Abstract
OBJECTIVE
To investigate whether enterovirus infections predict progression to type 1 diabetes in genetically predisposed children repeatedly positive for islet autoantibodies.
RESEARCH DESIGN AND METHODS
Since 1993, the Diabetes and Autoimmunity Study in the Young (DAISY) has followed 2,365 genetically predisposed children for islet autoimmunity and type 1 diabetes. Venous blood and rectal swabs were collected every 3–6 months after seroconversion for islet autoantibodies (against GAD, insulin, or insulinoma-associated antigen-2 [IA-2]) until diagnosis of diabetes. Enteroviral RNA in serum or rectal swabs was detected using reverse transcriptase PCR with primers specific for the conserved 5′ noncoding region, detecting essentially all enterovirus serotypes.
RESULTS
Of 140 children who seroconverted to repeated positivity for islet autoantibodies at a median age of 4.0 years, 50 progressed to type 1 diabetes during a median follow-up of 4.2 years. The risk of progression to clinical type 1 diabetes in the sample interval following detection of enteroviral RNA in serum (three diabetes cases diagnosed among 17 intervals) was significantly increased compared with that in intervals following a negative serum enteroviral RNA test (33 cases diagnosed among 1,064 intervals; hazard ratio 7.02 [95% CI 1.95–25.3] after adjusting for number of autoantibodies). Results remained significant after adjustment for ZnT8-autoantibodies and after restriction to various subgroups. Enteroviral RNA in rectal swabs was not predictive of progression to type 1 diabetes. No evidence for viral persistence was found.
CONCLUSIONS
This novel observation suggests that progression from islet autoimmunity to type 1 diabetes may increase after an enterovirus infection characterized by the presence of viral RNA in blood.
Type 1 diabetes results from destruction of the insulin-producing β-cells in the pancreatic islets (1). The majority of patients carry the HLA DRB1*03-DQB1*0201 or DRB1*04-DQB1*0302 susceptibility haplotype or both, but these are not sufficient for development of disease. For many years, viral infections have been suspected to play a role, but the specific etiologic agent(s) in human type 1 diabetes remains elusive. While several viruses have been linked to type 1 diabetes, seroepidemiology, histopathology, animal studies, and in vitro experiments have provided the strongest overall evidence for enteroviruses, although results have been somewhat conflicting and not conclusive (2–4).
Autoantibodies to islet autoantigens are present for years prior to diagnosis of type 1 diabetes (1), and prospective studies testing whether enterovirus could predict islet autoantibodies have yielded conflicting results, with positive results in Finnish studies (5–7) and no association found elsewhere (8,9).
Results from animal models suggested that viral infections usually cannot initiate the autoimmune disease process leading to diabetes but may accelerate an already initiated disease process. Studies in various strains of NOD mice have shown that enteroviral infections may accelerate the progression to diabetes only if they occur after autoreactive T-cells have already accumulated in the islets (10–13). In an attempt to evaluate for the first time whether such a general model of disease progression rather than initiation by enteroviruses applies to human type 1 diabetes, we tested whether enteroviral infections predict progression to type 1 diabetes in children repeatedly positive for islet autoantibodies.
RESEARCH DESIGN AND METHODS
From 1993 to 2004, children born at St. Joseph's Hospital in Denver carrying HLA genotypes that confer increased risk for type 1 diabetes and siblings or offspring of people with type 1 diabetes (regardless of their genotype), identified from the Barbara Davis Center for Childhood Diabetes, were enrolled in the Diabetes Autoimmunity Study in the Young (DAISY). Informed consent was obtained from parents of all children, and the study was approved by the Colorado Multiple Institutional Review Board. Children were followed longitudinally from soon after birth and screened for islet autoantibodies at ages 9, 15, and 24 months and annually thereafter. Siblings or offspring of individuals with type 1 diabetes were enrolled after 9 months of age (median age 1.33 years [range 0.02–7.9]). Children who tested positive for islet autoantibodies were scheduled for more frequent follow-up, with visits at 3–6 month intervals.
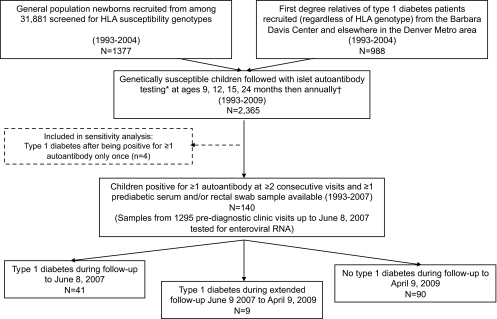
The current study is a cohort analysis of all children who tested positive for one or more islet autoantibody on two or more consecutive clinic visits and provided at least one sample for enterovirus testing during follow-up for type 1 diabetes. Figure 1 shows a flow chart illustrating how the study cohort was formed. A batch of samples collected 1993–2004 was sent for enterovirus testing in 2005, and another batch of samples collected during 2005–June 2007 was sent in 2008. The children were further followed for diagnosis of type 1 diabetes, and the current analysis includes information on autoantibody status and diabetes up to April 2009. Type 1 diabetes was clinically diagnosed based on American Diabetes Association criteria (14), and details of procedures and clinical characteristics have been described elsewhere (15,16).
FIG. 1.
Flow chart illustrating formation of the study cohort. *Samples were tested for the three islet autoantibodies: anti-GAD65, anti-insulin, and anti–IA-2. †If positive for ≥1 islet autoantibody at or after 12 months of age, frequency of blood sampling was increased to every 3–6 months.
