Abstract
OBJECTIVE
Childhood obesity strongly predisposes to some adult diseases. Recently, genome-wide association (GWA) studies in Caucasians identified multiple single nucleotide polymorphisms (SNPs) associated with BMI and obesity. The associations of those SNPs with BMI and obesity among other ethnicities are not fully described, especially in children. Among those previously identified SNPs, we selected six (rs7138803, rs1805081, rs6499640, rs17782313, rs6265, and rs10938397, in or near obesity-related genes FAIM2, NPC1, FTO, MC4R, BDNF, and GNPDA2, respectively) because of the relatively high minor allele frequencies in Chinese individuals and tested the associations of the SNPs with BMI and obesity in Chinese children.
RESEARCH DESIGN AND METHODS
We investigated the associations of these SNPs with BMI and obesity in school-aged children. A total of 3,503 children participated in the study, including 1,229 obese, 655 overweight, and 1,619 normal-weight children (diagnosed by the Chinese age- and sex-specific BMI cutoffs).
RESULTS
After age and sex adjustment and correction for multiple testing, the SNPs rs17782313, rs6265, and rs10938397 were associated with BMI (P = 1.0 × 10−5, 0.038, and 0.00093, respectively) and also obesity (P = 5.0 × 10−6, 0.043, and 0.00085, respectively) in the Chinese children. The SNPs rs17782313 and rs10938397 were also significantly associated with waist circumference, waist-to-height ratio, and fat mass percentage.
CONCLUSIONS
Results of this study support obesity-related genes in adults as important genes for BMI variation in children and suggest that some SNPs identified by GWA studies in Caucasians also confer risk for obesity in Chinese children.
In recent years, the prevalence of adult and childhood obesity has been increasing in most parts of the world, including China (1,2). Overweight children show a higher risk for severe adult obesity (3). Childhood obesity strongly predisposes to some adult diseases such as hypertension, type 2 diabetes, and cardiovascular disease (4–6).
Six single nucleotide polymorphisms (SNPs) (rs7138803, rs1805081, rs6499640, rs17782313, rs6265, and rs10938397, located in or near the fas apoptotic inhibitory molecule 2 [FAIM2] gene, the Niemann-Pick disease type C1 [NPC1] gene, the fat mass and obesity associated [FTO] gene, the melanocortin-4 receptor [MC4R] gene, the brain-derived neurotrophic factor [BDNF] gene, and the glucosamine-6-phosphate deaminase 2 [GNPDA2] gene, respectively) were reported to be associated with obesity by genome-wide association (GWA) studies in Caucasians (7–9). Among these SNPs, rs6265 was associated with BMI in European-American children (10), and rs17782313 and rs10938397 were associated with obesity in Chinese adults (11). However, no independent replication studies of the six SNPs were conducted in Chinese children.
To investigate the associations of these six SNPs with BMI and the risk of overweight and obesity in Chinese children, we genotyped the SNPs in children who participated in the population-based Beijing Child and Adolescent Metabolic Syndrome (BCAMS) study (12). We tested the associations of the six SNPs with BMI, waist circumference, fat mass percentage (FMP), waist-to-height ratio (WHtR), and birth weight as continuous traits. Furthermore, we analyzed the associations of the SNPs with risk of overweight or obesity as dichotomized traits.
RESEARCH DESIGN AND METHODS
Subjects were recruited from a cross-sectional population-based survey termed the BCAMS study (12). The survey included a questionnaire, medical examination, anthropometric measurement, and finger capillary blood tests among a representative sample (n = 19,593; boys 50%) of Beijing children aged 6–18 years in 2004. Anthropometric measurement included weight, height, waist circumference, and FMP by bioimpedance analysis. Birth weight was collected from self-report questionnaires that were completed by parents or guardians.
Within this large group of children, 1,229 obese, 655 overweight, and 1,619 normal-weight children were randomly recruited and diagnosed by the Chinese age- and sex-specific BMI cutoffs (supplementary Table 1 available at http://diabetes.diabetesjournals.org/cgi/content/full/db10-0273/DC1) (13). Also, venipuncture blood samples were collected. We obtained written informed consent from all participants. The BCAMS study was approved by ethics committees of the Capital Institute of Pediatrics.
