijc.10403. [DOI] [PubMed] [Google Scholar]
- 64.Kagesato Y, Mizushima H, Koshikawa N, Kitamura H, Hayashi H, Ogawa N, Tsukuda M, Miyazaki K. Jpn. J. Cancer Res. 2001;92:184. doi: 10.1111/j.1349-7006.2001.tb01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pyke C, Romer J, Kallunki P, Lund LR, Ralfkiaer E, Dano K, Tryggvason K. Am. J. Pathol. 1994;145:782. [PMC free article] [PubMed] [Google Scholar]
- 66.Katayama M, Sekiguchi K. J. Mol. Histol. 2004;35:277. doi: 10.1023/b:hijo.0000032359.35698.fe. [DOI] [PubMed] [Google Scholar]
- 67.Shinto E, Tsuda H, Ueno H, Hashiguchi Y, Hase K, Tamai S, Mochizuki H, Inazawa J, Matsubara O. Lab. Invest. 2005;85:257. doi: 10.1038/labinvest.3700199. [DOI] [PubMed] [Google Scholar]
- 68.Haas KM, Berndt A, Stiller KJ, Hyckel P, Kosmehl H. J. Histochem. Cytochem. 2001;49:1261. doi: 10.1177/002215540104901008. [DOI] [PubMed] [Google Scholar]
- 69.Hao J, Jackson L, Calaluce R, McDaniel K, Dalkin BL, Nagle RB. Am. J. Pathol. 2001;158:1129. doi: 10.1016/s0002-9440(10)64060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henning K, Berndt A, Katenkamp D, Kosmehl H. Histopathology. 1999;34:305. doi: 10.1046/j.1365-2559.1999.00634.x. [DOI] [PubMed] [Google Scholar]
- 71.Zinn M, Aumailley M, Krieg T, Smola H. Eur. J. Cell Biol. 2006;85:333. doi: 10.1016/j.ejcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Martin KJ, Kwan CP, Nagasaki K, Zhang X, o'Hare MJ, Kaelin CM, Burgeson RE, Pardee AB, Sager R. Mol. Med. 1998;4:602. [PMC free article] [PubMed] [Google Scholar]
- 73.Stallings-Mann M, Radisky D. Cells Tissues Organs. 2007;185:104. doi: 10.1159/000101310. [DOI] [PubMed] [Google Scholar]
- 74.Duffy M, McGowan P, Gallagher W. J. Pathol. 2008;214:283. doi: 10.1002/path.2282. [DOI] [PubMed] [Google Scholar]
- 75.Veitch DP, Nokelainen P, McGowan KA, Nguyen TT, Nguyen NE, Stephenson R, Pappano WN, Keene DR, Spong SM, Greenspan DS, Findell PR, Marinkovich MP. J. Biol. Chem. 2003;278:15661. doi: 10.1074/jbc.M210588200. [DOI] [PubMed] [Google Scholar]
- 76.Goldfinger LE, Jiang L, Hopkinson SB, Stack MS, Jones JC. J. Biol. Chem. 2000;275:34887. doi: 10.1074/jbc.M006652200. [DOI] [PubMed] [Google Scholar]
- 77.Tsubota Y, Mizushima H, Hirosaki T, Higashi S, Yasumitsu H, Miyazaki K. Biochem. Biophys. Res. Commun. 2000;278:614. doi: 10.1006/bbrc.2000.3851. [DOI] [PubMed] [Google Scholar]
- 78.Koshikawa N, Schenk S, Moeckel G, Sharabi A, Miyazaki K, Gardner H, Zent R, Quaranta V. FASEB J. 2004;18:364. doi: 10.1096/fj.03-0584fje. [DOI] [PubMed] [Google Scholar]
- 79.Decline F, Rousselle P. J. Cell. Sci. 2001;114:811. doi: 10.1242/jcs.114.4.811. [DOI] [PubMed] [Google Scholar]
- 80.Sadowski T, Dietrich S, Koschinsky F, Ludwig A, Proksch E, Titz B, Sedlacek R. Cell Mol. Life Sci. 2005;62:870. doi: 10.1007/s00018-005-4478-8. [DOI] [PubMed] [Google Scholar]
