Abstract
To mimic the dynamic regulation of signaling ligands immobilized on extracellular matrices or on the surfaces of neighboring cells for guidance of cell behavior and fate selection, we have harnessed biomolecular recognition in combination with polymer engineering to create dynamic surfaces on which the accessibility of immobilized ligands to cell surface receptors can be reversibly interconverted under physiological conditions. The cell-adhesive RGD peptide is chosen as a model ligand. RGD is fused to the C-terminus of a leucine zipper domain A and this fusion polypeptide is immobilized on surfaces through a residue at the N-terminus. The immobilized RGD can be converted from a cell-accessible to a cell-inaccessible state by addition of a conjugate of poly(ethylene) glycol (PEG) and another leucine zipper domain B (B-PEG). Heterodimerization between A and B allows co-immobilization of the PEG, which shields RGD from access by cells. The shielded RGD can be converted back to a cell-accessible state by addition of non-immobilized polypeptide A, which competes with the immobilized A for binding to B-PEG and removes B-PEG from the surface. This molecular design offers several advantages: the interconversion is reversible; the ligand remains immobilized during dynamic regulation so that cells are not exposed to the soluble form of the ligand that potentially has detrimental effects; the precision of the on/off states is assured by the molecular-level uniformity of the ligand and PEG co-immobilized through leucine zipper heterodimerization. The method can be readily adapted for dynamic regulation of other immobilized bioactive ligands of interest.
Using engineered materials to recapitulate the essential spatial and temporal characteristics of natural extracellular microenvironments during animal development is the key to successfully guiding cell behavior and fate selection in tissue engineering.1–6 However, it has remained a challenge to mimic the dynamic regulation of signaling ligands immobilized on extracellular matrices or on the surfaces of neighboring cells. Although soluble ligands can be added or removed readily during in vitro studies, the soluble form of a ligand that performs its function in an immobilized state in vivo potentially has opposite and detrimental effects because of competitive binding for cell surface receptors. This has been demonstrated by inhibition of Notch signaling in the presence of non-immobilized Notch ligands and death of anchorage-dependent cells in the presence of soluble RGD peptide.7–9 In recent years, materials capable of dynamically presenting immobilized bioactive ligands to cell surface receptors under physiological conditions have attracted increasing research interest.10–15 However, the methods developed to date have drawbacks including irreversible or imprecise transition between the on/off states and exposure of cells to the soluble counterparts of the ligands during in situ immobilization or cleavage. The limited ability to dynamically regulate immobilized bioactive ligands in engineered systems has hindered the progress of both technological development and fundamental understanding in tissue engineering.
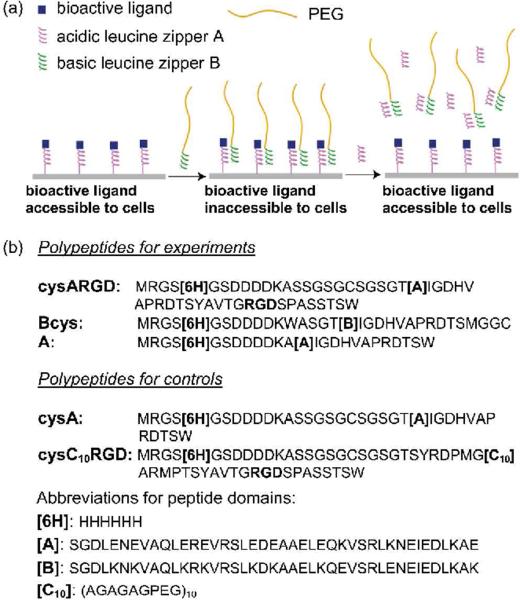
Here we report a novel method of harnessing the molecular recognition of intelligent biomolecules, in combination with polymer engineering, to create dynamic bioactive surfaces on which the accessibility of immobilized ligands to cell surface receptors is reversibly interconverted with high precision under physiological conditions (Figure 1a). We hypothesized that the accessibility of an immobilized bioactive ligand to cells could be dynamically regulated through reversible co-immobilization and removal of poly(ethylene) glycol (PEG) that, when coimmobilized, shields the surface-bound ligand from access by cells.16 We expected that reversible co-immobilization and removal of PEG could be achieved by exploiting the molecular recognition between a pair of complementary leucine zipper domains A and B, which heterodimerize through non-covalent, reversible interactions.17, 18 In the molecular design illustrated in Figure 1a, a bioactive ligand of interest is fused to one terminus of the leucine zipper A and immobilized on a surface through a residue at the other terminus. The modified surface allows co-immobilization of a B-PEG conjugate, which shields the surface-bound bioactive ligand and converts it to a cell-inaccessible state. To convert the ligand back to a cell-accessible state, non-immobilized polypeptide A is added in excess to compete with the immobilized A for binding to B-PEG so that B-PEG can be removed from the surface. This molecular design allows the immobilized ligand to be reversibly interconverted between cell-accessible and cell-inaccessible states under physiological conditions as many times as needed. The bioactive ligand remains immobilized during the dynamic regulation, so cells will not be exposed to the soluble counterpart of the ligand. An additional advantage of this design is that heterodimerization between A and B allows the bioactive ligand and PEG to be co-immobilized uniformly on the molecular level, preventing the formation of ligand islands free of PEG chains. Such islands would likely form when a conventional co-immobilization method is used, and they would compromise the precise switch of the ligand between cell-accessible and cell-inaccessible states.