Measurement of anthropometric parameters.
All instruments were validated following the standard methods of the manufacturers (14). BMI was calculated as weight in kilograms divided by the square of height in meters. Waist circumference was measured midway between the lowest rib and the superior border of the iliac crest at the end of normal expiration with an inelastic measuring tape to the nearest 0.1 cm. FMP was performed with the use of a body composition analyzer (TBF-300A; TANITA).
Selection of SNPs and genotyping.
To achieve a power of not less than 0.75, we selected only SNPs in obesity-related genes with minor allele frequencies >0.14 in Chinese individuals in the HapMap database. With the assumed effect size of 1.2 for an allele frequency of 0.14, the power for detecting positive association was greater than 0.75. We chose the six SNPs (rs7138803 [7], rs1805081 [9], rs6499640 [7], rs17782313 [8], rs6265 [7], and rs10938397 [8]) that have been shown to significantly associate with the risk of obesity.
Genomic DNA was isolated from peripheral blood white cells using the salt fractionation method. SNPs were genotyped by TaqMan (15) Allelic Discrimination Assays with the GeneAmp 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). TaqMan probes were used for genotyping rs7138803 (C__29248155_10), rs1805081 (C__25472673_10), rs6499640 (C__29387696_10), rs17782313 (C__32667060_10), rs6265 (C__11592758_10), and rs10938397 (C___1594245_10). Genotyping call rates for all SNPs were greater than 98%. In order to validate the accuracy of genotyping, we repeated 70 samples randomly for each SNP and observed 100% concordance between the results of the two tests.
Statistical analyses.
The allele increasing BMI was defined as the effect allele in our study. Genetic association between the genotypes at each given SNP and a given phenotype was assessed by multiple regression as implemented in the PLINK program (16). For qualitative and quantitative variables, logistic regression and linear regression were carried out, respectively, with age and sex as covariates. Quantitative variables were suitably transformed prior to analysis (log transformation for BMI, waist circumference, and WHtR; square root transformation for FMP). Genotypes were scored as 0, 1, and 2 reflecting the number of susceptibility alleles, where the latter are defined as alleles conferring risk to the phenotype. Empirical significance levels were computed by randomization tests (100,000 permutations) as implemented in the PLINK program. Nominal P value was computed in a one-sided manner, i.e., as one half of the two-sided P value (all susceptibility alleles were the same as in the published data). False discovery rate, which was computed based on the Benjamini and Hochberg (17) method, was applied for six SNPs, three models (dominant, recessive, and additive), and seven adiposity measures (BMI, waist circumference, WHtR, FMP, birth weight, overweight, and obesity) simultaneously (number of tests: 6 × 2.21 × 7). It has been shown that testing the three models is equivalent to a correction factor of 2.21 (18). Power calculation was performed using Quanto software (http://hydra.usc.edu/gxe/). Hardy-Weinberg equilibrium was assessed by χ2 tests. ANCOVA was applied for comparison of anthropometric indexes between genotypes of SNPs, and linear regression was used to estimate the effect of per allele on anthropometric indexes.
RESULTS
The study included 3,503 children diagnosed by the Chinese age- and sex-specific BMI cutoffs (supplementary Table 1) (13), of whom 1,229 were classified as obese and 655 as overweight. The mean age of subjects was 12.4 ± 3.1 years and 50.8% were boys (see supplementary Table 2 for additional characteristics). We genotyped six SNPs (rs7138803 near FAIM2, rs1805081 in exon 6 of NPC1, rs6499640 in intron 1 of FTO, rs17782313 near MC4R, rs6265 near BDNF, and rs10938397 near GNPDA2) in the cohort. The genotypes of the SNPs were in Hardy-Weinberg equilibrium in each group (P > 0.05) (supplementary Table 3).
After adjusting for multiple testing, statistically significant associations of the SNPs (rs7138803, rs17782313, rs6265, and rs10938397) with BMI (adjusted for age and sex, P = 0.0097, 1.0 × 10−5, 0.038, and 0.00093, respectively) were found under the additive model (Table 1). The data of three models of inheritance (dominant, recessive, and additive) are provided in supplementary Table 4.