Laboratory methods.
HLA genotyping was done at Roche Molecular Systems, Alameda, CA, as previously described (17). Children with genotypes DRB1*04-DQB1*0302/DRB1*03-DQB1*0201 were defined as high risk, and DRB1*04-DQB1*0302/DRB1*04-DQB1*0302 or DRB1*03/*03 or DRB1*04-DQB1*0302/X (where X is not DRB1*04, DQB1*0302, DRB1*03, or DR2,DQB1*0602) was categorized as conferring moderate risk for type 1 diabetes.
At each clinic visit, venous blood and rectal swabs were collected. Blood samples were immediately processed, aliquoted, and stored at −70°C until testing. Rectal swabs were immediately placed in 1 ml transport medium (veal infusion broth or M4−3 medium) and stored at −70°C as previously described (8). Radioimmunoassays were used to measure serum autoantibodies to insulin, GAD65, and IA-2 (BDC512) in George Eisenbarth's laboratory as previously described (18–21), with rigorous duplicate testing and confirmation of all positive and a subset of negative results (22). ZnT8 autoantibodies were measured in John Hutton's laboratory, as previously described, using a dimeric construct incorporating monomeric forms of the COOH-terminus with the polymorphic 325 Arg and Trp variants joined by a flexible linker (23,24). This autoantibody was measured in stored, available samples (81% of samples with valid serum enterovirus RNA measurements).
All enterovirus assays were carried out in Heikki Hyöty's laboratory at the University of Tampere. All virus analyses were done blindly, without knowledge of the disease status of the child. RNA was extracted from 140 μl serum and from 140 μl rectal swab solution according to the manufacturer's protocol (QIAamp viral RNA kit; Qiagen, Hilden, Germany). The presence of enterovirus RNA was detected with RT-PCR using primers specific for the 5′ noncoding region conserved among Picornaviridae and subsequent enterovirus-specific hybridization with lanthanide chelated probes, providing sensitive and specific detection of practically all known enterovirus serotypes (25). All samples with a RT-PCR signal five-fold or higher than a negative control were tested two more times, and a sample was interpreted as positive if at least two out of the three tests were five-fold or higher than the negative control. The 5′ noncoding region of detected enteroviruses was partially sequenced, and sequences were analyzed as described in detail in the online appendix (http://diabetes.diabetesjournals.org/cgi/content/full/db10-0866/DC1.
Enterovirus antibodies were measured in the batch of sera collected 1993–2004, using enzyme immunoassay as described previously (26–28). Antibodies tested were immunoglobulin M antibodies against an antigen cocktail containing coxsackievirus B3, A16, and echovirus 11, as well as IgA and IgG antibodies against purified coxsackievirus B4 and a synthetic enterovirus peptide antigen, KEVPALTVETGAT-C, derived from an immunodominant region of capsid protein VP1 (29), which is a common epitope for many enteroviruses (30). The purified viruses were heat treated to expose antigenic determinants common for various enterovirus serotypes (26).
Definition of infection.
Our primary, a priori definition of infection at a given visit was positivity (as defined above) for RT-PCR detection of enterovirus RNA in serum or rectal swab. Additional analyses were done separately for serum PCR and rectal PCR and for the subset of samples tested for enterovirus antibodies. A sample was defined as positive for serology if there was a twofold or higher increase in level (optical density value) of any of the measured antibodies in the subsequent visit (samples usually 3–6 months apart), with an additional requirement that the signal-to-background ratio should exceed three. Additional two-fold or higher increases in enterovirus antibodies in a third (or later) consecutive sample drawn within 9 months of a previous one were not counted as an additional infection. These criteria were the same as those used in our previous prospective studies (6).
Statistical analysis.
Using Cox regression, we compared the rate of progression to type 1 diabetes under two different models, which we have called the rapid effect model and the cumulative effect model. Both treat enterovirus infection as a time-dependent variable. In the rapid effect model, we estimated the rate of progression to diabetes in the sample interval following detection of enterovirus (median 4 months) compared with sample intervals where enterovirus was not detected. The exposure status returned to zero at the next clinic visit unless enterovirus was also found here. In the cumulative effect model, we estimated the rate of progression to diabetes according to the cumulative number of infections acquired during follow-up, which also allows for detection of delayed effects. Each individual first contributed follow-up time with zero infections, and the exposure variable increased by one at each visit when a new infection was detected. Because few individuals had repeated infections, the cumulative exposure variable had to be grouped (0 vs. ≥1 for serum RNA; 0, 1, and ≥2 for rectal swab RNA). The main time variable was time from the first clinic visit at which a child tested positive for islet autoantibodies to type 1 diabetes diagnosis or to the most recent visit (up to 9 April 2009) at which the child was known not to have diabetes. Because enteroviral RNA was relatively rarely detected in serum and, consequently, the number of events during the exposed periods were limited, we also carried out a Monte Carlo permutation test with 10,000 repeated permutations of the enterovirus variable to assess the validity of the standard inference based on the Cox regression model. All analyses were done using Stata, version 11 (StataCorp, College Station, TX). A 95% CI for the hazard ratio excluding the value 1.00 or a P value <0.05 was regarded as statistically significant.
RESULTS
A total of 140 children seroconverted for islet autoantibodies at a median age of 4.0 years. Of those, 50 developed type 1 diabetes at a median age of 8.7 years after a median follow-up of 4.1 years from the initial appearance of islet autoantibodies (Table 1). The samples tested for enterovirus were collected prior to June 2007, and 41 of the 50 children had developed type 1 diabetes by that time while another nine progressed to type 1 diabetes between June 2007 and April 2009 (Fig. 1).