Figure 1.
Illustration of the molecular design of bioactive surfaces capable of dynamically and reversibly regulating immobilized ligands (a) and the sequences of the polypeptides used in the study (b).
The polypeptides and their sequences used in this study are shown in Figure 1b. The complementary leucine zipper domains A and B were chosen because they could heterodimerize with an affinity (in the range between 10−8 M and 10−10 M) higher than that of homo-oligomerization (self-aggregation) of A or B.17, 18 The cell-adhesive RGD peptide was chosen as a model bioactive ligand. A cysteine residue was engineered at the N-terminus of A for surface immobilization. All the polypeptides were genetically engineered and biosynthesized. B-PEG conjugates were prepared by allowing Bcys to react with a large excess of PEG-diacrylate through a Michael-type addition reaction or with a large excess of PEG-maleimide through the thiol-maleimide reaction. After conjugation with PEG, B retained its ability to heterodimerize with A, as revealed from the mobility shift of cysARGD in native gel electrophoresis after cysARGD was incubated with B-PEG (SI, Figure S1).
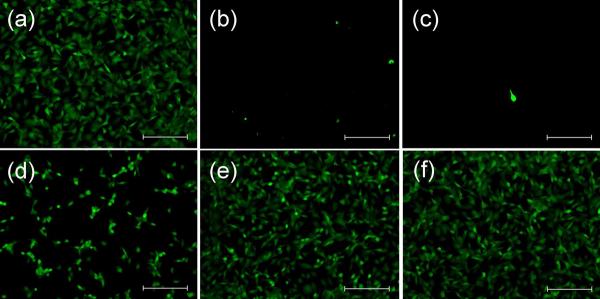
The ability of the surface-immobilized cysARGD to co-immobilize B-PEG through heterodimerization between A and B and the ability of the co-immobilized PEG to shield the surface-bound RGD from access by cells were examined. First, cysARGD and a control polypeptide cysA were each immobilized on piranha-solution-cleaned gold surfaces through the cysteine residue in the presence of 5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP). The surfaces modified with cysARGD supported the adhesion of fibroblasts (NIH 3T3) while those modified with cysA did not (Figure 2a and 2b), suggesting that cell adhesion on cysARGD-functionalized surfaces was regulated through the specific interaction between the RGD peptide and cell surface receptors. Then the cysARGD-functionalized surfaces were incubated with B-PEG conjugates of various PEG lengths (10 kDa, 3.4 kDa, 700 Da, and 258 Da), resulting in further modified surfaces exhibiting different cell adhesion properties depending on the PEG size. When the PEG was 10 kDa, the surfaces did not support cell adhesion (Figure 2c); when the PEG was 3.4 kDa, the surfaces partially supported cell adhesion (Figure 2d); and when the PEG was 700 Da or smaller, the surfaces were as adhesive as the cysARGD-functionalized surfaces without additional modification (Figure 2e, 2f). These results suggested that the immobilized cysARGD allowed co-immobilization of B-PEG through heterodimerization between A and B and the shielding effect provided by the co-immobilized PEG increased with its size.
Figure 2.
The shielding effect provided by co-immobilized PEG as revealed from a cell adhesion assay. (a) The cysARGD-functionalized surface. (b) The cysA-functionalized surface. (c–f) The cysARGD-functionalized surfaces further modified with B-PEG conjugates having PEG lengths of 10 kDa (c), 3.4 kDa (d), 700 Da (e), and 258 Da (f), respectively. The scale bars are 200 μm.