TABLE 1.
BMI according to genotypes and per effect allele change in BMI after age and sex adjustment
| SNP | Position | Nearest gene | Effect allele (1) | Other allele (2) | BMI |
P (additive model) |
Per effect allele change in BMI (β) | Lower bound of one-sided 95% CI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | Nominal P | Permuted P | |||||||
| rs7138803 | 48533735 | FAIM2 | A | G | 22.4 ± 0.2 | 22.0 ± 0.1 | 21.7 ± 0.1 | 0.0016 | 0.0097 | 0.35 | 0.15 |
| rs1805081 | 19394430 | NPC1 | A | G | 22.0 ± 0.1 | 21.9 ± 0.1 | 21.4 ± 0.3 | 0.063 | 0.28 | 0.21 | −0.0047 |
| rs6499640 | 52327178 | FTO | A | G | 22.1 ± 0.4 | 22.1 ± 0.1 | 21.8 ± 0.1 | 0.023 | 0.12 | 0.29 | 0.044 |
| rs17782313 | 56002077 | MC4R | C | T | 23.1 ± 0.3 | 22.2 ± 0.1 | 21.6 ± 0.1 | 3.6 × 10−7 | 1.0 × 10−5 | 0.63 | 0.42 |
| rs6265 | 27636492 | BDNF | G | A | 22.2 ± 0.1 | 21.9 ± 0.1 | 21.6 ± 0.2 | 0.0067 | 0.038 | 0.27 | 0.093 |
| rs10938397 | 45023455 | GNPDA2 | G | A | 22.6 ± 0.2 | 22.0 ± 0.1 | 21.6 ± 0.1 | 0.00016 | 0.00093 | 0.45 | 0.26 |
Data are means ± SE. Nominal P values were adjusted for age and sex, and permuted P values were further corrected for multiple testing. All P values were one-sided.
We also tested the associations between the six SNPs and anthropometric indexes, including waist circumference, WHtR, FMP, and birth weight (supplementary Table 5). Our results showed that the SNPs rs17782313 and rs10938397 were significantly associated with waist circumference, WHtR, and FMP, and the SNP rs7138803 was significantly associated with waist circumference and WHtR, but none of the six SNPs were associated with birth weight.
All six SNPs, including rs1805081 (borderline significance, P = 0.048), were significantly associated with obesity in the Chinese children after age and sex adjustment (Table 2). After adjusting for multiple testing, the significant associations of the SNPs rs17782313, rs6265, and rs10938397 with obesity remained (P = 5.0 × 10−6, 0.043, and 0.00085, respectively). The power for rs1805081 was 0.994, assuming the effect size of 1.33 for the effect allele (9) with the frequency of 0.744. In the analysis of overweight status, the only statistically significant association observed was with rs17782313 after age and sex adjustment and correction for multiple testing (Table 2). The data of three models (dominant, recessive, and additive) with power calculations are provided in supplementary Table 6.
TABLE 2.
Associations of SNPs with overweight and obesity in multinomial logistic regression with age and sex adjustment
| SNP | Chr. | Nearest gene | Effect allele | Other allele | Overweight (n = 655) vs. normal weight (n = 1,619) |
Obese (n = 1,229) vs. normal weight (n = 1,619) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (additive model) | Lower bound of one-sided 95% CI | Nominal P | Permuted P | OR (additive model) | Lower bound of one-sided 95% CI | Nominal P | Permuted P | Reported OR (95% CI) | |||||
| rs7138803 | 12 | FAIM2 | A | G | 1.12 | 0.99 | 0.056 | 0.26 | 1.15 | 1.04 | 0.020 | 0.11 | 1.14 (1.09–1.19) |
| rs1805081 | 18 | NPC1 | A | G | 0.95 | 0.83 | 0.20 | 0.48 | 1.12 | 1.01 | 0.048 | 0.23 | 1.33 (1.07–1.75) |
| rs6499640 | 16 | FTO | A | G | 1.03 | 0.89 | 0.38 | 0.50 | 1.19 | 1.05 | 0.016 | 0.088 | 1.16 (1.10–1.21) |
| rs17782313 | 18 | MC4R | C | T | 1.26 | 1.10 | 0.0021 | 0.013 | 1.37 | 1.23 | 8.2 × 10−7 | 5.0 × 10−6 | 1.20 (1.09–1.31) |
| rs6265 | 11 | BDNF | G | A | 1.05 | 0.94 | 0.21 | 0.48 | 1.16 | 1.06 | 0.0075 | 0.043 | 1.12 (1.06–1.19) |
| rs10938397 | 4 | GNPDA2 | G | A | 1.12 | 1.00 | 0.057 | 0.26 | 1.24 | 1.13 | 0.00014 | 0.00085 | 1.20 (1.09–1.31) |
Nominal P values were adjusted for age and sex, and permuted P values were further corrected for multiple testing. All P values were one-sided. Obese, overweight, and normal-weight children were diagnosed by the Chinese age- and sex-specific BMI cutoffs (supplementary Table 1) (13). ORs and 95% CIs were calculated using multinomial logistic regression with genotypes, age, and sex as the independent variables. Chr., chromosome.
