Abstract
The site-specific motion of Arg residues in a membrane-bound disulfide-linked antimicrobial peptide, protegrin-1 (PG-1), is investigated using magic-angle spinning solid-state NMR, to better understand the membrane insertion and lipid interaction of this cationic membrane-disruptive peptide. C-H and N-H dipolar couplings and 13C chemical shift anisotropies were measured in the anionic POPE/POPG membrane and found to be reduced from the rigid-limit values by varying extents, indicating the presence of segmental motion. An Arg residue at the β-turn region of the peptide shows much weaker spin interactions, indicating larger amplitudes of motion, than an Arg residue in the β-strand region of the peptide. This is consistent with the exposure of the β-turn to the membrane surface and the immersion of the β-strand in the hydrophobic middle of the membrane, and supports the previously proposed oligomerization of the peptide into β-barrels in the anionic membrane. 13C T2 and 1H T1ρ relaxation times indicate that the β-turn backbone undergoes large-amplitude intermediate-timescale motion in the fluid phase of the membrane, causing significant line broadening and loss of spectral intensity. This study illustrates the strong correlation between the dynamics and the structure of membrane proteins and the capability of solid-state NMR spectroscopy for providing detailed information on site-specific dynamics in complex membrane protein assemblies.
Keywords: membrane protein dynamics, molecular dynamics, antimicrobial peptides, guanidinium-phosphate complexation, order parameters, solid-state NMR, arginine
Introduction
Molecular motion is common in membrane proteins and is often intimately related to the function and lipid-interaction of these molecules. Solid-state NMR (SSNMR) spectroscopy is a versatile tool to characterize molecular dynamics on a wide range of timescales (picoseconds to seconds) and to determine the amplitude of anisotropic motion. Large-amplitude segmental motion has been reported, for example, for a bacterial toxin that spontaneously inserts into the lipid membrane as a result of its intrinsic conformational plasticity [1], a lipidated Ras signaling protein [2], the catalytic domain of a membrane-bound enzyme [3], and the loops of the seven-transmembrane-helix protein rhodopsin [4]. In addition to internal segmental motion, whole-body reorientation has been discovered for many small membrane peptides of both β-sheet and α-helical secondary structures [5–7].
Protegrin-1 (PG-1) is a broad-spectrum antimicrobial peptide found in porcine leukocytes [8, 9]. It is a β-hairpin molecule stabilized by two disulfide bonds and contains six Arg residues (RGGRLCYCRRRFCVCVGR). PG-1 achieves its antimicrobial function by forming non-selective pores in the microbial cell membrane that disrupt the membrane’s barrier function [10, 11]. Recently, the high-resolution oligomeric structure of PG-1 at the pores was determined using 1H and 19F spin diffusion NMR techniques [12]. The peptide was found to self-assemble into a transmembrane β-barrel in bacteria-mimetic anionic POPE/POPG membranes. 13C-31P distance constraints indicate that the Arg residues in these transmembrane β-barrels are complexed with lipid phosphates [13], suggesting that the charge neutralization by ion pairing reduces the free energy of peptide insertion into the hydrophobic part of the membrane, and the consequent tethering of lipid headgroups may be the cause for toroidal pore formation.
The experiments that yielded the equilibrium oligomeric structure of PG-1 and the toroidal pore morphology of the lipid membrane were carried out at low temperatures of about −40°C, in the gel phase of the membrane, to eliminate any motion that would average the distance-dependent dipolar couplings. On the other hand, PG-1 carries out its antimicrobial action in the liquid-crystalline (LC) phase of the membrane, where it is expected to be more mobile. How the dynamics of PG-1 and its Arg sidechains affect toroidal pore formation has not yet been studied. If Arg-phosphate complex formation is true, then the functional groups involved in the complex – the guanidinium ions, the lipid phosphates, and possibly water – should be less mobile than in their respective bulk environments. Thus, understanding Arg motion in PG-1 in the lipid membrane may provide additional insight into guanidinium-phosphate interaction. More generally, although the motion of long-chain amino acid residues has begun to be investigated in microcrystalline proteins [14–16], motion of the same residues in membrane proteins is still scarcely studied by NMR. Arg is particularly common in many medically important membrane peptides and proteins such as antimicrobial peptides (AMPs) [17], cell-penetrating peptides [18, 19], and voltage-sensing domains of ion channels [20].
In this work we report the amplitudes of microsecond timescale motions of Arg and other residues in PG-1 bound to the POPE/POPG membrane. We found that an Arg in the β-strand part of the molecule, which is embedded in the hydrophobic interior of the membrane, is much less mobile than an Arg in the β-turn part of the molecule, which is exposed to the membrane surface. This is consistent with the oligomeric structure and lipid interaction of this antimicrobial peptide.
