Abstract
OBJECTIVE
Several single nucleotide polymorphisms (SNPs) in diabetes risk genes reduce glucose- and/or incretin-induced insulin secretion. Here, we investigated interactions between glycemia and such diabetes risk polymorphisms.
RESEARCH DESIGN AND METHODS
Insulin secretion was assessed by insulinogenic index and areas under the curve of C-peptide/glucose in 1,576 subjects using an oral glucose tolerance test (OGTT). Participants were genotyped for 10 diabetes risk SNPs associated with β-cell dysfunction: rs5215 (KCNJ11), rs13266634 (SLC30A8), rs7754840 (CDKAL1), rs10811661 (CDKN2A/2B), rs10830963 (MTNR1B), rs7903146 (TCF7L2), rs10010131 (WFS1), rs7923837 (HHEX), rs151290 (KCNQ1), and rs4402960 (IGF2BP2).
Furthermore, the impact of the interaction between genetic variation in TCF7L2 and glycemia on changes in insulin secretion was tested in 315 individuals taking part in a lifestyle intervention study.
RESULTS
For the SNPs in TCF7L2 and WFS1, we found a significant interaction between glucose control and insulin secretion (all P ≤ 0.0018 for glucose × genotype). When plotting insulin secretion against glucose at 120 min OGTT, the compromising SNP effects on insulin secretion are most apparent under high glucose. In the longitudinal study, rs7903146 in TCF7L2 showed a significant interaction with baseline glucose tolerance upon change in insulin secretion (P = 0.0027). Increased glucose levels at baseline predicted an increase in insulin secretion upon improvement of glycemia by lifestyle intervention only in carriers of the risk alleles.
CONCLUSIONS
For the diabetes risk genes TCF7L2 and WFS1, which are associated with impaired incretin signaling, the level of glycemia determines SNP effects on insulin secretion. This indicates the increasing relevance of these SNPs during the progression of prediabetes stages toward clinically overt type 2 diabetes.
Type 2 diabetes is a disorder characterized by chronically elevated blood glucose levels due to insulin resistance and a relative lack of compensatory pancreatic insulin secretion. Environmental triggers such as a sedentary lifestyle, physical inactivity, and increased body weight play an important role in the development of the disease. In this regard, genetics and especially gene-environment interactions play an important role. Recent research revealed more than 25 gene variants leading to a higher risk for the development of type 2 diabetes (1). Interestingly, most of the diabetes risk genes alter β-cell function (1). This supports the hypothesis that the main genetic effect in the development of type 2 diabetes could be impaired insulin secretion. Neither environmental triggers nor genetics alone can explain the multifactorial disease type 2 diabetes, thus a close interaction between both is presumed (2–4). Hence, environmental influences may determine an individual's susceptibility for single nucleotide polymorphism (SNP) effects, or vice versa genotype may designate a person's susceptibility toward environmental factors.
One “environmental” factor that plays a role early in the pathogenesis of type 2 diabetes is elevated glucose. It is well known that years before type 2 diabetes occurs, glucose control is altered, as reflected by higher fasting glucose and/or higher postprandial glucose (5). High glucose exerts unfavorable effects on insulin sensitivity and secretion, known as glucotoxicity (6,7). On the other hand, elevated glucose levels are needed for the incretin effect. Glucagon-like peptide 1–induced insulin secretion becomes fully active only in the hyperglycemic range (8,9). Incretin-dependent insulin secretion might therefore be of particular importance when compensatory insulin hypersecretion is required.
The aim of this study was to investigate whether glycemia influences the effects of genetic variation associated with type 2 diabetes on insulin secretion. We therefore studied 10 genome-wide association study–derived variants that were furthermore found to influence β-cell function in subsequent studies (rev. in 1,10). Of these, 2 (in the TCF7L2 and WFS1 loci) are associated with incretin-stimulated insulin secretion (1). As the magnitude of incretin-stimulated insulin secretion is dependent on elevated glucose levels (8,9), we hypothesized that glucose levels specifically interact with the effect of those SNPs on insulin secretion both in cross-sectional and longitudinal intervention studies.