For the 126 (six SNPs, three genetic models, and seven phenotypes) resulting P values, associated false discovery rate levels were performed (0.05 as criteria). After multiple testing, the significant associations of the SNPs rs17782313, rs6265, and rs10938397 with BMI and obesity remained (data not shown).
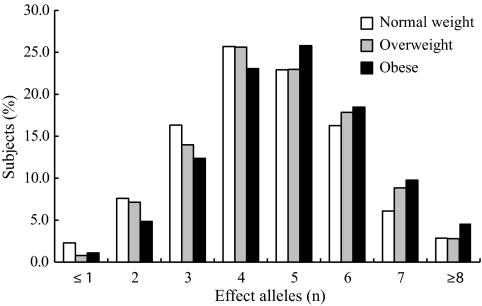
Figure 1 shows the frequency distribution of the number of effect alleles of the six SNPs in normal-weight, overweight, and obese groups. The associations of the number of effect alleles with overweight and obesity are shown in supplementary Table 7. Most subjects of each group carry four to six effect alleles. In the obese group, compared with subjects that carry three or fewer effect alleles, the risk of obesity of the subjects that carry four to six and greater than seven effect alleles was 1.54 (one-sided 95% CI >1.31; one-sided P = 5.9 × 10−6) and 2.50 (one-sided 95% CI >1.97; one-sided P = 1.3 × 10−10), respectively. There were significant associations of the number of effect alleles with anthropometric indexes (except birth weight) including BMI, waist circumference, WHtR, and FMP (supplementary Table 8).
FIG. 1.
Frequency distribution of the number of effect alleles in normal-weight, overweight, and obese groups.
DISCUSSION
Recently, GWA studies have discovered several genetic variants in obesity-related genes that modulate body weight in Caucasians (7–9) ([9] was a GWA study for extreme obesity, not BMI in population-based samples as the other GWA studies were). Although the associations of the SNPs rs6265, rs17782313, and rs10938397 with BMI or obesity were investigated in European-American children (10) and subsequently replicated in an adult Chinese population (11), independent replication studies have not been carried out in Chinese children. In the present study, we examined six SNPs in obesity-related genes in Chinese children. There were significant associations of the SNPs rs7138803, rs17782313, rs6265, and rs10938397 with BMI after age and sex adjustment and correction for multiple testing (Table 1). Our results also showed that the SNPs rs17782313, rs6265, and rs10938397 increased the risk of obesity, and the odds ratios (ORs) of these SNPs were higher than those in Europeans (7–9,19) (Table 2). We demonstrated the associations with obesity and BMI as well as other anthropometric indexes including waist circumference, WHtR, and FMP. Our results showed that none of these SNPs were associated with birth weight, suggesting that the genetic influence of body weight occurs after birth.
In our study, no statistically significant association of rs1805081 with BMI and obesity in Chinese children was found (Tables 1 and 2); however, the trend of association is in the same direction as the initial finding, and the A allele is also the risk allele for obesity. Association of rs1805081 with BMI and obesity in our study showed borderline significance after age and sex adjustment. The differences in obesity prevalence between adults and children or the differences in effect size between Caucasians and Chinese might be the reasons why the effect of SNP could not be replicated in our study.