Results
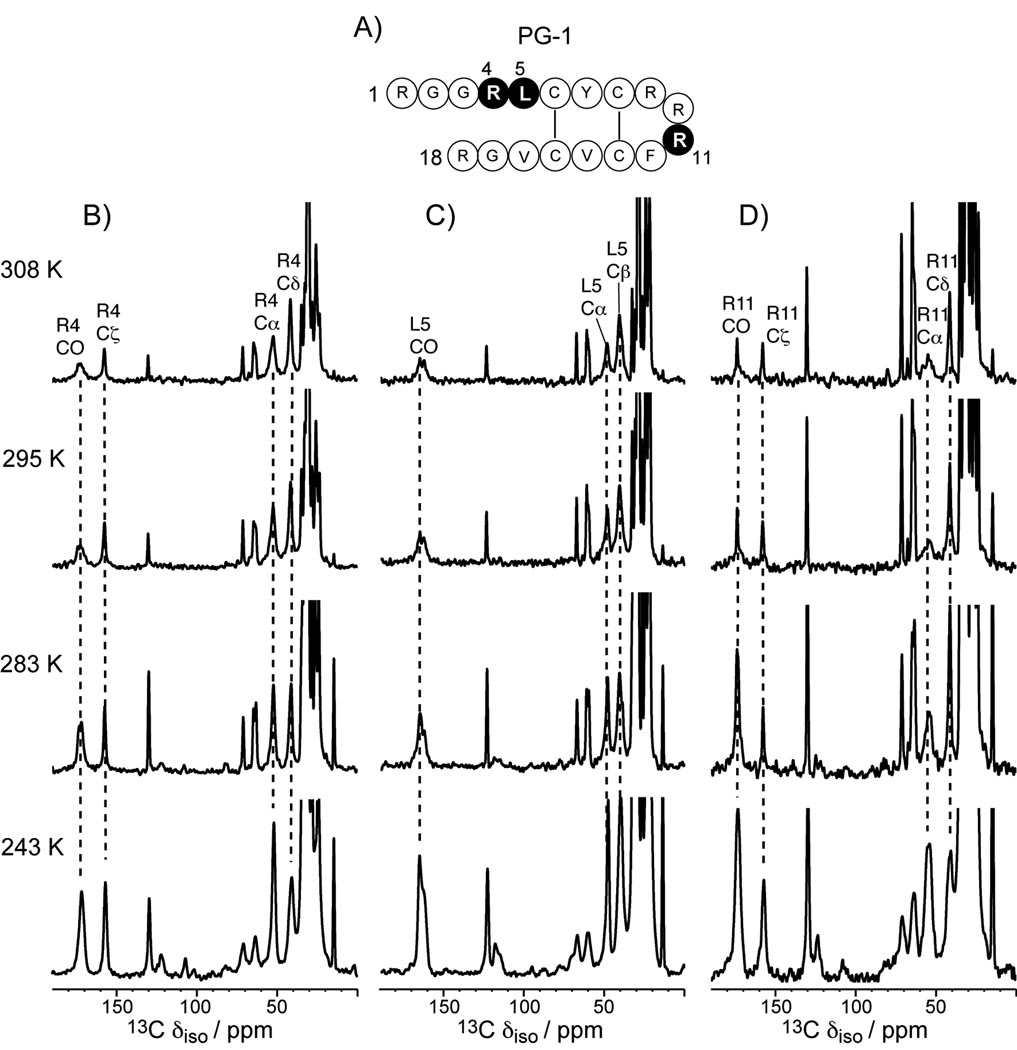
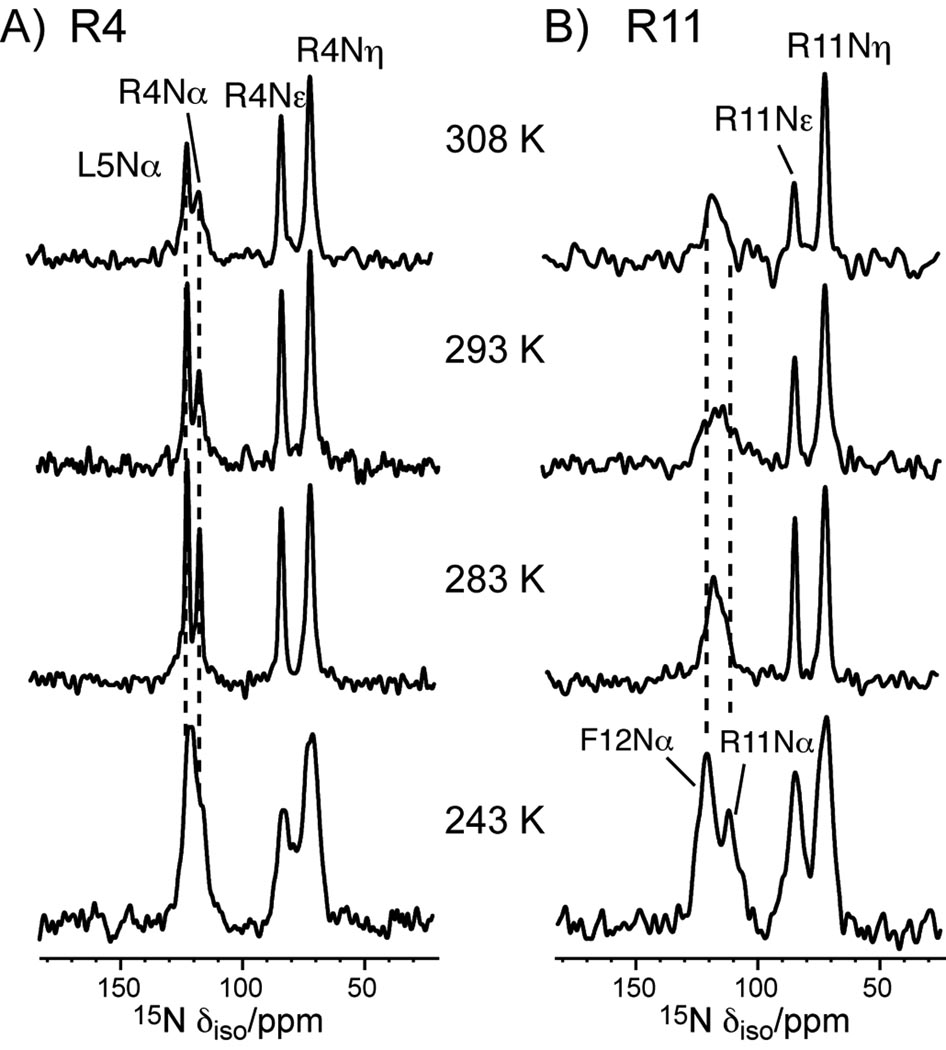
We first characterized the dynamic structure of PG-1 in POPE/POPG bilayers by variable-temperature 13C and 15N CP-MAS experiments. A series of CP spectra were collected between 243 K and 308 K for PG-1 containing U-13C, 15N-labeled Arg4, Leu5, and Arg11, and 15N-labeled Phe12. As shown in Figure 1, the Cα peaks of Arg4 and Leu5 are much sharper and higher than the Cα peak of Arg11. At 295 K, the full widths at half-maximum (FWHM) of Arg4 and Leu5 Cα’s are ~3 ppm, compared to 6 ppm for Arg11 Cα. As the temperature decreases, the Arg11 Cα intensity increases significantly. This suggests that in the liquid-crystalline phase of the membrane Arg11 backbone undergoes large-amplitude intermediate-timescale motion that becomes frozen in the gel phase of the membrane, while the Arg4 and Leu5 Cα sites are more rigid. In other words, the β-turn backbone is more mobile than the β-strand backbone. A similar trend is observed in the 15N CP-MAS spectra (Figure 2). The backbone Nα peaks of Arg4 and Leu5 are sharp and well resolved, with FWHM of 2 – 3 ppm at 283 K, while the Arg11 Nα peak is broad and overlaps with Phe12 Nα, giving a FWHM of 9 ppm for the combined peak at 283 K. Only at 243 K do the Arg11 Nα and Phe12 Nα peaks become resolved. We assigned the Nα peaks by 13C-15N 2D correlation experiments (data not shown) [21].
Figure 1.
13C CP-MAS spectra in PG-1 bound to the POPE/POPG membrane (P/L = 1:12.5) from 243 K to 308 K. A) Amino acid sequence of PG-1. Labeled residues are shaded. B) 13C CP-MAS spectra of Arg4, C) 13C CP-MAS spectra of Leu5, D) 13C CP-MAS spectra of Arg11. Peptide peaks are assigned and annotated.
Figure 2.