- 81.Pirila E, Sharabi A, Salo T, Quaranta V, Tu H, Heljasvaara R, Koshikawa N, Sorsa T, Maisi P. Biochem. Biophys. Res. Commun. 2003;303:1012. doi: 10.1016/s0006-291x(03)00452-2. [DOI] [PubMed] [Google Scholar]
- 82.Udayakumar TS, Chen ML, Bair EL, Von Bredow DC, Cress AE, Nagle RB, Bowden GT. Cancer Res. 2003;63:2292. [PubMed] [Google Scholar]
- 83.Tsuji T, Kawada Y, Kai-Murozono M, Komatsu S, Han SA, Takeuchi K, Mizushima H, Miyazaki K, Irimura T. Clin. Exp. Metastasis. 2002;19:127. doi: 10.1023/a:1014573204062. [DOI] [PubMed] [Google Scholar]
- 84.Gonzales M, Haan K, Baker SE, Fitchmun M, Todorov I, Weitzman S, Jones JC. Mol. Biol. Cell. 1999;10:259. doi: 10.1091/mbc.10.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou H, Kramer RH. J. Biol. Chem. 2005;280:10624. doi: 10.1074/jbc.M411900200. [DOI] [PubMed] [Google Scholar]
- 86.Choma DP, Milano V, Pumiglia KM, DiPersio CM. J. Invest. Dermatol. 2007;127:31. doi: 10.1038/sj.jid.5700505. [DOI] [PubMed] [Google Scholar]
- 87.DeHart GW, Jones JC. Cell Motil. Cytoskeleton. 2004;57:107. doi: 10.1002/cm.10161. [DOI] [PubMed] [Google Scholar]
- 88.Hamelers IH, Olivo C, Mertens AE, Pegtel DM, van der Kammen RA, Sonnenberg A, Collard JG. J. Cell Biol. 2005;171:871. doi: 10.1083/jcb.200509172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giancotti FG. Trends Pharmacol. Sci. 2007;28:506. doi: 10.1016/j.tips.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 90.Manohar A, Shome SG, Lamar J, Stirling L, Iyer V, Pumiglia K, DiPersio CM. J. Cell Sci. 2004;117:4043. doi: 10.1242/jcs.01277. [DOI] [PubMed] [Google Scholar]
- 91.Mainiero F, Pepe A, Wary KK, Spinardi L, Mohammadi M, Schlessinger J, Giancotti FG. EMBO J. 1995;14:4470. doi: 10.1002/j.1460-2075.1995.tb00126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kippenberger S, Loitsch S, Muller J, Guschel M, Kaufmann R, Bernd A. J. Invest. Dermatol. 2004;123:444. doi: 10.1111/j.0022-202X.2004.23323.x. [DOI] [PubMed] [Google Scholar]
- 93.Nguyen BP, Gil SG, Carter WG. J. Biol. Chem. 2000;275:31896. doi: 10.1074/jbc.M006379200. [DOI] [PubMed] [Google Scholar]
- 94.Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Giancotti FG. Cancer Cell. 2004;6:471. doi: 10.1016/j.ccr.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 95.Sehgal BU, DeBiase PJ, Matzno S, Chew TL, Claiborne JN, Hopkinson SB, Russell A, Marinkovich MP, Jones JC. J. Biol. Chem. 2006;281:35487. doi: 10.1074/jbc.M606317200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kligys K, Claiborne JN, DeBiase PJ, Hopkinson SB, Wu Y, Mizuno K, Jones JC. J. Biol. Chem. 2007;282:32520. doi: 10.1074/jbc.M707041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Franz M, Hansen T, Richter P, Borsi L, Bohmer FD, Hyckel P, Schleier P, Katenkamp D, Zardi L, Kosmehl H, Berndt A. Histochem. Cell Biol. 2006;126:125. doi: 10.1007/s00418-005-0126-5. [DOI] [PubMed] [Google Scholar]