TABLE 1.
Characteristics of the cohort and results of Cox regression survival analysis of progression from islet autoimmunity to type 1 diabetes
| Progressed to type 1 diabetes (n = 50) | No type 1 diabetes (n = 90) | Unadjusted HR (95% CI) | Adjusted HR (95% CI)† | |
|---|---|---|---|---|
| Age (years) at diagnosis of type 1 diabetes | 8.7 (1.9–15) | |||
| Follow-up (years) from onset of islet autoimmunity | 4.1 (0.2–11) | 4.6 (1.6–14) | ||
| Positive for >2 islet autoantibodies at the first and/or second positive visit | 36 (72%) | 21 (23.3%) | 4.57 (2.46–8.51) | 4.24 (2.26–7.95) |
| Female sex | 26 (52%) | 48 (53.3%) | 1.18 (0.67–2.06) | 1.41 (0.79–2.50) |
| First-degree relative with type 1 diabetes‡ | 35 (70%) | 53 (58.9%) | 1.23 (0.67–2.26) | 1.13 (0.61–2.10) |
| HLA DRB1*04-DQB1*0302/DRB1*03-DQB1*0201 | 26 (52.0%) | 27 (30.0%) | 1.84 (1.06–3.21) | 1.51 (0.86–2.67) |
| Non–Hispanic white ethnicity§ | 46 (92.0%) | 72 (80.0%) | 1.94 (0.70–5.39) | 1.45 (0.51–4.13) |
| Age (years) when first islet autoantibody positive¶ | 3.1 (0.7–12) | 5.2 (0.7–13) | 0.93 (0.85–1.02) | 1.01 (0.091–1.11) |
Data are median (range) or n (%) unless otherwise indicated.
†Estimates from Cox regression model simultaneously adjusting for multiple autoantibodies in first two visits, HLA high risk genotype, presence of first-degree relative with type 1 diabetes, and age when first positive for islet autoantibodies.
‡Of these, 35 had an affected father only, 16 had an affected mother only, 34 had an affected sibling, and 3 had a sibling and a parent with type 1 diabetes.
§Ethnic group was self-reported. There were 118 non–Hispanic whites, 19 Hispanics, one African American, and two children of mixed ethnicity in the cohort.
¶HRs per year increase in age when first positive for islet autoantibodies.
Positivity for two or more islet autoantibodies at the first or second positive visit strongly predicted progression to type 1 diabetes, independent of other factors (Table 1). Those who progressed to type 1 diabetes tended to more often carry the high-risk HLA genotype, to have a first-degree relative with type 1 diabetes, and to seroconvert for islet autoantibodies at an earlier age, but these factors did not significantly predict progression to type 1 diabetes independently of positivity for multiple islet autoantibodies in at least one of the two first positive visits (Table 1). The number of positive islet autoantibodies treated as a time-dependent variable was also highly predictive of progression to type 1 diabetes, and positivity for ZnT8-autoantibodies significantly predicted progression both before and after adjusting for the other three islet autoantibodies (supplemental Table 1).
Enterovirus infections.
Enteroviral RNA results were available from serum and rectal swabs collected at 1,081 and 1,242 prediagnostic clinic visits, respectively. Results were available for either serum or rectal swab at 1,295 visits and from both types of specimens at 1,028 visits. The median interval between the visits was 4 months. Enteroviral RNA was detected at a total of 54 of 1,295 visits (4.2%). At eight of these 54 visits, enteroviral RNA was detected in both serum and rectal swab. Of the 140 children in the cohort, 31 (22.1%) had at least one serum or rectal swab sample positive for enteroviral RNA. While 19 of these 31 were positive only once, some had up to six positive visits. Only two children were ever positive twice for serum enteroviral RNA.
The prevalence of enterovirus RNA in serum or rectal swabs declined with age from nearly 10% for the age-group <2.5 years to ∼1% for the age-group ≥7.5 years (supplemental Fig. 1). Enteroviral RNA tended to be more frequent in boys and at visits positive for multiple islet autoantibodies, but these differences were mostly of borderline statistical significance and not consistent among serum and rectal swab samples (supplemental Table 2). Of the 17 serum samples and 14 rectal swab samples collected on the day of the diabetes diagnosis, none were positive for enteroviral RNA.
Viral sequence was obtained from 8 of 17 positive serum samples and from 33 of 45 positive rectal samples. The sequences were deposited in the GenBank sequence database under accession no. HM746666–HM746706 (supplemental Table 3). Sequences are shown in supplemental Fig. 2 together with reference strain sequences listed in supplemental Table 3. Samples on which sequencing was not successful contained low concentrations of viral RNA. All samples but one clustered into enterovirus genogroup II, which contains, among others, the coxsackie B viruses (31) (supplemental Fig. 3). The sequence data indicated that viruses detected simultaneously in serum and rectal samples represented the same virus strain and only a single nucleotide substitution was once observed between such strains. Viruses detected in successive samples taken from the same individual represented different enterovirus strains. Thus, no evidence of viral persistence was found.
Progression to type 1 diabetes following enterovirus infections.
