Abstract
Nanoparticles are regarded as promising transfection reagents for effective and safe delivery of nucleic acids into specific type of cells or tissues providing an alternative manipulation/therapy strategy to viral gene delivery. However, the current process of searching novel delivery materials is limited due to conventional low-throughput and time-consuming multistep synthetic approaches. Additionally, conventional approaches are frequently accompanied with unpredictability and continual optimization refinements, impeding flexible generation of material diversity creating a major obstacle to achieving high transfection performance. Here we have demonstrated a rapid developmental pathway toward highly efficient gene delivery systems by leveraging the powers of a supramolecular synthetic approach and a custom-designed digital microreactor. Using the digital microreactor, broad structural/functional diversity can be programmed into a library of DNA-encapsulated supramolecular nanoparticles (DNA⊂SNPs) by systematically altering the mixing ratios of molecular building blocks and a DNA plasmid. In vitro transfection studies with DNA⊂SNPs library identified the DNA⊂SNPs with the highest gene transfection efficiency, which can be attributed to cooperative effects of structures and surface chemistry of DNA⊂SNPs. We envision such a rapid developmental pathway can be adopted for generating nanoparticle-based vectors for delivery of a variety of loads.
Keywords: supramolecular nanoparticle, gene delivery, digital microreactor, combinaltorial library, cyclodextrin, molecular recognition
Introduction
Gene delivery constitutes one of the most critical steps in gene manipulation and therapy.1, 2 By mimicking the size and function of viral vectors, numerous non-viral gene delivery systems based on biocompatible nanostructured materials,3–5 e.g., inorganic nanoparticles,6–9 carbon nanotubes,10 liposomes,11 cationic polymers12, 13 and dendrimers,14 have been developed to provide an alternative approach to the problems in viral gene delivery. However, an emerging challenge of nanoparticle-based delivery reagents is their low transfection performance.3 In order to overcome this problem researchers have attempted to modulate15 the structural (sizes16 and shapes) and functional (surfaces chemistry and charges) properties of nanoparticles,15 often requiring multiple optimization cycles to gradually improve the performance of nanoparticle-based delivery reagents. Such procedures have proven to be time-intensive and have demonstrated limited diversity over the past decades. Alternatively, it is believed that exploration of a new developmental pathway capable of rapid and parallel programming of a combinatorial library of nanoparticle-based delivery systems could lead to revolutionary breakthroughs in nanoparticle-based delivery systems.
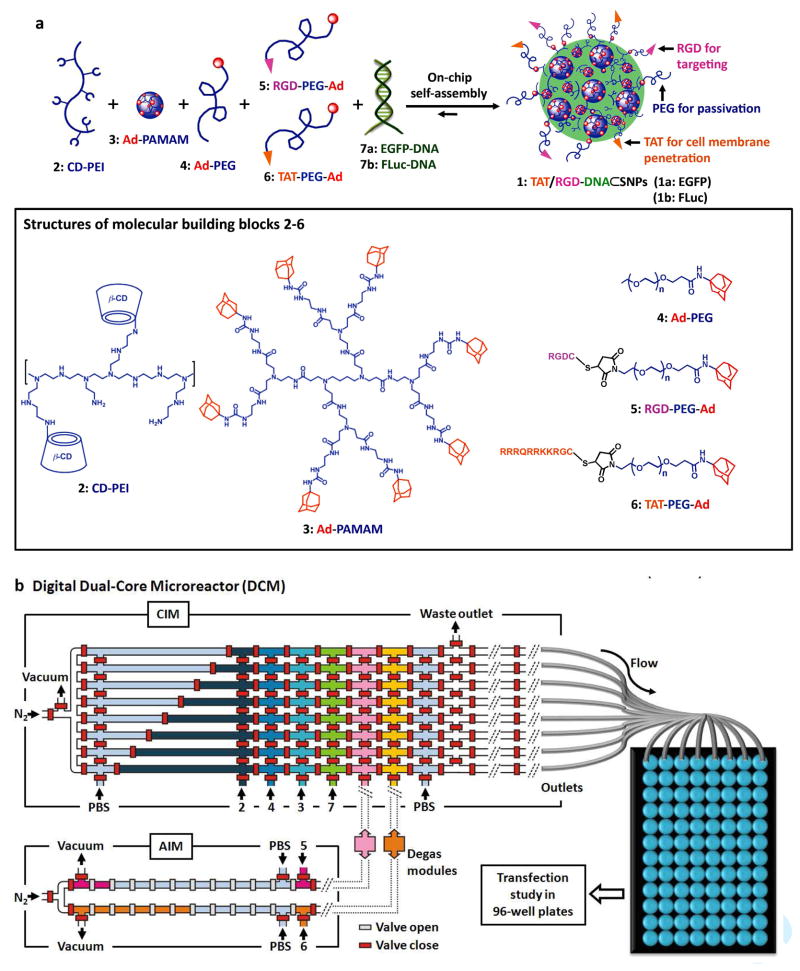