15N CP-MAS spectra of PG-1 in the POPE/POPG membrane at various temperatures. A) Arg4, Leu5. B) Arg11 and Phe12. Assignments were obtained from 13C-15N 2D correlation spectra (not shown).
To distinguish the contribution of static structural heterogeneity versus dynamic disorder to the linewidths, we measured the 13C T2 of Arg4 and Arg11 at two different temperatures, 283 K and 243 K, using the Hahn echo experiment. Table 1 shows the 13C apparent linewidths, Δ*, read off from the CP spectra, and the 13C homogeneous linewidths, Δ, obtained from the T2 values according to Δ = 1/πT2. At 243 K, the homogeneous linewidths of Arg4 and Arg11 are similar, indicating that motion is largely frozen. However, the apparent linewidth of Arg11 backbone Cα (604 Hz, or 6.0 ppm) is much larger than Arg4 Cα (222 Hz, or 2.2 ppm), indicating that there is much larger conformational disorder at the β-turn backbone than at the β-strand. In comparison, the sidechains of Arg4 and Arg11 at 243 K exhibit similar homogeneous linewidths as well as similar apparent linewidths, indicating that both the static and dynamic heterogeneities are comparable for the two sidechains. At 283 K, Arg11 Cα exhibits both larger Δ and larger Δ* than Arg4 Cα, indicating that the β-turn backbone has greater dynamic as well as static disorder than the β-strand backbone. In contrast, the sidechain of Arg11 has narrower Δ and Δ* than the Arg4 sidechain, indicating that Arg11 sidechain undergoes faster motions than Arg4.
Table 1.
13C apparent linewidths (Δ*) and homogeneous linewidths (Δ) of PG-1 in POPE/POPG membrane at 283 K and 243 K. The apparent linewidths are read off from 1D CP spectra. The homogeneous linewidths are obtained from T2 measurements as Δ = 1/πT2. The linewidths were measured at a 13C Larmor frequency of 100 MHz.