The progression to type 1 diabetes in the 17 intervals following detection of enteroviral RNA in serum was significantly more rapid (three type 1 diabetes cases diagnosed) than that in the 1,064 intervals following negative enteroviral RNA serum test (33 type 1 diabetes cases diagnosed; hazard ratio [HR]6.36) (Table 2). Further adjustment for number of positive conventional islet autoantibodies did not alter the result (Table 2). Because only three children were diagnosed in the interval after being positive for enteroviral RNA in serum, we employed a permutation test to make sure that the results of the standard inference based on Cox regression were valid. With 10,000 permutations, the (Monte Carlo) P value was 0.0075, thus confirming the highly significant result. After restricting the analysis to the 81% of samples with available data on ZnT8 autoantibodies, there was still a significant relation between serum enteroviral RNA and progression to type 1 diabetes (HR 6.21 [95% CI 1.82–21.2]), and this essentially was not affected by adjustment for ZnT8-autoantibody positivity in models without (8.50 [2.21–32.6]) or with (9.08 [2.30–35.8]) additional adjustment for the number of other islet autoantibodies.
TABLE 2.
Progression from islet autoimmunity to clinical type 1 diabetes in sample interval (median ∼4 months) following infection detected by enterovirus RNA in serum or rectal swab sample
| Type of sample | Person-years of follow-up | Cases progressing to type 1 diabetes in interval* | Unadjusted HR (95% CI) | HR (95% CI) adjusted for islet autoantibodies |
|---|---|---|---|---|
| Serum | ||||
| No enterovirus RNA in previous sample | 494 | 33 | 1.00 (ref.) | 1.00 (ref.) |
| Enterovirus RNA in previous sample | 6.5 | 3 | 6.36 (1.89–21.4)† | 7.02 (1.95–25.3) |
| Rectal swab | ||||
| No enterovirus RNA in previous sample | 537.1 | 32 | 1.00 (ref.) | 1.00 (ref.) |
| Enterovirus RNA in previous sample | 21.2 | 1 | 0.93 (0.12–6.90) | 0.79 (0.10–5.92) |
Data are n unless otherwise indicated.
*Forty-one of 140 children in the study cohort progressed to type 1 diabetes during the period wherein collected samples were tested for enterovirus, of which serum enterovirus RNA results were available from the clinic visit preceding diagnosis in 36 (of which 3 were positive) and rectal swab enterovirus RNA was available in 33 (of which 1 was positive, and serum from the same visit was also positive for enterovirus RNA). Enterovirus exposure variables coded according to the rapid effect model described in research design and methods.
†Cox regression model: P = 0.003. Permutation test based on Cox regression model with 10,000 permutations of the enterovirus variable: P = 0.0075.
The three children who progressed to type 1 diabetes in the interval following a positive serum enteroviral RNA test all had typical characteristics of high risk for progression to type 1 diabetes. They had an early age at seroconversion for multiple islet autoantibodies and an affected sibling, and two of three carried the HLA DR3/4 genotype (supplemental Table 4). They also had a near-average interval length between clinic visits, and all were male and of non–Hispanic white ethnicity. Results were similar and remained statistically significant after restriction of the analysis to these respective subgroups (supplemental Table 5). Furthermore, the results were essentially unchanged after including four children who progressed to type 1 diabetes after being positive only once for islet autoantibodies (all four were negative for enteroviral RNA, adjusted HR 6.56 [95% CI 1.84–23.5]).
Presence of enteroviral RNA in rectal swabs did not predict progression to type 1 diabetes in the following sample interval (adjusted HR 0.79 [95% CI 0.10–5.92]) (Table 2).
Analysis of progression to type 1 diabetes according to the cumulative number of enterovirus infections during follow-up, which allows for delayed effect, showed no significant relation with progression to type 1 diabetes for either serum or rectal swab enteroviral RNA or for serologically defined infections (supplemental Table 6). We also ran a Cox regression model simultaneously including variables modeling enterovirus according to the rapid effect model and the cumulative effect model. The results confirmed that the nonsignificant tendency toward an association for the cumulative effect variable was entirely due to the rapid effect, while the rapid effect of serum enteroviral RNA was unaltered and still significant (5.79 [1.23–27.3] for rapid effects model and 1.07 [0.37–3.11] for cumulative effect model).
There was also no relation between infections defined as increases in enterovirus antibodies and progression to type 1 diabetes according to the rapid effect model (supplemental Table 7). (Note that antibodies were only tested in the subset of samples collected during 1993–2004).
Finally, there were 19 children (61.3%) who progressed to type 1 diabetes among the 31 with one or more enteroviral RNA–positive serum or rectal swab samples compared with 31 (28.4%) among the 109 children in whom enteroviral RNA were not detected during follow-up (P = 0.001). The proportion of visits where both serum and rectal swabs were positive for enteroviral RNA was higher among those who progressed to type 1 diabetes (6 of 425 prediagnostic visits [1.4%]) than among nonprogressors (2 of 603 visits [0.3%]), but this difference was not statistically significant.
DISCUSSION
To our knowledge, this is the first study to specifically assess the role of viral infections in the progression from islet autoimmunity to clinical type 1 diabetes in humans. We found that the rate of progression from islet autoimmunity to diabetes was significantly increased in sample intervals (of an average of 4 months) after the detection of enteroviral RNA in serum but not after detection of enteroviral RNA in rectal swab samples.
Strengths and limitations.