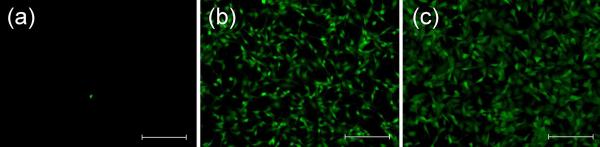
The reversible switch of the surface-bound RGD between cell-accessible and cell-inaccessible states was performed in the presence of cells under physiological conditions. To demonstrate the accessible-to-inaccessible conversion of the immobilized RGD peptide, fibroblasts were allowed to adhere on a surface as shown in Figure 2a for 3 h. Then B-PEG (PEG: 10 kDa, 300 μM) was added into the culture medium, followed by incubation under shaking (at a speed at which the liquid just started to move to enhance mass transfer) for 2 h. Cell detachment was observed (Figure 3a). A control performed on a cysC10RGD-functionalized surface (C10 is a hydrophilic random coil17) under the same condition did not result in cell detachment (Figure 3b), suggesting that the cell detachment from the cysARGD-functionalized surface was caused by co-immobilization of B-PEG through heterodimerization between A and B. These results not only suggested that the surface-bound cysARGD allowed co-immobilization of B-PEG in the presence of cells under physiological conditions but also showed that even the ligands that had already been engaged in the interactions with cell surface receptors could be converted to the cell-inaccessible state due to the physical, reversible, and dynamic nature of the interactions between ligands and cell surface receptors.
Figure 3.
Reversible switch of the accessibility of immobilized RGD to cell surface receptors under physiological conditions. (a) The cells adhered to the cysARGD-functionalized surface as shown in Figure 2a detached when B-PEG (PEG: 10 kDa) was added in the culture medium. (b) The cells adhered to a cysC10RGD-functionalized control surface did not detach when B-PEG (PEG: 10 kDa) was added. (c) The non-adhesive surface as shown in Figure 2c became cell-adhesive when non-immobilized polypeptide A was added in the culture medium. The scale bars are 200 μm.
To demonstrate that the cell-inaccessible RGD shielded by PEG could be converted back to the cell-accessible state in the presence of cells, a cysARGD-functionalized surface was further modified with B-PEG (PEG: 10 kDa); cells were seeded on the resulting non-adhesive surface. Non-immobilized polypeptide A (300 μM) was added in the cell culture medium, followed by shaking incubation (to enhance mass transfer) for 2 h and static incubation (to allow cell adhesion) for 3 h sequentially. The surface was converted from non-adhesive (Figure 2c) to cell-adhesive (Figure 3c), suggesting that the non-immobilized A competed with the immobilized cysARGD for binding with B-PEG and most B-PEG molecules could be removed from the surface when the amount of non-immobilized A was sufficiently greater than that of immobilized cysARGD. The RGD peptide in immobilized cysARGD was consequently converted from the cell-inaccessible to cell-accessible state. After the medium was changed, the cell-accessible RGD could be reconverted to be cell-inaccessible by addition of B-PEG.
Because surface immobilization of an A-ligand can be achieved through various chemical approaches, such as those enabled by the unique functional groups introduced through non-natural amino acid analogs,19 the ligands are not limited to cysteine-free peptides and the substrates are not limited to gold surfaces. For example, a thiol-containing A-ligand can be immobilized to the surfaces of hydrogels functionalized with maleimide or ene,20, 21 so that temporal control of immobilized ligands can be coupled with regulation of substrate stiffness to further optimize cell microenvironments for desired outcomes. In addition, this method potentially allows both temporal and spatial control of ligands. For example, a step gradient of a ligand can be created via micro/nano-printing, even though creation of a continuous gradient might be difficult. One limitation of the method is that the interconversion is not instant, so it cannot be used for applications such as guidance of rapid cell migration. However, this method will find use in many other applications such as regulation of stem cell differentiation, which is stepwise and often involved with changes in microenvironmental signals on the order of hours or days. Even though shaking incubation is necessary to overcome the mass transfer limitation and allow the interconversion to occur at 1 or 2 h (the experiment described in Figure 3a was also tested at 1 h and the same result was obtained) and exposure of cells to the shear stress associated with the convective flow may introduce some complexity, it is expected that the fluid velocity required for convective mass transfer to dominate over diffusive mass transfer is small. The scaling analysis based on the length scale of cells and the time scale required for the interconversion to occur under static conditions (ca. 10 h) suggests that a velocity on the order of 10−6 cm/s is enough to result in a Peclet number (the ratio of convective mass transfer to diffusive mass transfer) of 100.22 The shear stress associated with such small velocity is low.