| Residue | Sites | 283 K | 243 K | ||
|---|---|---|---|---|---|
| Δ* / Hz | Δ / Hz | Δ * / Hz | Δ / Hz | ||
| Arg4 | Cα | 272 | 199 | 222 | 118 |
| Cδ | 222 | 187 | 493 | 289 | |
| Cζ | 111 | 84 | 201 | 80 | |
| Arg11 | Cα | 473 | 289 | 604 | 133 |
| Cδ | 161 | 106 | 534 | 265 | |
| Cζ | 81 | 53 | 222 | 94 | |
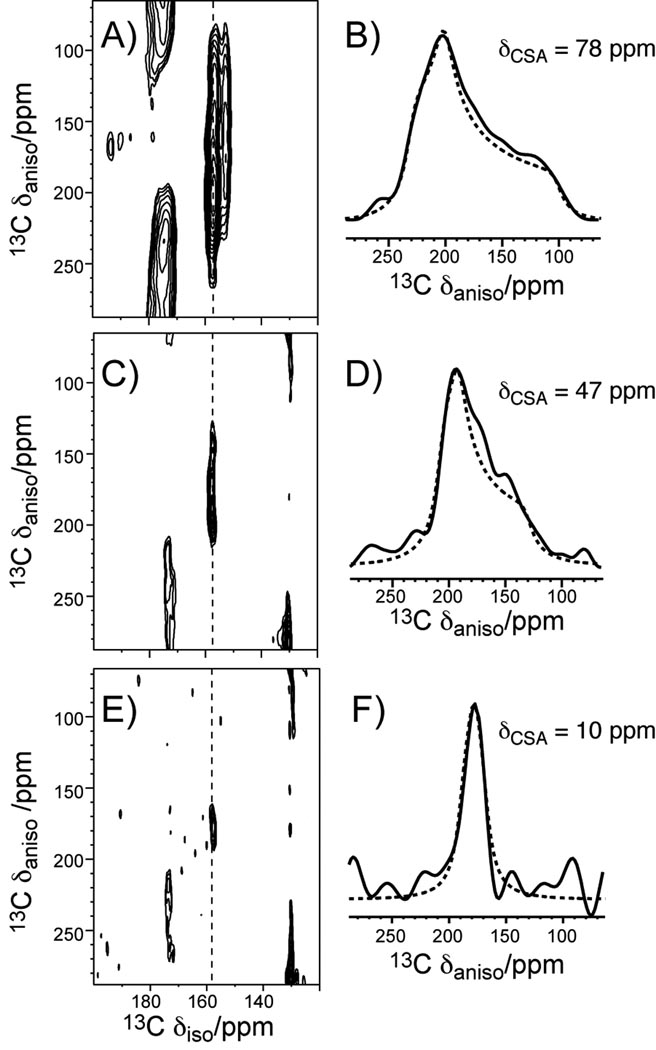
To obtain information on the motional amplitudes of the Arg sidechains, especially the guanidinium group, we measured the 13C chemical shift anisotropy (CSA) of Cζ, the center of the guanidinium ion. We chose the intermediate temperature of 283 K for the CSA and the subsequent dipolar coupling experiments, since at this temperature the spectra have the best overall combination of resolution and sensitivity. The theoretical phase transition temperature of the POPE/POPG (3:1) membrane is 291 K, thus the spectra theoretically correspond to the gel-phase membrane, but the phase transition is likely broadened by the peptide. The peptide mobility closer to the physiological temperature may be extrapolated from the 283 K data and is expected to be higher, but the differences between residues should be similar. We used the 2D separation of undistorted powder patterns by effortless recoupling (SUPER) experiment [22] to recouple the CSA interaction and correlate it with the isotropic 13C chemical shift. Figure 3 shows the 2D SUPER spectra and 1D cross sections of the model compound Fmoc-Arg(MTR)-OH, and Arg4 and Arg11 in PG-1 bound to the POPE/POPG membrane. For the dry powder sample Fmoc-Arg(MTR)-OH, the Cζ cross section yielded a CSA anisotropy parameter δ, defined as the difference between the largest principal value δzz and the isotropic shift δiso, of 78 ppm. This CSA is the rigid-limit value, since C-H dipolar couplings of the sidechain carbons in this model compound have nearly rigid-limit values (Table 2). In comparison, PG-1 Arg4 and Arg11 Cζ both give reduced CSA’s: the Arg4 Cζ δ̄ is 47.3 ppm whereas the Arg11 Cζ CSA is much smaller, 10.3 ppm. These correspond to a motional scaling factor of 0.13 for Arg11 and 0.61 for Arg4. Thus, the Arg11 sidechain has larger-amplitude motion than Arg4. Since T2 data indicate narrower homogeneous linewidths of Arg11 Cδ and Cζ than Arg4, the Arg11 sidechain motion is both faster and larger in amplitude than the Arg4 sidechain.
Figure 3.
Arg Cζ chemical shift anisotropies from the SUPER experiment. The 2D SUPER spectra are shown in A), C), E) and the corresponding Cζ 1D cross sections are shown in B), D), F). A, B) Fmoc-Arg(MTR)-OH. C, D) PG-1 Arg4. E, F) PG-1 Arg11. The PG-1 data were measured at 283 K in the POPE/POPG membrane.
Table 2.
Dipolar order parameters and CSA motional scaling factors a of PG-1 residues at 283 K and of three crystalline model compounds at 295 K.