Given the amount of data available and many possible ways of analyzing data, we took great care to make all decisions a priori regarding algorithms for defining infections and methods of analysis. We used a formal cohort design and employed two main models (rapid effect and cumulative effect) to analyze two main indicators of enterovirus infections: enterovirus RNA in serum or in rectal swabs. Admittedly, our a priori–defined main exposure, presence of enterovirus RNA in either serum or rectal swabs, did not significantly predict progression to type 1 diabetes (supplemental Table 7). However, in preplanned subanalyses of serum and rectal swab enteroviral RNA examined separately, we found the presence of enterovirus RNA in serum to be a highly significant predictor of progression. Also, in the Finnish studies of enterovirus as a risk factor for islet autoimmunity, enteroviral RNA in serum samples have been more predictive than enteroviral RNA in stool samples (4). The number of children who progressed in sample intervals after the detection of viral RNA in serum was limited. However, rather than relying on standard inference alone, we confirmed the highly significant result using a permutation test, which is not susceptible to bias with small sample sizes. Furthermore, the result was consistent and remained significant in subgroups defined by characteristics of those who progressed to type 1 diabetes after enteroviral RNA was found in serum.
As a marker of islet autoimmunity, we used repeated presence of at least one islet autoantibody. This probably does not always reflect insulitis or activation of autoreactive T-cells, but autoantibodies are currently the best way of predicting type 1 diabetes in humans (1).
Interpretation.
Approximately 8% of the children progressing to type 1 diabetes had enteroviral RNA in their serum a few months prior to diagnosis. While our finding supports the hypothesis that infections resulting in enteroviral RNA in serum lead to a more rapid progression to clinical disease in some high-risk individuals, it may also suggest that enterovirus infection is a relatively uncommon cause of progression to type 1 diabetes. These observations may be explained by at least three potential scenarios.
First, we may be seeing only the tip of an iceberg because enterovirus is normally present in blood for only a few days during infection in immunocompetent hosts (5,32). Thus, the sampling intervals (median 4 months) are probably too wide to catch most of the causal infections, and enterovirus infections could turn out to be a major cause of progression from islet autoimmunity to diabetes. On the other hand, while viral shedding in feces rarely lasts more than 1 or 2 months (33), the prevalence of enteroviral RNA in rectal swab samples collected at ages <2.5 years in the current study was of a magnitude (8.7%) similar to that seen in other longitudinal studies with stool samples collected monthly from healthy children aged 3–28 months in Norway (11.5%) (33) and 3–22 months in Finland (6.0%) (34). To explain the lack of association between enteroviral RNA in rectal swabs, we may speculate that not all instances of gut infection are associated with a period with enteroviral RNA in the blood.
Second, enterovirus may establish low-grade persistent infection in children with islet autoimmunity, but the quantity of viral RNA in serum and feces may be below the detection limit in most such cases. Some studies have indicated presence of enterovirus in pancreatic tissue in a sizeable proportion of patients dying soon after onset of type 1 diabetes (35–38). Although the results varied depending on methodology and quality of specimens, detection of enterovirus in β-cells clearly strengthens the case for its role in the pathogenesis. In addition, a recent study suggests that the virus is present in the intestinal mucosa of diabetic patients (39). Enteroviral RNA was only detected at one time point in children diagnosed with type 1 diabetes during the sample interval following a positive serum enteroviral RNA test, which does not support the hypothesis of viral persistence. Sequence analysis did not give support for persistent infection because all sequences obtained from children with multiple infections were from different genotypes. Furthermore, it is notable that none of the samples collected at the day of diabetes diagnosis were positive for enteroviral RNA. This is consistent with a previous study of serum samples from Finland (5) but apparently inconsistent with the majority of studies of enterovirus RNA in plasma or serum taken from patients soon after diagnosis, which have found ∼30% of patients to be positive (40–42). We have no explanation for this except a suggestion that an international laboratory standardization workshop could shed more light on these differences.
Third, enterovirus infection may be just one of many factors that can accelerate progression to diabetes, e.g., through nonspecific activation of autoreactive T-cells. Additional host and environmental factors are also likely to play a role. It is currently unclear whether certain enterovirus serotypes are more diabetogenic in humans than others. The most frequently implicated serotype, coxsackievirus B4 (43), was responsible for 2.4% of the enterovirus infectious episodes in the Norwegian study of healthy children (33). There may also be differences within serotypes because enteroviruses are known to mutate rapidly (4,32).
Another possibility that cannot be discarded is that progression to type 1 diabetes is enhanced because the viral infection induced insulin resistance sufficient to precipitate clinical disease. Nonspecific febrile illness or other infectious symptoms in a period prior to diagnosis seems to be quite commonly reported, but few studies have been able to obtain comparable data in age-matched controls (44) and the large majority of enterovirus infections are asymptomatic (32). Furthermore, while biopsy studies and previous cross-sectional or retrospective studies of enterovirus infections in patients with type 1 diabetes cannot exclude the possibility that the disease influenced the risk of infection, our longitudinal design allowed us to draw stronger inference in this regard. The fact that none of the children who were tested on the day of diagnosis were positive for enteroviral RNA, including those who were enterovirus positive in the interval before diagnosis, shows that reverse causation was unlikely.
A number of potential mechanisms for how viral infections may induce or accelerate autoimmune diabetes have been proposed, mostly based on animal models or in vitro studies (43,45). Mechanisms in humans are likely to be complex, but may initially involve, for example, activation of the innate immune system, secretion of interferon-α, and perhaps upregulation of major histocompatibility complex molecules on β-cells (46). Results from animal models cannot automatically be generalized to humans, but studies in strains of NOD mice have indicated a requirement for preceding β-cell damage and release of β-cell antigens taken up by antigen-presenting cells (13,47), as previously reviewed (43). The “fertile field hypothesis” proposes that different viruses may increase the risk of diabetes in susceptible time windows after an infection, while outside this window a similar viral infection would be resolved with no further consequences for the host (3).