Herein, we describe a rapid developmental pathway (Figure 1) that leverages the powers of (i) a combinatorial synthetic approach (Figure 1a) based on supramolecular assembly17–19 and (ii) a digital microreactor20, 21 (Figure 1b–d), towards the generation of a highly efficient nanoparticle-based gene delivery system. Unlike the slow, multistep syntheses employed for producing existing gene-delivery materials,3 our supramolecular method (Figure 1a) enables a convenient, flexible, and modular method22 for generating a combinatorial library of DNA⊂SNPs, in which a broad structural/functional diversity covering the size variation, surface chemistry and DNA loading capacity was programmed into individual DNA⊂SNPs by systematically altering the mixing ratios of five functional molecular building blocks (2–6), DNA plasmid (7a: enhanced green fluorescent protein (EGFP), and 7b: firefly luciferase (FLuc)). In order to reduce human operational errors, accelerate handling procedures, enhance experimental fidelity, and achieve economical use of reagents, a digital Dual-Core Microreactor (DCM, Figure 1b–d) was designed and implemented to allow automated sampling, dilution, metering and mixing of 2–7, resulting in a combinatorial library composed of 648 different DNA⊂SNPs within 2.5 h. The structural/functional diversity of the DNA⊂SNPs library can be translated into diversity in performance by conducting transfection studies of individual DNA⊂SNPs in 96 well plates containing mouse fibroblast cells. A small group of DNA⊂SNPs that facilitates high levels of delivery performance was identified. We then carried out comprehensive characterizations on these DNA⊂SNPs revealing that improved transfection performance can be attributed to the defined size, surface chemistry, zeta potential, uniformity and dynamic stability. Compared to the leading gene transfection reagents, such as lipofectamine 2000 and RGD-jet-PEI, the identified 40-nm TAT/RGD-DNA⊂SNPs (1) with defined surface chemistry (RGD:23–26 a αvβ3 binding peptide and TAT27–29 a cell-penetrating peptide) exhibited significantly improved gene transfection efficiency and low toxicity in a number of cancer cell lines and fibroblast cells.
Figure 1.
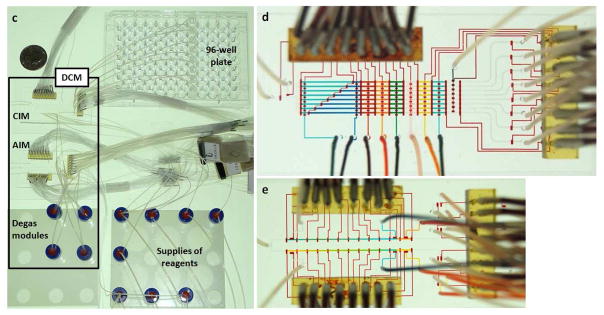
A rapid developmental pathway that leverages the powers of (i) a combinatorial synthetic approach based on supramolecular assembly and (ii) an automated microreactor has been demonstrated for screening and production of a highly efficient nanoparticle-based gene delivery system. a) Graphical schematic representations of the self-assembly approach for producing a combinatorial library of DNA encapsulated supramolecular nanoparticles (DNA⊂SNPs), in which a broad structural/functional diversity can be programmed into individual DNA⊂SNPs (1) by systematically altering the mixing ratios of the five functional molecular building blocks i.e., CD-PEI (2), Ad-PAMAM(3), Ad-PEG (4), RGD-PEG-Ad (5) and TAT-PEG-Ad (6), as well as DNA plasmid (7a: enhanced green fluorescent protein (EGFP) and 7b: firefly luciferase (FLuc)). b) Graphical illustration of a digital Dual-Core Microreactor (DCM). The device settings are composed of a central integrated mixer (CIM) and an Auxiliary Integrated Mixer (AIM), and degas modules bridges. The operation of the circuit was computer controlled using color-coded pressure-driven valves: red – positive pressure, off/on; yellow – peristaltic pumping; green – vacuum. Eight slugs containing eight different formulated DNA encapsulated DNA⊂SNPs were generated in parallel and transferred to a 96-well plate through PTFE tubes. The gene transfection performance for each combination was evaluated by a plate reader. c) The picture of the whole set of actual instrument. d and e) Optical images of the CIM and AIM, respectively. The various channels were loaded with dyes to visualize the different components.