This study is the first to show the associations of SNPs (rs7138803, rs1805081, rs6499640, rs17782313, rs6265, and rs10938397) with BMI and risk of obesity in children with Chinese ancestry. It is important to clarify the impact of genetic susceptibility by investigating the association between genetic variants on obesity-related genes and childhood adiposity.
In conclusion, our study showed the associations of six SNPs in obesity-related genes with BMI and risk of obesity in the Chinese children. Results of this study support obesity-related genes in adults as important genes for BMI variation in children. It suggests that some SNPs identified by GWA studies in Caucasians also confer risk for obesity in Chinese children. The functional studies are needed to investigate the roles of these genes in children to prevent adult obesity and other related diseases.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants to J.M. from the National Natural Science Foundation of China (30872165), the Beijing Health System Leading Talent Fund (2009-1-08), and the Beijing Municipal Science & Technology Commission (H030930030031 and D08050700320801); grants to J.O. from the National Natural Science Foundation of China (30730057 and 30700442); and a grant to X.W. from the Beijing Hypertension League Institute.
No potential conflicts of interest relevant to this article were reported.
L.W. wrote the manuscript, researched data, contributed to the discussion, and reviewed/edited the manuscript. B.X. and M.Z. analyzed and interpreted data, and reviewed/edited the manuscript. Y.S. researched data, contributed to the discussion, and reviewed/edited the manuscript. X.Z., H.C., and D.H. reviewed/edited the manuscript. D.S. analyzed data. J.O. analyzed data and reviewed/edited the manuscript. X.W. contributed to the design and discussion and made critical revisions of the manuscript. J.M. designed the study; collected, analyzed, and interpreted data; and made critical revisions of the manuscript.
The authors thank Weili Yan, Xinjiang Medical University, Xinjiang, China; Weihua Zhang, Public Health and Primary Care, Imperial College Faculty of Medicine, London, U.K.; Maolian Gong, Max-Delbrück-Centrum Für Molekulare Medizin, Berlin-Buch, Germany; Franz Rüschendorf, Max-Delbrück-Centrum Für Molekulare Medizin, Berlin-Buch, Germany; and Gaifen Liu, Beijing Tiantan Hospital affiliated with Capital Medical University, for constructive comments on this manuscript. The authors also acknowledge Xuejun Ma, Chinese Center for Disease Control and Prevention, Beijing, China, for providing the genotyping facilities for their work; and Miao Wang, Chinese Center for Disease Control and Prevention, Beijing, China, for the excellent technical assistance.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Chen CM: Overview of obesity in Mainland China. Obes Rev 2008;9(Suppl. 1):S14–S21 [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Lobstein T: Worldwide trends in childhood overweight and obesity. Int J Pediatr Obes 2006;1:11–25 [DOI] [PubMed] [Google Scholar]
- 3.Singh AS, Mulder C, Twisk JW, van Mechelen W, Chinapaw MJ: Tracking of childhood overweight into adulthood: a systematic review of the literature. Obes Rev 2008;9:474–488 [DOI] [PubMed] [Google Scholar]
- 4.Virdis A, Ghiadoni L, Masi S, Versari D, Daghini E, Giannarelli C, Salvetti A, Taddei S: Obesity in the childhood: a link to adult hypertension. Curr Pharm Des 2009;15:1063–1071 [DOI] [PubMed] [Google Scholar]
- 5.Thearle MS, Bunt JC, Knowler WC, Krakoff J: Childhood predictors of adult acute insulin response and insulin action. Diabetes Care 2009;32:938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beilin L, Huang RC: Childhood obesity, hypertension, the metabolic syndrome and adult cardiovascular disease. Clin Exp Pharmacol Physiol 2008;35:409–411 [DOI] [PubMed] [Google Scholar]
- 7.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K: Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009;41:18–24 [DOI] [PubMed] [Google Scholar]