- 98.Berndt A, Borsi L, Hyckel P, Kosmehl H. J. Cancer Res. Clin. Oncol. 2001;127:286. doi: 10.1007/s004320000205. [DOI] [PubMed] [Google Scholar]
- 99.Salmela MT, Karjalainen-Lindsberg ML, Puolakkainen P, Saarialho-Kere U. Br. J. Cancer. 2001;85:383. doi: 10.1054/bjoc.2001.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okamura N, Yoshida M, Shibuya A, Sugiura H, Okayasu I, Ohbu M. Pathol. Int. 2005;55:724. doi: 10.1111/j.1440-1827.2005.01891.x. [DOI] [PubMed] [Google Scholar]
- 101.Baba Y, Iyama K, Honda S, Ishikawa S, Miyanari N, Baba H. Oncology. 2006;71:221. doi: 10.1159/000106426. [DOI] [PubMed] [Google Scholar]
- 102.Ortiz-Urda S, Garcia J, Green CL, Chen L, Lin Q, Veitch DP, Sakai LY, Lee H, Marinkovich MP, Khavari PA. Science. 2005;307:1773. doi: 10.1126/science.1106209. [DOI] [PubMed] [Google Scholar]
- 103.Bienstock RJ, Barrett JC. Mol. Carcinog. 2001;32:139. doi: 10.1002/mc.1073. [DOI] [PubMed] [Google Scholar]
- 104.Hemler ME. Nat. Rev. Mol. Cell Biol. 2005;6:801. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 105.Hasegawa H, Nomura T, Kishimoto K, Yanagisawa K, Fujita S. J. Immunol. 1998;161:3087. [PubMed] [Google Scholar]
- 106.Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK. J. Cell Sci. 1999;112:833. doi: 10.1242/jcs.112.6.833. [DOI] [PubMed] [Google Scholar]
- 107.Nishiuchi R, Sanzen N, Nada S, Sumida Y, Wada Y, Okada M, Takagi J, Hasegawa H, Sekiguchi K. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1939. doi: 10.1073/pnas.0409493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lammerding J, Kazarov AR, Huang H, Lee RT, Hemler ME. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7616. doi: 10.1073/pnas.1337546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sawada K, Mitra AK, Radjabi AR, Bhaskar V, Kistner EO, Tretiakova M, Jagadeeswaran S, Montag A, Becker A, Kenny HA, Peter ME, Ramakrishnan V, Yamada SD, Lengyel E. Cancer Res. 2008;68:2329. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. J. Cell Biol. 2003;163:165. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. Cancer Cell. 2008;13:221. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cowin AJ, Adams D, Geary SM, Wright MD, Jones JC, Ashman LK. J. Invest. Dermatol. 2006;126:680. doi: 10.1038/sj.jid.5700142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Amano S, Akutsu N, Ogura Y, Nishiyama T. Br. J. Dermatol. 2004;151:961. doi: 10.1111/j.1365-2133.2004.06175.x. [DOI] [PubMed] [Google Scholar]
- 114.Lee HK, Lee JH, Kim M, Kariya Y, Miyazaki K, Kim EK. Invest. Ophthalmol. Vis. Sci. 2006;47:873. doi: 10.1167/iovs.05-0826. [DOI] [PubMed] [Google Scholar]
- 115.Maschler S, Wirl G, Spring H, Bredow DV, Sordat I, Beug H, Reichmann E. Oncogene. 2005;24:2032. doi: 10.1038/sj.onc.1208423. [DOI] [PubMed] [Google Scholar]