| Sites | Arg4 | Arg11 | Fmoc-Arg | Arg-HCl | Leu5 | Leu |
|---|---|---|---|---|---|---|
| Nα | 1.05 | 0.70 | - | - | 0.95 | - |
| Cα | 0.93 | 0.70 | 0.91 | 0.91 | 0.93 | 0.95 |
| Cβ | 0.61 | - | 0.86 | 0.91 | 0.56 | 0.93 |
| Cγ | 0.63 | - | 0.91 | 0.91 | 0.44 | - |
| Cδ | 0.48 | 0.21 | 0.91 | 1.02 | 0.43 | 0.34 |
| Nε | 0.48 | 0.24 | - | - | - | - |
| Cζa | 0.61 | 0.13 | - | - | - | - |
| Nη | 0.36 | 0.28 | - | - | - | - |
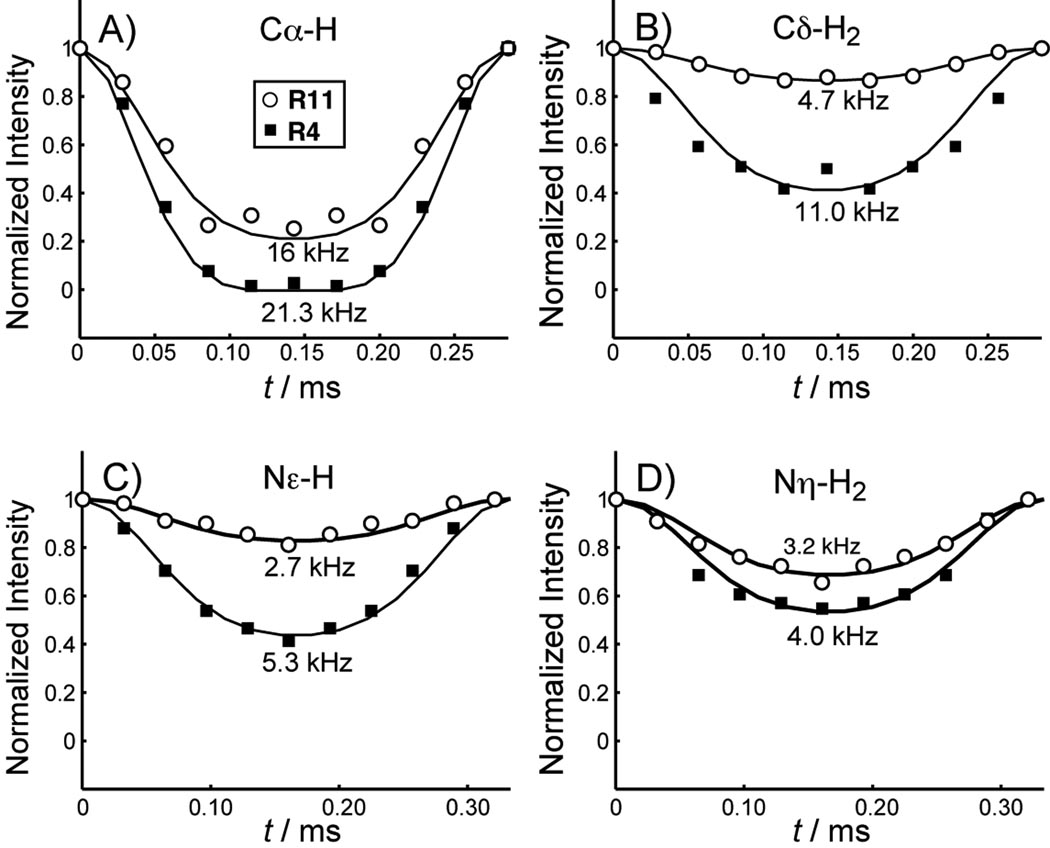
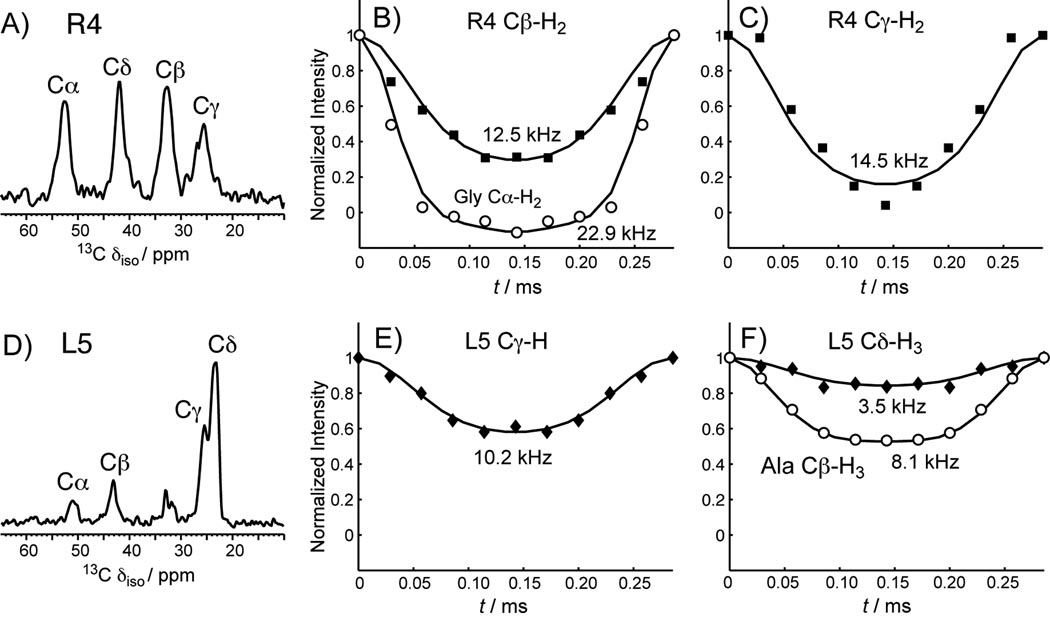
To obtain more quantitative information on the motional amplitude, we measured C-H and N-H dipolar couplings, whose tensor orientation and rigid-limit coupling strength are exactly known. The dipolar couplings were readily measured using the 2D dipolar-chemical shift correlation (DIPSHIFT) experiment to yield the bond order parameter, S = δ̄/δ. Figure 4 shows representative DIPSHIFT curves of Arg4 and Arg11 in POPE/POPG-bound PG-1. Cα-H represents the backbone, while Cδ-H2, Nε-H and Nη-H2 represent the sidechains. The order parameters are compiled in Table 2. Both the backbone Nα and Cα of Arg4 and Leu5 exhibit nearly rigid-limit couplings, with order parameters of 0.93–1.00. In contrast, the Arg11 Cα and Nα have significantly lower order parameters of 0.70. Thus, the β-strand backbone of the peptide is immobilized in the POPE/POPG membrane at this temperature, while the Arg11 backbone retains significant local segmental motion. For resolved sites (Cδ, Nε, and Nη) in the sidechains, Arg4 and Leu5 also have stronger dipolar couplings than those of Arg11, indicating that the β-strand sidechains have smaller amplitudes of motion, consistent with the variable-temperature spectra and the CSA results. Some 13C sites in the sidechain, such as Arg Cβ, Cγ, Leu Cγ and Cδ, overlap with the lipid peaks, so we used a double-quantum (DQ) filtered DIPSHIFT experiment to suppress the lipid signals and measure the C-H couplings of these Arg sites [1]. Figure 5 shows representative 1D DQ spectra and DQ-DIPSHIFT dephasing curves of Arg4 and Leu5. Arg11 has prohibitively low sensitivity in the DQ-DIPSHIFT experiment due to unfavorable motional rates at this temperature and is thus not measured. Table 2 shows that in general, the sidechain order parameters decrease with increasing distance from the backbone. Arg11 at the β-turn, which is close to the membrane surface, has much higher amplitudes of motion, or much lower order parameters, than Arg4 and Leu5 in the β-strand part of the peptide, which is embedded in the membrane [12].