Future studies and final conclusion.
Despite the huge undertaking of screening and prospectively following a large number of children for several years, the number of end points was still limited and independent replication in future studies would strengthen the results. Children who progressed to type 1 diabetes immediately after detection of enterovirus RNA all had clinical characteristics consistent with high risk of progression such as early development of multiple islet autoantibodies. Future studies could investigate further the potential role of additional host and viral factors in this process. Prospective studies are challenging, and up to now such studies have mainly focused on the initiation of autoimmunity as the end point (7–9,26), with mixed results. The Environmental Determinants of Diabetes in the Young (TEDDY) study (48) has the potential to provide answers concerning the role of enterovirus and progression to type 1 diabetes with greater power and avoiding some of the limitations of the study presented here.
In conclusion, the rate of progression from islet autoimmunity to type 1 diabetes was significantly increased in the approximately 4-month interval following detection of enteroviral RNA in serum.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants DK-32493, DK-32083, and DK-50979; Diabetes Endocrinology Research Center Clinical Investigation and Bioinformatics Core P30 DK 57516; and the Children's Diabetes Foundation. L.C.S. was supported in part by a grant from the National Research Council of Norway (148359/330). Virus analyses in the laboratory of H.H. were partly supported by a grant from the Academy of Finland.
H.H. is a shareholder (<5%) in Vactech LTD, Tampere, Finland, a company that develops vaccines against picornaviruses. No other potential conflicts of interest relevant to this article were reported.
L.C.S. planned the present study with assistance from K.J.B., designed the analysis strategy, did the statistical analyses, and wrote the manuscript with input from all authors. H.H. and S.O. were responsible for the enterovirus testing and enteroviral sequence analysis. K.B. was responsible for managing and preparing the databases. G.K. was responsible for the clinical evaluation and diagnosis of type 1 diabetes. J.C.H. was responsible for measuring the ZnT8 autoantibodies. H.A.E. was responsible for the HLA genotyping. G.S.E. was responsible for measuring the anti-insulin, -GAD, and –IA-2 autoantibodies. M.R. was the principal investigator and developed the general protocol for the DAISY study with input from J.M.N., G.K., H.A.E., and G.S.E.
The authors thank the DAISY and Barbara Davis Center staff for their help, Liping Yu at the Barbara Davis Center for running the autoantibody assays, and Dory Bugawan at Roche Molecular Systems for help with the HLA genotyping.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Atkinson MA, Eisenbarth GS: Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet 2001;358:221–229 [DOI] [PubMed] [Google Scholar]
- 2.Jun HS, Yoon JW: A new look at viruses in type 1 diabetes. Diabete Metab Res Rev 2003;19:8–31 [DOI] [PubMed] [Google Scholar]
- 3.von Herrath M: Can we learn from viruses how to prevent type 1 diabetes? The role of viral infections in the pathogenesis of type 1 diabetes and the development of novel combination therapies. Diabetes 2009;58:2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tauriainen S, Oikarinen S, Oikarinen M, Hyöty H: Enteroviruses in the pathogenesis of type 1 diabetes. Semin Immunopathol. 28April2010[Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 5.Lönnrot M, Salminen K, Knip M, Savola K, Kulmala P, Leinikki P, Hyypiä T, Åkerblom HK, Hyöty Hthe Childhood Diabetes in Finland (DiMe) Study Group Enterovirus RNA in serum is a risk factor for beta-cell autoimmunity and clinical type 1 diabetes: a prospective study. J Med Virol 2000;61:214–220 [PubMed] [Google Scholar]
- 6.Salminen K, Sadeharju K, Lönnrot M, Vähäsalo P, Kupila A, Korhonen S, Ilonen J, Simell O, Knip M, Hyöty H: Enterovirus infections are associated with the induction of β-cell autoimmunity in a prospective birth cohort study. J Med Virol 2003;69:91–98 [DOI] [PubMed] [Google Scholar]
- 7.Sadeharju K, Hämäläinen AM, Knip M, Lönnrot M, Koskela P, Virtanen SM, Ilonen J, Åkerblom HK, Hyöty Hthe Finnish TRIGR Study Group Enterovirus infections as a risk factor for type I diabetes: virus analyses in a dietary intervention trial. Clin Exp Immunol 2003;132:271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graves PM, Rotbart HA, Nix WA, Pallansch MA, Erlich HA, Norris JM, Hoffman M, Eisenbarth GS, Rewers M: Prospective study of enteroviral infections and development of beta-cell autoimmunity: Diabetes Autoimmunity Study in the Young (DAISY). Diabetes Res Clin Pract 2003;59:51–61 [DOI] [PubMed] [Google Scholar]