RESEARCH DESIGN AND METHODS
The participants were selected from the ongoing Tübingen Family Study for type 2 diabetes (11). In short, the aim of this study is to investigate individuals at high risk but yet without known type 2 diabetes. We selected 1,576 subjects for which complete oral glucose tolerance test (OGTT) data and genotypes were available. According to the World Health Organization diagnostic criteria, 1,079 subjects were identified as normal glucose tolerant (NGT), 143 showed impaired fasting glucose (IFG), 138 had impaired glucose tolerance (IGT), and 112 had both IFG and IGT. In 82 subjects, type 2 diabetes was newly diagnosed. Individuals on medication affecting glucose metabolism were excluded. An additional 22 subjects were excluded because of implausible insulin and/or glucose values.
The clinical characteristics of the study population are shown in supplementary Table 1 in the online appendix available at http://diabetes.diabetesjournals.org/cgi/content/full/db10-0674/DC1. Informed written consent was obtained from all participants, and the local ethics committee approved the protocol.
We also analyzed longitudinal data from a subgroup of 342 individuals who were metabolically reanalyzed after a follow-up period of 9 months, during which they took part in a lifestyle intervention program (see the online appendix for additional information).
Selection of loci/SNPs.
We selected representative SNPs in type 2 diabetes risk loci previously reported to affect distinct aspects of β-cell function: KCNJ11 (rs5215), SLC30A8 (rs13266634), CDKAL1 (rs7754840), CDKN2A/2B (rs10811661), MTNR1B (rs10830963), TCF7L2 (rs7903146), WFS1 (rs10010131), HHEX (rs7923837), KCNQ1 (rs151290), and IGF2BP2 (rs4402960). All SNPs were genotyped in the whole cohort in the course of earlier studies (12–18) using TaqMan assays (Applied Biosystems, Foster City, CA) and passed the quality controls. Details on this as well as on minor allele frequencies, genotyping success rates, and Hardy-Weinberg equilibrium are reported in the aforementioned references.
OGTT.
After an overnight fast, subjects ingested a standard solution containing 75 g of glucose at 08:00 h. Plasma glucose, insulin, and C-peptide concentrations were determined in venous blood samples obtained at 0, 30, 60, 90, and 120 min.
Plasma insulin and C-peptide were determined by commercial chemiluminescence assays for ADVIA Centaur (Siemens Medical Solutions, Fernwald, Germany). Blood glucose was measured using a bedside glucose analyzer (glucose oxidase method; Yellow Springs Instruments, Yellow Springs, OH).
Calculations.
Insulinogenic index (IGI) was calculated from the OGTT ([insulin30 − insulin0]/[glucose30 − glucose0]). Areas under the curve (AUCs) were calculated according to the trapezoid method as: 0.5 × (c0/2 + c30 + c60 + c90 +c120/2) where c is concentration. OGTT-derived insulin sensitivity index was estimated as described earlier (10).
Statistical analyses.
Unless otherwise stated, data are given as means ± SD. Data that were not normally distributed were logarithmically transformed prior to statistical analysis. To identify covariates interacting with SNP effects on insulin secretion, we used ANCOVA. P values < 0.05 were considered to indicate nominal associations, after correction for multiple comparisons taking into account the 10 genes tested (according to Bonferroni correction). P values < 0.0051 were considered statistically significant. Differences between genotype groups were tested using multivariate linear regression analysis. P values were determined using the additive and the dominant inheritance models. After correction for multiple comparisons taking into account the 2 genes tested further (according to Bonferroni correction), P values < 0.025 were considered statistically significant. For statistical analysis, JMP 7.0 (SAS Institute, Cary, NC) statistical software package was used.
RESULTS
We screened for the interaction of 120 min postload glucose (glucose120), AUC of glucose, and A1C with effects of the 10 SNPs on the insulin secretion parameters AUCC-peptide/AUCGlucose and IGI by ANCOVA. After correction for multiple comparisons, TCF7L2 SNP rs7903146 and WFS1 SNP rs10010131 showed significant interaction with AUCGlucose and glucose120 concerning their effect on insulin secretion measured by C-peptide– or insulin-derived secretion indexes. In addition, both variants showed nominal interaction with A1C (Table 1). For the other 8 SNPs, only a few interactions with secretion parameters were detected (Table 1), which did not pass correction for multiple comparisons.
TABLE 1.