Figure 4.
13C-1H and 15N-1H DIPSHIFT curves of several sites of Arg4 (closed squares) and Arg11 (open circles) in PG-1 at 283 K. A) Cα-H. B) Cδ-H2. C) Nε-H. D) Nη-H2. Arg11 gives weaker couplings than Arg4, indicating larger motional amplitudes.
Figure 5.
1D 13C DQ filtered spectra and DQ-DIPSHIFT curves of Arg4 and Leu5 in PG-1 in the POPE/POPG membrane. A) 1D DQ spectrum of Arg4. The Cβ and Cγ peaks no longer overlap with the lipid peaks. B) DIPSHIFT curves of Arg4 Cβ (squares) and the crystalline amino acid Gly Cα (circles). The Gly Cα data give the rigid-limit coupling for CH2 groups, which is 22.9 kHz. C) DIPSHIFT curve of Arg4 Cγ. D) 1D DQ spectrum of Leu5. The Cγ and Cδ peaks no longer overlap with the lipid peaks. E) DIPSHIFT curve of Leu5 Cγ. F) DIPSHIFT curves of Leu5 Cδ (diamonds) and the crystalline amino acid Ala Cβ (circles). The Ala Cβ data give the rigid-limit coupling for methyl groups, which is 8.1 kHz. This is one-third of the one bond C-H coupling due to the three-site jump of the CH3 group.
To obtain further information on the rates of motions of these residues, we measured the 1H T1ρ relaxation times, listed in Table 3. Most sites in Arg4, Leu5 and Arg11 have similar 1H T1ρ values (1.6 – 2.6 ms), except for Arg11 Hα, which has a much shorter T1ρ (0.83 ms) than Arg4 Hα (2 ms). This is consistent with the 13C T2 data indicating more pronounced intermediate-timescale motion of the β-turn backbone compared to the β-strand backbone.
Table 3.
1H T1ρ (ms) of POPE/POPG-bound PG-1 at 283 K and of crystalline Arg˙HCl at 295 K. Experimental uncertainties are given in the parentheses. The 1H spin-lock field strengths were 50 kHz in the 15N-detected experiment and 62.5 kHz in the 13C-detected experiments.
| Sites | Arg4 | Arg11 | Arg ˙ HCl |
|---|---|---|---|
| HN | 2.6 (0.2) | 2.2 (0.3) | - |
| Hα | 2.0 (0.1) | 0.8 (0.1) | 8.8 |
| Hδ | 1.6 (0.1) | 1.9 (0.1) | 9.5 |
| Hε | 2.2 (0.2) | 2.6 (0.2) | 8.8 |
| Hη | 1.9 (0.2) | 1.8 (0.1) | 8.8 |
Discussion
The solid-state NMR data shown here indicate that the β-turn backbone undergoes large-amplitude segmental motion on the microsecond timescale, while the β-strand backbone is mostly immobilized in the POPE/POPG membrane in the liquid-crystalline phase. The latter is consistent with the previously reported immobilization of PG-1 strand residues in POPC/POPG membranes [23]. Concomitant with the backbone mobility difference, the sidechains also exhibit dynamic differences: Arg11 has much lower order parameters than Arg4 (Table 2), indicating large motional amplitudes. Both membrane-associated Arg’s are much more mobile than the crystalline compound Arg ˙ HCl.
The dynamic difference between Arg4 and Arg11 can be understood in terms of the self-assembly of PG-1 and the peptide-lipid interactions. The β-strands containing Arg4 and Leu5 are involved in intermolecular association with other PG-1 molecules through N–H…O=C hydrogen bonds to form β-barrels [12, 24], thus these residues should experience hindered motion. The strand aggregation is important to PG-1 antimicrobial activity. Mutation of Val14 to N-methyl-Val, which disrupted hydrogen bonding of the Val14 backbone to its intermolecular partner, resulted in much lower antimicrobial activity [25]. In contrast, the β-turn Arg11 is not involved in intermolecular hydrogen bonding and is located near the membrane surface, thus has more motional freedom.