- 116.Decline F, Okamoto O, Mallein-Gerin F, Helbert B, Bernaud J, Rigal D, Rousselle P. Cell Motil. Cytoskeleton. 2003;54:64. doi: 10.1002/cm.10086. [DOI] [PubMed] [Google Scholar]
- 117.Kimmel KA, Carey TE. Cancer Res. 1986;46:3614. [PubMed] [Google Scholar]
- 118.Yamamoto T, Kamata N, Kawano H, Shimizu S, Kuroki T, Toyoshima K, Rikimaru K, Nomura N, Ishizaki R, Pastan I, Gamou S, Shimizu N. Cancer Res. 1986;46:414. [PubMed] [Google Scholar]
- 119.Tennenbaum T, Belanger AJ, Quaranta V, Yuspa SH. J. Investig. Dermatol. Symp. Proc. 1996;1:157. [PubMed] [Google Scholar]
- 120.Mainiero F, Pepe A, Yeon M, Ren Y, Giancotti FG. J. Cell Biol. 1996;134:241. doi: 10.1083/jcb.134.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boccaccio C, Gaudino G, Gambarotta G, Galimi F, Comoglio PM. J. Biol. Chem. 1994;269:12846. [PubMed] [Google Scholar]
- 122.Trusolino L, Bertotti A, Comoglio PM. Cell. 2001;107:643. doi: 10.1016/s0092-8674(01)00567-0. [DOI] [PubMed] [Google Scholar]
- 123.Falcioni R, Antonini A, Nistico P, Di Stefano S, Crescenzi M, Natali PG, Sacchi A. Exp. Cell Res. 1997;236:76. doi: 10.1006/excr.1997.3695. [DOI] [PubMed] [Google Scholar]
- 124.Mariotti A, Kedeshian PA, Dans M, Curatola AM, Gagnoux-Palacios L, Giancotti FG. J. Cell Biol. 2001;155:447. doi: 10.1083/jcb.200105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Santoro MM, Gaudino G, Marchisio PC. Dev. Cell. 2003;5:257. doi: 10.1016/s1534-5807(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 126.Wang H, Fu W, Im JH, Zhou Z, Santoro SA, Iyer V, DiPersio CM, Yu QC, Quaranta V, Al-Mehdi A, Muschel RJ. J. Cell Biol. 2004;164:935. doi: 10.1083/jcb.200309112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Homan SM, Mercurio AM, LaFlamme SE. J. Cell Sci. 1998;111:2717. doi: 10.1242/jcs.111.18.2717. [DOI] [PubMed] [Google Scholar]
- 128.Guo P, Imanishi Y, Cackowski FC, Jarzynka MJ, Tao HQ, Nishikawa R, Hirose T, Hu B, Cheng SY. Am. J. Pathol. 2005;166:877. doi: 10.1016/s0002-9440(10)62308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Seftor RE, Seftor EA, Koshikawa N, Meltzer PS, Gardner LM, Bilban M, Stetler-Stevenson WG, Quaranta V, Hendrix MJ. Cancer Res. 2001;61:6322. [PubMed] [Google Scholar]
- 130.Tran M, Rousselle P, Nokelainen P, Tallapragada S, Nguyen NT, Fincher EF, Marinkovich MP. Cancer Res. 2008;68:2885. doi: 10.1158/0008-5472.CAN-07-6160. [DOI] [PubMed] [Google Scholar]
- 131.Seftor RE, Seftor EA, Kirschmann DA, Hendrix MJ. Mol. Cancer Ther. 2002;1:1173. [PubMed] [Google Scholar]
- 132.Sroka TC, Pennington ME, Cress AE. Carcinogenesis. 2006;27:1748. doi: 10.1093/carcin/bgl005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Akita N, Maruta F, Seymour LW, Kerr DJ, Parker AL, Asai T, Oku N, Nakayama J, Miyagawa S. Cancer Sci. 2006;97:1075. doi: 10.1111/j.1349-7006.2006.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]