Interaction of glucose120, AUCGlucose, and A1C with SNPs on insulin secretion
| Glucose120 |
AUCGlucose |
A1C |
||||
|---|---|---|---|---|---|---|
| IGI | AUCC-peptide/AUCGlucose | IGI | AUCC-peptide/AUCGlucose | IGI | AUCC-peptide/AUCGlucose | |
| KCNJ11 rs5215 | 0.5 | 0.3 | 0.9 | 0.3 | 0.5 | 0.2 |
| KCNQ1 rs151290 | 0.0498 | 0.3 | 0.4 | 0.4 | 0.8 | 0.5 |
| SLC30A8 rs13266634 | 0.1 | 0.3 | 0.2 | 0.3 | 0.7 | 1.0 |
| CDKAL1 rs7754840 | 0.6 | 0.1 | 0.4 | 0.05 | 0.6 | 0.3 |
| CDKN2B rs10811661 | 0.7 | 0.08 | 0.6 | 0.3 | 0.6 | 0.3 |
| IGF2BP2 rs4402960 | 0.5 | 0.4 | 0.2 | 0.5 | 0.4 | 0.6 |
| MTNR1B rs10830963 | 1.0 | 0.7 | 0.3 | 0.9 | 0.6 | 0.5 |
| TCF7L2 rs7903146 | 0.0017 | 0.0018 | 0.0006 | <0.0001 | 0.0438 | 0.0487 |
| WFS1 rs10010131 | 0.0076 | 0.0002 | 0.0241 | 0.0003 | 0.05 | 0.0162 |
| HHEX rs7923837 | 0.0055 | 0.2 | 0.0186 | 0.2 | 0.3 | 0.4 |
Data are P values determined using the additive inheritance model. Data in bold represent statistical significance.
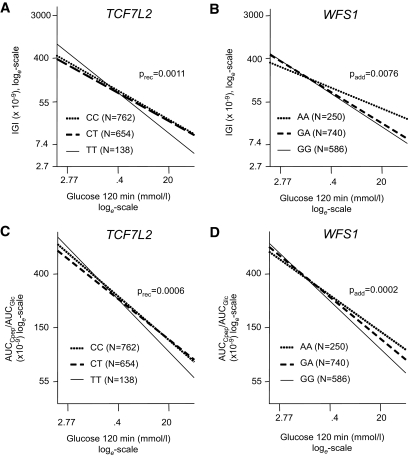
The two SNPs that were affected by glucose control were investigated in detail. Therefore, we stratified the cohort for genotypes and plotted insulin secretion values against glucose120 as a proxy for glycemia. Insulin secretion was adjusted for possible confounders (sex, age, BMI, and insulin sensitivity) by multivariate linear regression analysis. Regression lines of these analyses are shown in Fig. 1. The compromising effect of the risk alleles on insulin secretion is most apparent under high glucose conditions for the TCF7L2 SNP rs7903146 as well as for the WFS1 SNP rs10010131. By contrast, when glucose levels are low, no SNP effect on insulin secretion is obvious or the risk allele may even be beneficial. In this respect, the T-allele of TCF7L2 SNP rs7903146 appears to affect insulin secretion in a recessive way, whereas the mode of action of the G-allele of WFS1 SNP rs10010131 is less clear. Performing similar analyses for the other eight SNPs resulted in no significant interactions (supplementary Fig. 2).
FIG. 1.
Association between IGI and glucose 120 min by TCF7L2 SNP rs7903146 (A) and WFS1 SNP rs10010131 (B). Association between AUCC-peptide/AUCGlucose and glucose120 by TCF7L2 SNP rs7903146 (C) and WFS1 SNP rs10010131 (D). Lines represent regression lines. Data were loge-transformed prior to statistical analysis and adjusted for sex, age, BMI, and OGTT-derived insulin sensitivity index by multivariate linear regression analysis.
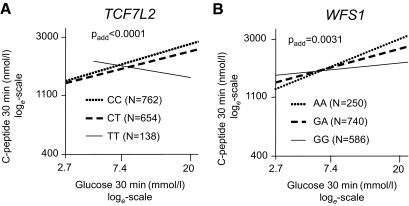
To further evaluate the interaction of TCF7L2 rs7903146 and WFS1 rs10010131 with acute glucose-dependent insulin secretion, we plotted C-peptide levels at 30 min of the OGTT against glucose levels at the same time point. At this time, variation of glucose is most pronounced and C-peptide levels are hardly influenced by clearance. C-peptide was adjusted for possible confounders (sex, age, and BMI) by multivariate linear regression analysis. In carriers of the nonrisk alleles of the TCF7L2 and WFS1 SNPs, the C-peptide rises with higher glucose concentrations. By contrast, this relation is blunted in carriers of the risk alleles (Fig. 2). Using insulin instead of C-peptide yielded comparable results (supplementary Fig. 1).
FIG. 2.