In summary, the molecular engineering approach reported here represents a platform that can be readily adapted to dynamically and reversibly regulate the accessibility of a variety of immobilized bioactive ligands to cell surface receptors under physiological conditions. The materials having such dynamic bioactive surfaces can be used to temporally control the properties of engineered extracellular microenvironments.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by a seed grant in the MRSEC Program of the National Science Foundation (DMR-0212302) and an NIH Biotechnology Training Grant (GM008347).
Footnotes
Supporting Information Available: Experimental procedures and gel electrophoresis data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- (2).Lee KY, Peters MC, Anderson KW, Mooney DJ. Nature. 2000;408:998–1000. doi: 10.1038/35050141. [DOI] [PubMed] [Google Scholar]
- (3).Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Campbell PG, Miller ED, Fisher GW, Walker LM, Weiss LE. Biomaterials. 2005;26:6762–6770. doi: 10.1016/j.biomaterials.2005.04.032. [DOI] [PubMed] [Google Scholar]
- (5).DeLong SA, Moon JJ, West JL. Biomaterials. 2005;26:3227–3234. doi: 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- (6).DeForest CA, Polizzotti BD, Anseth KS. Nat. Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Vas V, Szilagyi L, Paloczi K, Uher F. J. Leukoc. Biol. 2004;75:714–720. doi: 10.1189/jlb.1003462. [DOI] [PubMed] [Google Scholar]
- (8).Varnum-Finney B, Wu LZ, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. J. Cell Sci. 2000;113:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- (9).Michel JB. Arterioscler. Thromb. Vasc. Biol. 2003;23:2146–2154. doi: 10.1161/01.ATV.0000099882.52647.E4. [DOI] [PubMed] [Google Scholar]
- (10).Salinas CN, Anseth KS. Biomaterials. 2008;29:2370–2377. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yeo WS, Yousaf MN, Mrksich M. J. Am. Chem. Soc. 2003;125:14994–14995. doi: 10.1021/ja038265b. [DOI] [PubMed] [Google Scholar]
- (12).Chan EWL, Park S, Yousaf MN. Angew. Chem., Int. Ed. 2008;47:6267–6271. doi: 10.1002/anie.200800166. [DOI] [PubMed] [Google Scholar]
- (13).Todd SJ, Farrar D, Gough JE, Ulijn RV. Soft Matter. 2007;3:547–550. doi: 10.1039/b618256a. [DOI] [PubMed] [Google Scholar]
- (14).Petersen S, Alonso JM, Specht A, Duodu P, Goeldner M, del Campo A. Angew. Chem., Int. Ed. 2008;47:3192–3195. doi: 10.1002/anie.200704857. [DOI] [PubMed] [Google Scholar]
- (15).Auernheimer J, Dahmen C, Hersel U, Bausch A, Kessler H. J. Am.Chem. Soc. 2005;127:16107–16110. doi: 10.1021/ja053648q. [DOI] [PubMed] [Google Scholar]
- (16).Dori Y, Bianco-Peled H, Satija SK, Fields GB, McCarthy JB, Tirrell M. J. Biomed. Mater. Res. 2000;50:75–81. doi: 10.1002/(sici)1097-4636(200004)50:1<75::aid-jbm11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- (17).Shen W, Zhang KC, Kornfield JA, Tirrell DA. Nat. Mater. 2006;5:153–158. doi: 10.1038/nmat1573. [DOI] [PubMed] [Google Scholar]
- (18).McGrath KP, Butler MM, DiGirolamo CM, Kaplan DL, Petka WA, Laue TM. J. Bioact. Compat. Polym. 2000;15:334–356. [Google Scholar]
- (19).Conor RE, Tirrell DA. Polym. Rev. 2007;47:9–28. [Google Scholar]
- (20).Kosif I, Park E, Sanyal R, Sanyal A. Macromolecules. 2010;43:4140–4148. [Google Scholar]
- (21).Aimetti AA, Machen AJ, Anseth KS. Biomaterials. 2009;30:6048–6054. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Deen WM. Analysis of Transport Phenomena. Oxford University Press; New York: 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.