A second contributing factor to the different sidechain dynamics of Arg11 and Arg4 may be the guanidinium-phosphate interaction. 13C-31P distance data indicated that both sidechains lie within hydrogen-bonding distance to lipid phosphates [13]. However, while the Arg4 guanidinium group interacts with the phosphate groups that have moved to the middle of the membrane as part of the toroidal pore, the Arg11 guanidinium ion interacts with phosphates at the membrane surface with much higher mobility. Thus, the motional restriction caused by the lipid phosphate groups is more severe for Arg4 than for Arg11. We note that at the temperature of 283 K where most dynamics data were obtained, the lipid molecules are much more mobile than at ~230 K where the 13C-31P distances were measured. Thus, the guanidinium-phosphate association at 283 K is likely to be transient rather than permanent.
The high mobility of the β-hairpin tip of PG-1 dovetails the observation of an analogous β-hairpin antimicrobial peptide, TP-I [26]. There, G10 at the β-turn exhibited an order of magnitude shorter 1H T1ρ than the β-strand residues. Field-dependent T1ρ analysis indicated that the shorter T1ρ of G10 results from larger motional amplitudes of the β-turn and not to rate differences from the rest of the peptide [26].
Molecular dynamics simulations of the S4 helix of the voltage-gated potassium channel KvAP [27] suggested that lipid headgroups and water stabilize Arg insertion by forming a hydrogen-bonded network. The effective lipid bilayer thickness was reduced to a remarkably small 10 Å near the inserted S4 helix so that water and phosphate groups can stabilize the Arg’s in the middle of the S4 helix by hydrogen bonds [28]. Based on the comparison of the mean-square displacement of phosphate groups near the peptide with those far away from the peptide and the analysis of the survival function of water molecules in the system, it was found that both phosphate groups and water molecules are much less mobile in the vicinity of the guanidinium groups than in their respective bulk environments. In particular, the mean residence times for water molecules hydrogen-bonded to Arg9 and Arg12 in the S4 helix, which are close to the bilayer surface, are much shorter than those hydrogen-bonded to Arg15 and Arg18, which lie in the hydrophobic core of the membrane (90–300 ps versus 1000–2000 ps). This different residence time suggests that the water molecules near Arg in the hydrophobic core are less mobile than those near Arg at the membrane surface. This in turn suggests that Arg’s in the hydrophobic part of the membrane are less mobile than those close to the bilayer surface. These are consistent with the different mobility observed between Arg4 and Arg11 in PG-1.
In summary, we have measured the dipolar couplings, CSA’s, and T2 and T1ρ relaxation times of key Arg residues in PG-1 in the bacteria-mimetic anionic POPE/POPG membrane. The linewidths and motional scaling factors show that the β-turn Arg11 near the membrane surface is significantly more mobile than the β-strand Arg4 and Leu5 in the hydrophobic part of the membrane. The different mobility is consistent with the location of the residues with respect to the membrane, the intermolecular aggregation of this peptide, and the strong Arg-phosphate interaction. Thus, the site-specific dynamics of PG-1 correlate well with its topological and oligomeric structure. Solid-state NMR is shown to be a useful tool for elucidating the relation between membrane protein dynamics and its structure.
Experimental section
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine (POPE), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) were purchased from Avanti Polar Lipids (Alabaster, AL). PG-1 (NH2-RGGRLCYCRRRFCVCVGR-CONH2) was synthesized using Fmoc chemistry as previously described [7]. Three PG-1 samples were synthesized, containing U-13C, 15N-Arg4 and 15N-Leu5, U-13C, 15N-Arg11 and 15N-Phe12, U-13C, 15N-Leu5. U-13C, 15N-labeled Arg was obtained from Spectra Stable Isotopes (Columbia, MD) as Fmoc-Arg(MTR)-OH.
POPE and POPG lipids (3:1) were mixed in chloroform and blown dry under N2 gas. The mixture was then redissolved in cyclohexane and lyophilized. The dry lipid powder was dissolved in water and subjected to five cycles of freeze-thawing to form uniform vesicles. An appropriate amount of PG-1 to reach a peptide-lipid molar ratio (P/L) of 1 : 12.5 was dissolved in water and mixed with the lipid vesicle solution, incubated at 303 K overnight, then centrifuged at 55,000 rpm for 2.5 hours. The pellet was packed into a MAS rotor, giving a fully hydrated membrane sample.
NMR experiments were carried out on a Bruker DSX-400 (9.4 Tesla) spectrometer (Karlsruhe, Germany). Triple-resonance magic-angle spinning (MAS) probes with a 4 mm spinning module was used. Temperatures were controlled by a Kinetics Thermal Systems XR air-jet sample cooler (Stone Ridge, NY) on the 400 MHz system. Typical 90° pulse lengths were 5 – 6 µs for 13C and 15N, and 1H decoupling fields of 50–80 kHz were used. 13C chemical shifts were referenced externally to the α-Gly 13C’ signal at 176.49 ppm on the TMS scale. 15N chemical shifts were referenced externally to the N-acetyl-Val 15Nα signal at 121.72 ppm.
13C-1H and 15N-1H dipolar couplings were measured using the 2D DIPSHIFT experiment [29] at 3.0–3.5 kHz MAS with MREV-8 for 1H homonuclear decoupling. Pulse lengths of 3.5 µs were used in the MREV-8 pulse train. The N-H DIPSHIFT experiments were performed with dipolar doubling [30, 31] to increase the precision of the measured couplings. Some 13C sites overlap with lipid peaks, so the double-quantum-filtered (DQ) DIPSHIFT experiments [1] were used to measure these dipolar couplings. The DQ filter used SPC5 homonuclear dipolar recoupling sequence [32]. The 13C CSA was measured using the 2D SUPER experiment [22] under 3.5 kHz MAS. The corresponding 13C field strength was 42 kHz. 1H rotating-frame spin-lattice relaxation times (T1ρ) was measured using spin-lock field strengths of 50–62.5 kHz. The 13C and 15N 1D spectra were measured between 243 and 308 K. All DIPSHIFT, SUPER and T1ρ experiments were carried out at 283 K.