Association between C-peptide levels at 30 min of the OGTT and glucose levels at 30 min during the OGTT by TCF7L2 SNP rs7903146 (A) and WFS1 SNP rs10010131 (B). Lines represent regression lines. Data were loge-transformed prior to statistical analysis. C-peptide data are adjusted for sex, age, and BMI by multivariate linear regression analysis.
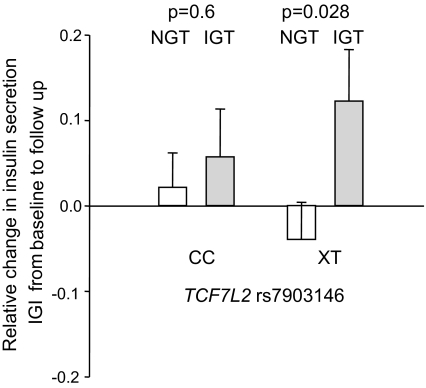
Furthermore, we tested whether glycemia influences the impact of genetic variance in TCF7L2 on changes in insulin secretion during a longitudinal lifestyle intervention study. We found a significant interaction between TCF7L2 genotype and glycemia (glucose at 120 min of the OGTT) at baseline on change in insulin secretion (P value using the dominant inheritance model = 0.0027) in a model including sex, age, insulin secretion at baseline, change in BMI, and change in glucose at 120 min of the OGTT. Participants with IGT at baseline who carried the T risk allele showed a significant increase in insulin secretion during lifestyle intervention compared with participants with NGT at baseline. No differences in change of insulin secretion between subjects with NGT and IGT were present in carriers of the wild-type C-allele (Fig. 3). For WFS1 rs10010131, we found no significant interaction using the same model as described above (P = 0.5).
FIG. 3.
Associations between fold-change in insulin secretion (measured by IGI) during 9-month lifestyle intervention and at baseline glucose tolerance status. Participants with IGT at baseline who carried the T risk allele showed a significant increase in insulin secretion compared with participants with NGT at baseline. No differences in change of insulin secretion between subjects with NGT and IGT were present in carriers of the wild-type C-allele. Padd, P values determined using the additive inheritance model.
DISCUSSION
In the present study, we demonstrated an interaction between SNP effects on insulin secretion and glucose levels for 2 of the 10 tested SNPs, i.e., in TCF7L2 rs7903146 and WFS1 rs10010131. Both SNPs were found to impair incretin-stimulated insulin secretion (13,15). It is well known that incretins are biologically effective only under high glucose conditions (9). Therefore, it seems plausible that the effects of these variants are more pronounced when glucose levels are elevated. The genetic variation in the other diabetes risk genes associated with reduced insulin secretion (KCNJ11, SLC30A8, CDKAL1, CDKN2A/2B, MTNR1B, HHEX, KCNQ1, and IGF2BP2) did not show such an interaction. Therefore, their impact is constantly present over the whole range of glucose levels from NGT to IGT and overt diabetes.
Insulin secretion is markedly impaired in carriers of the risk alleles when glucose levels reach values defining IGT. Thus, it may be important to lower glucose levels early in subjects with these risk variants as a primary prevention in subjects in the prediabetic and pre-IGT states.
To test this hypothesis, we analyzed the change in insulin secretion during a lifestyle intervention with improving glycemia in relation to baseline glycemia and genetic variation in TCF7L2. We took advantage of the Tübingen Lifestyle Intervention Program (TULIP), which uses an intervention similar to those of the Finnish Diabetes Prevention Study (DPS) and U.S. Diabetes Prevention Program (DPP) (19,20). This intervention, based on dietary advice and increase of physical activity, improves mainly insulin sensitivity (11). We previously reported that the risk allele of TCF7L2 rs7903146 is associated with less weight loss in response to this lifestyle intervention (21). Improvement in insulin sensitivity and glucose levels was not dependent on the genotype. Furthermore, insulin secretion did not change during lifestyle intervention (21). However, in our present analysis, we found that increased glucose levels before lifestyle intervention predicted an increase in insulin secretion in carriers of the risk alleles of TCF7L2 rs7903146. This was not the case in carriers of the nonrisk allele. Thus, specifically subjects with IGT on the eve of diabetes who carry the TCF7L2 rs790314 risk allele profit from dietary and exercise interventions designed to lower glucose levels by improving insulin sensitivity. A plausible explanation may be that the relative impairment of the incretin effect in carriers of the risk allele is secondary to chronic hyperglycemia, and thus the lifestyle intervention, by normalizing the hyperglycemia in these subjects, would also restore the insulinotropic effect of incretins.