- 9.Füchtenbusch M, Irnstetter A, Jäger G, Ziegler AG: No evidence for an association of Coxsackie virus infections during pregnancy and early childhood with development of islet autoantibodies in offspring of mothers or fathers with type 1 diabetes. J Autoimmun 2001;17:333–340 [DOI] [PubMed] [Google Scholar]
- 10.Serreze DV, Ottendorfer EW, Ellis TM, Gauntt CJ, Atkinson MA: Acceleration of type 1 diabetes by a coxsackievirus infection requires a preexisting critical mass of autoreactive T-cells in pancreatic islets. Diabetes 2000;49:708–711 [DOI] [PubMed] [Google Scholar]
- 11.Serreze DV, Wasserfall C, Ottendorfer EW, Stalvey M, Pierce MA, Gauntt C, O'Donnell B, Flanagan JB, Campbell-Thompson M, Ellis TM, Atkinson MA: Diabetes acceleration or prevention by a coxsackievirus B4 infection: critical requirements for both interleukin-4 and gamma interferon. J Virol 2005;79:1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drescher KM, Kono K, Bopegamage S, Carson SD, Tracy S: Coxsackievirus B3 infection and type 1 diabetes development in NOD mice: insulitis determines susceptibility of pancreatic islets to virus infection. Virology 2004;329:381–394 [DOI] [PubMed] [Google Scholar]
- 13.Horwitz MS, Fine C, Ilic A, Sarvetnick N: Requirements for viral-mediated autoimmune diabetes: β-cell damage and immune infiltration. J Autoimmun 2001;16:211–217 [DOI] [PubMed] [Google Scholar]
- 14.Silverstein J, Klingensmith G, Copeland K, Plotnick L, Kaufman F, Laffel L, Deeb L, Grey M, Anderson B, Holzmeister LA, Clark N: Care of children and adolescents with type 1 diabetes: a statement of the American Diabetes Association. Diabetes Care 2005;28:186–212 [DOI] [PubMed] [Google Scholar]
- 15.Barker JM, Goehrig SH, Barriga K, Hoffman M, Slover R, Eisenbarth GS, Norris JM, Klingensmith GJ, Rewers M: Clinical characteristics of children diagnosed with type 1 diabetes through intensive screening and follow-up. Diabetes Care 2004;27:1399–1404 [DOI] [PubMed] [Google Scholar]
- 16.Stene LC, Barriga K, Hoffman M, Kean J, Klingensmith G, Norris JM, Erlich HA, Eisenbarth GS, Rewers M: Normal but increasing hemoglobin A1c levels predict progression from islet autoimmunity to overt type 1 diabetes: Diabetes Autoimmunity Study in the Young (DAISY). Pediatr Diabetes 2006;7:247–253 [DOI] [PubMed] [Google Scholar]
- 17.Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, McDuffie RS, Jr, Hamman RF, Klingensmith G, Eisenbarth GS, Erlich HA: Newborn screening for HLA markers associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 1996;39:807–812 [DOI] [PubMed] [Google Scholar]
- 18.Grubin CE, Daniels T, Toivola B, Landin-Olsson M, Hagopian WA, Li L, Karlsen AE, Boel E, Michelsen B, Lernmark Å: A novel radioligand binding assay to determine diagnostic accuracy of isoform-specific glutamic acid decarboxylase antibodies in childhood IDDM. Diabetologia 1994;37:344–350 [DOI] [PubMed] [Google Scholar]
- 19.Gianani R, Rabin DU, Verge CF, Yu L, Babu SR, Pietropaolo M, Eisenbarth GS: ICA512 autoantibody radioassay. Diabetes 1995;44:1340–1344 [DOI] [PubMed] [Google Scholar]
- 20.Yu L, Rewers M, Gianani R, Kawasaki E, Zhang Y, Verge C, Chase P, Klingensmith G, Erlich H, Norris J, Eisenbarth GS: Antiislet autoantibodies usually develop sequentially rather than simultaneously. J Clin Endocrinol Metab 1996;81:4264–4267 [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Robles DT, Abiru N, Kaur P, Rewers M, Kelemen K, Eisenbarth GS: Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc Natl Acad Sci U S A 2000;97:1701–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker JM, Barriga KJ, Yu L, Miao D, Erlich HA, Norris JM, Eisenbarth GS, Rewers M: Prediction of autoantibody positivity and progression to type 1 diabetes: Diabetes Autoimmunity Study in the Young (DAISY). J Clin Endocrinol Metab 2004;89:3896–3902 [DOI] [PubMed] [Google Scholar]
- 23.Wenzlau JM, Juhl K, Yu L, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW, Hutton JC: The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci U S A 2007;104:17040–17045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenzlau JM, Liu Y, Yu L, Moua O, Fowler KT, Rangasamy S, Walters J, Eisenbarth GS, Davidson HW, Hutton JC: A common nonsynonymous single nucleotide polymorphism in the SLC30A8 gene determines ZnT8 autoantibody specificity in type 1 diabetes. Diabetes 2008;57:2693–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lönnrot M, Sjöroos M, Salminen K, Maaronen M, Hyypiä T, Hyöty H: Diagnosis of enterovirus and rhinovirus infections by RT-PCR and time-resolved fluorometry with lanthanide chelate labeled probes. J Med Virol 1999;59:378–384 [PubMed] [Google Scholar]
- 26.Lönnrot M, Korpela K, Knip M, Ilonen J, Simell O, Korhonen S, Savola K, Muona P, Simell T, Koskela P, Hyöty H: Enterovirus infection as a risk factor for β-cell autoimmunity in a prospectively observed birth cohort: the Finnish Diabetes Prediction and Prevention Study. Diabetes 2000;49:1314–1318 [DOI] [PubMed] [Google Scholar]