Supplementary Material
Acknowledgement
This work is supported by the National Institutes of Health grant GM-066976 to M. H.
References
- 1.Huster D, Xiao LS, Hong M. Biochemistry. 2001;40:7662–7674. doi: 10.1021/bi0027231. [DOI] [PubMed] [Google Scholar]
- 2.Reuther G, Tan KT, Vogel A, Nowak C, Arnold K, Kuhlmann J, Waldmann H, Huster D. J. Am. Chem. Soc. 2006;128:13840–13846. doi: 10.1021/ja063635s. [DOI] [PubMed] [Google Scholar]
- 3.Williams JC, McDermott AE. Biochemistry. 1995;34:8309–8319. doi: 10.1021/bi00026a012. [DOI] [PubMed] [Google Scholar]
- 4.Etzkorn M, Martell S, Andronesi OC, Seidel K, Engelhard M, Baldus M. Angew. Chem. Int. Ed. 2007;46:459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- 5.Cady SD, Goodman C, DeGrado WF, Hong M. J. Am. Chem. Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 6.Park SH, Mrse AA, Nevzorov AA, De Angelis AA, Opella SJ. J. Magn. Reson. 2006;178:162–165. doi: 10.1016/j.jmr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- 8.Bellm L, Lehrer RI, Ganz T. Exp. Opin. Invest. Drugs. 2000;9:1731–1742. doi: 10.1517/13543784.9.8.1731. [DOI] [PubMed] [Google Scholar]
- 9.Kokryakov VN, Harwig SS, Panyutich EA, Shevchenko AA, Aleshina GM, Shamova OV, Korneva HA, Lehrer RI. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 10.Mangoni ME, Aumelas A, Charnet P, Roumestand C, Chiche L, Despaux E, Grassy G, Calas B, Chavanieu A. FEBS Lett. 1996;383:93–98. doi: 10.1016/0014-5793(96)00236-0. [DOI] [PubMed] [Google Scholar]
- 11.Sokolov Y, Mirzabekov T, Martin DW, Lehrer RI, Kagan BL. Biochim. Biophys. Acta. 1999;1420:23–29. doi: 10.1016/s0005-2736(99)00086-3. [DOI] [PubMed] [Google Scholar]
- 12.Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M, Waring AJ, Hong M. J. Am. Chem. Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 14.Lorieau J, McDermott AE. Magn. Reson. Chem. 2006;44:334–347. doi: 10.1002/mrc.1773. [DOI] [PubMed] [Google Scholar]
- 15.Lorieau JL, McDermott AE. J. Am. Chem. Soc. 2006;128:11505–11512. doi: 10.1021/ja062443u. [DOI] [PubMed] [Google Scholar]
- 16.Wylie BJ, Franks WT, Graesser DT, Rienstra CM. J. Am. Chem. Soc. 2005;127:11946–11947. doi: 10.1021/ja053862e. [DOI] [PubMed] [Google Scholar]
- 17.Hancock RE, Lehrer R. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 18.Vives E, Brodin P, Lebleu B. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 19.Jarver P, Langel U. Biochim. Biophys. Acta. 2006;1758:260–263. doi: 10.1016/j.bbamem.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Long SB, Campbell EB, Mackinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 21.Hong M, Griffin RG. J. Am. Chem. Soc. 1998;120:7113–7114. [Google Scholar]
- 22.Liu SF, Mao JD, Schmidt-Rohr K. J. Magn. Reson. 2002;155:15–28. doi: 10.1006/jmre.2002.2503. [DOI] [PubMed] [Google Scholar]
- 23.Buffy JJ, Waring AJ, Lehrer RI, Hong M. Biochemistry. 2003;42:13725–13734. doi: 10.1021/bi035187w. [DOI] [PubMed] [Google Scholar]
- 24.Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M. Biochemistry. 2006;45:8341–8349. doi: 10.1021/bi060305b. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Falla TJ, Liu HJ, Hurst MA, Fujii CA, Mosca DA, Embree JR, Loury DJ, Radel PA, Chang CC, Gu L, Fiddes JC. Biopolymers. 2000;55:88–98. doi: 10.1002/1097-0282(2000)55:1<88::AID-BIP80>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Doherty T, Waring AJ, Hong M. Biochemistry. 2008;47:1105–1116. doi: 10.1021/bi701390t. [DOI] [PubMed] [Google Scholar]
- 27.Hessa T, White SH, von Heijne G. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 28.Freites JA, Tobias DJ, von Heijne G, White SH. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munowitz MG, Griffin RG, Bodenhausen G, Huang TH. J. Am. Chem. Soc. 1981;103:2529–2533. [Google Scholar]
- 30.Hong M, Gross JD, Rienstra CM, Griffin RG, Kumashiro KK, Schmidt-Rohr K. J. Magn. Reson. 1997;129:85–92. doi: 10.1006/jmre.1997.1242. [DOI] [PubMed] [Google Scholar]
- 31.Huster D, Yamaguchi S, Hong M. J. Am. Chem. Soc. 2000;122:11320–11327. [Google Scholar]
- 32.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. J. Chem. Phys. 1999;110:7983–7992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.