These findings are consistent with the findings of the DPS and DPP studies, in which the TCF7L2 genotype has an effect on the progression from IGT to diabetes in the control group but not in the lifestyle intervention group (22,23). This indicates that lifestyle intervention can mitigate the risk conferred by genetic background. On the basis of our data, we hypothesize that improving blood glucose by improving insulin sensitivity as was achieved by lifestyle intervention in the DPS, DPP, and TULIP studies reduces the need for compensatory insulin hypersecretion. Therefore, the contribution of incretin action on insulin secretion that is impaired in risk allele carriers becomes less important.
The interaction of glycemia with genetic variation in genes associated with insulin secretion also has further implications for gentopye-phenotype association studies. The interaction between genetic and environmental factors such as glucose control may hinder the detection of other SNPs in cross-sectional studies (24). To avoid this, knowledge of such environmental factors might be of great value when analyzing case-control data, and additional prospective studies controlling for them should be performed.
A question raised by our data is whether SNPs with stronger effects under high glucose conditions determine the further progression toward the disease. One might hypothesize that effects of the risk allele suppress insulin secretion even stronger in the IGT state and therefore might accelerate conversion toward type 2 diabetes. To test this assumption further prospective studies are needed.
Among the limitations of our study is the fact that we could only detect relatively large effect sizes, and the study was therefore underpowered to detect smaller effect sizes. Indeed, because we tested 10 SNPs in parallel, only 2 of them reached statistical significance after Bonferroni correction. Thus, to verify our findings, additional studies in larger cohorts including subjects with other ethnic backgrounds are needed. For example, we did not detect an interaction effect of WFS1 and glucose levels at baseline on change in insulin secretion in the longitudinal lifestyle intervention study. This might be due to the relative low number of subjects that could be included in this analysis. Furthermore, our cohort was also not balanced between sexes and a single OGTT was used to measure glucose levels for each participant, thus not taking into account day-to-day variability. The fact that we observed interactions with 120 min glucose but not with A1C may be due to the relatively low power of the study or to glycemia-independent factors that alter A1C levels.
Taken together, we found that glucose control determines the effects of certain SNPs on insulin secretion. The effects of both TCF7L2 rs7903146 and WFS1 rs10010131 worsen with rising glucose levels, and both SNPs are associated with impaired incretin action on insulin secretion. This indicates the relevance of SNPs in different stages of the pathogenesis toward type 2 diabetes (7,25) as well as prevention strategies for type 2 diabetes.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the German Research Foundation (FR1561/5-1) and the German Federal Ministry of Education and Research (DLR 01GI0925) and a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V.).
No potential conflicts of interest relevant to this article were reported.
M.H. researched data, wrote the manuscript, and contributed to discussion. C.K. researched data and wrote the manuscript. C.T. contributed to discussion and reviewed and edited the manuscript. S.A.H.-S., M.G., and N.S. contributed to discussion. F.M. researched data and reviewed and edited the manuscript; H.S. contributed to discussion and reviewed and edited the manuscript; A.F. researched data, wrote the manuscript, and reviewed and edited the manuscript; H.-U.H. contributed to discussion and reviewed and edited the manuscript.