- 27.Hyöty H, Hiltunen M, Knip M, Laakkonen M, Vähäsalo P, Karjalainen J, Koskela P, Roivainen M, Leinikki P, Hovi T, Åkerblom HKthe Childhood Diabetes in Finland Study Group A prospective study of the role of Coxsackie B and other enterovirus infections in the pathogenesis of IDDM. Diabetes 1995;44:652–657 [DOI] [PubMed] [Google Scholar]
- 28.Samuelson A, Glimåker M, Skoog E, Cello J, Forsgren M: Diagnosis of enteroviral meningitis with IgG-EIA using heat-treated virions and synthetic peptides as antigens. J Med Virol 1993;40:271–277 [DOI] [PubMed] [Google Scholar]
- 29.Hovi T, Roivainen M: Peptide antisera targeted to a conserved sequence in poliovirus capsid VP1 cross-react widely with members of the genus Enterovirus. J Clin Microbiol 1993;31:1083–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roivainen M, Närvänen A, Korkolainen M, Huhtala ML, Hovi T: Antigenic regions of poliovirus type 3/Sabin capsid proteins recognized by human sera in the peptide scanning technique. Virology 1991;180:99–107 [DOI] [PubMed] [Google Scholar]
- 31.Pöyry T, Kinnunen L, Hyypiä T, Brown B, Horsnell C, Hovi T, Stanway G: Genetic and phylogenetic clustering of enteroviruses. J Gen Virol 1996;77:1699–1717 [DOI] [PubMed] [Google Scholar]
- 32.Pallansch MA, Roos RP: Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In Fields Virology. 5th ed Knipe DM, Howley PM. Eds. Eds. Philadelphia, Lippincott Williams & Wilkins, 2007, p. 839–893 [Google Scholar]
- 33.Witsø E, Palacios G, Cinek O, Stene LC, Grinde B, Janowicz D, Lipkin WI, Rønningen KS: Natural circulation of human enteroviruses: high prevalence of human enterovirus A infections. J Clin Microbiol 2006;44:4095–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salminen KK, Vuorinen T, Oikarinen S, Helminen M, Simell S, Knip M, Ilonen J, Simell O, Hyöty H: Isolation of enterovirus strains from children with preclinical type 1 diabetes. Diabet Med 2004;21:156–164 [DOI] [PubMed] [Google Scholar]
- 35.Tauriainen S, Oikarinen M, Keim J, Oikarinen S, Hyöty Hthe nPOD Study Group Detection of enterovirus in pancreatic tissues of cadaver organ donors: results from the Network for Pancreatic Organ Donors with Diabetes (nPOD) study. Abstract presented at the 10th International Congress of the Immunology of Diabetes Society, 17–20 May 2009, Malmö, Sweden [Google Scholar]
- 36.Dotta F, Censini S, van Halteren AG, Marselli L, Masini M, Dionisi S, Mosca F, Boggi U, Muda AO, Del Prato S, Elliott JF, Covacci A, Rappuoli R, Roep BO, Marchetti P: Coxsackie B4 virus infection of β cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci U S A 2007;104:5115–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG: The prevalence of enteroviral capsid protein vp1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia 2009;52:1143–1151 [DOI] [PubMed] [Google Scholar]
- 38.Ylipaasto P, Klingel K, Lindberg AM, Otonkoski T, Kandolf R, Hovi T, Roivainen M: Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia 2004;47:225–239 [DOI] [PubMed] [Google Scholar]
- 39.Oikarinen M, Tauriainen S, Honkanen T, Oikarinen S, Vuori K, Kaukinen K, Rantala I, Mäki M, Hyöty H: Detection of enteroviruses in the intestine of type 1 diabetic patients. Clin Exp Immunol 2008;151:71–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nairn C, Galbraith DN, Taylor KW, Clements GB: Enterovirus variants in the serum of children at the onset of type 1 diabetes mellitus. Diabet Med 1999;16:509–513 [DOI] [PubMed] [Google Scholar]
- 41.Craig ME, Howard NJ, Silink M, Rawlinson WD: Reduced frequency of HLA DRB1*03-DQB1*02 in children with type 1 diabetes associated with enterovirus RNA. J Infect Dis 2003;187:1562–1570 [DOI] [PubMed] [Google Scholar]
- 42.Sarmiento L, Cabrera-Rode E, Lekuleni L, Cuba I, Molina G, Fonseca M, Heng-Hung L, Borroto AD, Gonzalez P, Mas-Lago P, Diaz-Horta O: Occurrence of enterovirus RNA in serum of children with newly diagnosed type 1 diabetes and islet cell autoantibody-positive subjects in a population with a low incidence of type 1 diabetes. Autoimmunity 2007;40:540–545 [DOI] [PubMed] [Google Scholar]
- 43.Richer MJ, Horwitz MS: Coxsackievirus infection as an environmental factor in the etiology of type 1 diabetes. Autoimmun Rev 2009;8:611–615 [DOI] [PubMed] [Google Scholar]
- 44.Gamble DR: Relation of antecedent illness to development of diabetes in children. Br Med J 1980;281:99–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wucherpfennig KW: Mechanisms for the induction of autoimmunity by infectious agents. J Clin Invest 2001;108:1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foulis AK: Pancreatic pathology in type 1 diabetes in human. Novartis Found Symp 2008;292:2–13 [PubMed] [Google Scholar]
- 47.Horwitz MS, Ilic A, Fine C, Balasa B, Sarvetnick N: Coxsackieviral-mediated diabetes: induction requires antigen-presenting cells and is accompanied by phagocytosis of beta cells. Clin Immunol 2004;110:134–144 [DOI] [PubMed] [Google Scholar]
- 48.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann N Y Acad Sci 2008;1150:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.