- 8.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O'Rahilly S, Purmann C, Rees MG, Ridderstråle M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins NWellcome Trust Case Control Consortium, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JNGenetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009;41:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyre D, Delplanque J, Chèvre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proença C, Gaget S, Körner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Järvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P: Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet 2009;41:157–159 [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Bradfield JP, Li M, Wang K, Zhang H, Kim CE, Annaiah K, Glessner JT, Thomas K, Garris M, Frackelton EC, Otieno FG, Shaner JL, Smith RM, Chiavacci RM, Berkowitz RI, Hakonarson H, Grant SF: The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity (Silver Spring) 2009;17:2254–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung CY, Tso AW, Cheung BM, Xu A, Ong KL, Fong CH, Wat NM, Janus ED, Sham PC, Lam KS: Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a Chinese population. J Clin Endocrinol Metab 2010;95:1395–1403 [DOI] [PubMed] [Google Scholar]
- 12.Mi J, Cheng H, Hou DQ, Duan JL, Teng HH, Wang YF: Prevalence of overweight and obesity among children and adolescents in Beijing in 2004. Zhonghua Liu Xing Bing Xue Za Zhi 2006;27:469–474[in Chinese] [PubMed] [Google Scholar]
- 13.Ji CYWorking Group on Obesity in China Report on childhood obesity in China (1)–body mass index reference for screening overweight and obesity in Chinese school-age children. Biomed Environ Sci 2005;18:390–400 [PubMed] [Google Scholar]
- 14.World Health Organization Physical Status: the Use and Interpretation of Anthropometry: Report of a WHO Expert Committee. Geneva, World Health Org.,1995, p. 1–452(Tech. Rep. Ser., no. 854) [PubMed] [Google Scholar]
- 15.Heid CA, Stevens J, Livak KJ, Williams PM: Real time quantitative PCR. Genome Res 1996;6:986–994 [DOI] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y: Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 1995;57:289–300 [Google Scholar]
- 18.González JR, Carrasco JL, Dudbridge F, Armengol L, Estivill X, Moreno V: Maximizing association statistics over genetic models. Genet Epidemiol 2008;32:246–254 [DOI] [PubMed] [Google Scholar]
- 19.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, Berndt SIProstate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Jacobs KB, Chanock SJ, Hayes RB, Bergmann S, Bennett AJ, Bingham SA, Bochud M, Brown M, Cauchi S, Connell JM, Cooper C, Smith GD, Day I, Dina C, De S, Dermitzakis ET, Doney AS, Elliott KS, Elliott P, Evans DM, Sadaf Farooqi I, Froguel P, Ghori J, Groves CJ, Gwilliam R, Hadley D, Hall AS, Hattersley AT, Hebebrand J, Heid IM, KORA, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Herrera B, Hinney A, Hunt SE, Jarvelin MR, Johnson T, Jolley JD, Karpe F, Keniry A, Khaw KT, Luben RN, Mangino M, Marchini J, McArdle WL, McGinnis R, Meyre D, Munroe PB, Morris AD, Ness AR, Neville MJ, Nica AC, Ong KK, O'Rahilly S, Owen KR, Palmer CN, Papadakis K, Potter S, Pouta A, Qi LNurses' Health Study, Randall JC, Rayner NW, Ring SM, Sandhu MS, Scherag A, Sims MA, Song K, Soranzo N, Speliotes EKDiabetes Genetics Initiative, Syddall HE, Teichmann SA, Timpson NJ, Tobias JH, Uda MSardiNIA Study, Vogel CI, Wallace C, Waterworth DM, Weedon MNWellcome Trust Case Control Consortium, Willer CJ, FUSION, Wraight, Yuan X, Zeggini E, Hirschhorn JN, Strachan DP, Ouwehand WH, Caulfield MJ, Samani NJ, Frayling TM, Vollenweider P, Waeber G, Mooser V, Deloukas P, McCarthy MI, Wareham NJ, Barroso I, Jacobs KB, Chanock SJ, Hayes RB, Lamina C, Gieger C, Illig T, Meitinger T, Wichmann HE, Kraft P, Hankinson SE, Hunter DJ, Hu FB, Lyon HN, Voight BF, Ridderstrale M, Groop L, Scheet P, Sanna S, Abecasis GR, Albai G, Nagaraja R, Schlessinger D, Jackson AU, Tuomilehto J, Collins FS, Boehnke M, Mohlke KL: Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet 2008;40:768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.