We thank all study participants for their cooperation. We gratefully acknowledge the excellent technical assistance of Anna Bury, Alke Guirguis, Andreas Vosseler, Melanie Weisser, and Roman Werner (all study nurses or medical-technical assistants from the Eberhard Karls University Tübingen).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Staiger H, Machicao F, Fritsche A, Häring HU: Pathomechanisms of type 2 diabetes genes. Endocr Rev 2009;30:557–585 [DOI] [PubMed] [Google Scholar]
- 2.Wareham NJ, Franks PW, Harding AH: Establishing the role of gene-environment interactions in the etiology of type 2 diabetes. Endocrinol Metab Clin North Am 2002;31:553–566 [DOI] [PubMed] [Google Scholar]
- 3.Grarup N, Andersen G: Gene-environment interactions in the pathogenesis of type 2 diabetes and metabolism. Curr Opin Clin Nutr Metab Care 2007;10:420–426 [DOI] [PubMed] [Google Scholar]
- 4.Weyrich P, Stefan N, Häring HU, Laakso M, Fritsche A: Effect of genotype on success of lifestyle intervention in subjects at risk for type 2 diabetes. J Mol Med 2007;85:107–117 [DOI] [PubMed] [Google Scholar]
- 5.Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR: Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 2009;373:2215–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poitout V, Robertson RP: Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocr Rev 2008;29:351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stumvoll M, Goldstein BJ, van Haeften TW: Type 2 diabetes: principles of pathogenesis and therapy. Lancet 2005;365:1333–1346 [DOI] [PubMed] [Google Scholar]
- 8.Kjems LL, Holst JJ, Vølund A, Madsbad S: The influence of GLP-1 on glucose-stimulated insulin secretion: effects on beta-cell sensitivity in type 2 and nondiabetic subjects. Diabetes 2003;52:380–386 [DOI] [PubMed] [Google Scholar]
- 9.Drucker DJ: The biology of incretin hormones. Cell Metab 2006;3:153–165 [DOI] [PubMed] [Google Scholar]
- 10.McCarthy MI, Zeggini E: Genome-wide association studies in type 2 diabetes. Curr Diab Rep 2009;9:164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schäfer S, Kantartzis K, Machann J, Venter C, Niess A, Schick F, Machicao F, Häring HU, Fritsche A, Stefan N: Lifestyle intervention in individuals with normal versus impaired glucose tolerance. Eur J Clin Invest 2007;37:535–543 [DOI] [PubMed] [Google Scholar]
- 12.Staiger H, Machicao F, Schafer SA, Kirchhoff K, Kantartzis K, Guthoff M, Silbernagel G, Stefan N, Haring HU, Fritsche A: Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine beta-cell function. PLoS One 2008;3:e3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schäfer SA, Müssig K, Staiger H, Machicao F, Stefan N, Gallwitz B, Häring HU, Fritsche A: A common genetic variant in WFS1 determines impaired glucagon-like peptide-1-induced insulin secretion. Diabetologia 2009;52:1075–1082 [DOI] [PubMed] [Google Scholar]
- 14.Müssig K, Staiger H, Machicao F, Kirchhoff K, Guthoff M, Schäfer SA, Kantartzis K, Silbernagel G, Stefan N, Holst JJ, Gallwitz B, Häring HU, Fritsche A: Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes 2009;58:1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schäfer SA, Tschritter O, Machicao F, Thamer C, Stefan N, Gallwitz B, Holst JJ, Dekker JM, 't Hart LM, Nijpels G, van Haeften TW, Häring HU, Fritsche A: Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia 2007;50:2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchhoff K, Machicao F, Haupt A, Schäfer SA, Tschritter O, Staiger H, Stefan N, Häring HU, Fritsche A: Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia 2008;51:597–601 [DOI] [PubMed] [Google Scholar]
- 17.Tschritter O, Stumvoll M, Machicao F, Holzwarth M, Weisser M, Maerker E, Teigeler A, Häring H, Fritsche A: The prevalent Glu23Lys polymorphism in the potassium inward rectifier 6.2 (KIR6.2) gene is associated with impaired glucagon suppression in response to hyperglycemia. Diabetes 2002;51:2854–2860 [DOI] [PubMed] [Google Scholar]
- 18.Staiger H, Machicao F, Stefan N, Tschritter O, Thamer C, Kantartzis K, Schafer SA, Kirchhoff K, Fritsche A, Haring HU: Polymorphisms within novel risk loci for type 2 diabetes determine beta-cell function. PLoS One 2007;2:e832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuomilehto J, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa MFinnish Diabetes Prevention Study Group Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med 2001;344:1343–1350 [DOI] [PubMed] [Google Scholar]
- 20.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DMDiabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haupt A, Thamer C, Heni M, Ketterer C, Machann J, Schick F, Machicao F, Stefan N, Claussen CD, Häring HU, Fritsche A, Staiger H: Gene variants of TCF7L2 influence weight loss and body composition during lifestyle intervention in a population at risk for type 2 diabetes. Diabetes 2010;59:747–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Kuusisto J, Vänttinen M, Kuulasmaa T, Lindström J, Tuomilehto J, Uusitupa M, Laakso M: Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia 2007;50:1192–1200 [DOI] [PubMed] [Google Scholar]
- 23.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler DDiabetes Prevention Program Research Group TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreasen CH, Andersen G: Gene-environment interactions and obesity–further aspects of genomewide association studies. Nutrition 2009;25:998–1003 [DOI] [PubMed] [Google Scholar]
- 25.Kahn SE, Hull RL, Utzschneider KM: Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006;444:840–846 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.