Results and Discussion
The five molecular building blocks (Figure 1a) – CD-PEI (2), Ad-PAMAM (3), Ad-PEG (4), RGD-PEG-Ad (5) and TAT-PEG-Ad (6) were prepared according to literatures,17, 22 otherwise see Supporting Information. The functions of the five molecular building blocks are summarized below. First, the complementary CD-PEI (2) and Ad-PAMAM (3) are responsible for constructing cationic hydrogel networks that can encapsulate anionic DNA (7) forming the cores of DNA⊂SNPs. Therefore, the DNA loading capacity of DNA⊂SNPs is dependent on the net positive charges embedded in the hydrogel networks. By using electrophoresis analysis and ethidium bromide exclusion assay,22 the nitrogen/phosphate (N/P) ratio above five was determined. Second, Ad-PEG (4) serves as a capping/solvation reagent that not only constrains continuous growth of the DNA-encapsulated hydrogel networks, but also confers desired water solubility, structural stability, and passivation performance to the resulting DNA⊂SNPs. Third, the two functional ligands (5 and 6) can be incorporated onto the surfaces of DNA⊂SNPs via dynamic exchange in order to enable delivery specificity (to recognize a certain population of cells with αvβ3-integrin receptors) and cell transfusion capability (to foster internalization through membrane) of the resulting DNA⊂SNPs, respectively. By systematically altering the mixing ratios among the five molecular building blocks (2–6) and DNA (7), distinct structural/functional properties (i.e., sizes and surface chemistry) can be programmed into individual DNA⊂SNPs in the combinatorial library. It is important to point out that, in contrast to the lipid-like gene delivery system where the diversity was built upon the binary combination of a plethora of molecular precursors,30, 31 the diversity of our DNA⊂SNPs library was generated by ratiometric combination of only the five molecular building blocks and DNA loading.
It is feasible to manually prepare the DNA⊂SNPs library by pipetting and mixing individual molecular building blocks and DNA plasmid at different ratios, while potential operation errors, slow handling speed, and a significant amount of sample consumption might compromise the throughput, fidelity and efficiency of the study. Digital microfluidic reactors32–35 are promising platforms to overcome these challenges. In our case, an oil-free, digital DCM (Figure 1b–d) composed of (i) a Central Integrated Mixer (CIM) and (ii) an Auxiliary Integrated Mixer (AIM), was designed and implemented to systematically program the structural/functional diversity into the DNA⊂SNPs libraries.
As illustrated in Figure 1b, there are eight parallel microchannels in CIM, capable of parallel generation of eight slugs with different mixing ratios of 2–7. Each microchannel was partitioned by hydraulic-actuated microvalves into nine confined plug regions, which were assigned for precise metering of five molecular building blocks (2–6), a DNA plasmid (7a: EGFP or 7b: FLuc) and phosphate buffered saline (PBS). AIM incorporates a pair of fractionally partitioned microchannels, capable of synchronized supply of functional ligands (5 and 6) at different concentrations. CIM and AIM were coupled together by a pair of degas modules (Supporting Information) that remove gas through liquid transferring processes.
By using the digital DCM, the automated preparation of DNA⊂SNPs library was achieved by systematically modulating the mixing ratios of CD-PEI (2), RGD-PEG-Ad (5) and TAT-PEG-Ad (6) against fixed amounts of Ad-PAMAM (3), Ad-PEG (4) and plasmid DNA (7). In CIM, constant concentrations of 3 (3.0 μM), 4 (60.0 μM) and 7 (100 ng/μL) were assigned to fill into plug regions (ca. 20 nL per region). To adjust the quantities of CD-PEI (2), the eight uneven partitioned plug regions on the west side of CIM were utilized to accommodate different volumes of 2 (4.0 μM, 10–80 nL). The concentration modulation of the two functional ligands (5 and 6) was accomplished in AIM, where each fractionally partitioned microchannel is responsible for ratiometric mixing of the respective ligand (5: 0–9.6 μM and 6: 0–12.0 μM) with PBS. In each operation cycle, CIM generate eight 200-nL slugs containing different precursor mixtures, which were introduced into 96-well plates by nitrogen through eight poly(tetrafluoroethylene) (PTFE) tubes attached at the east side of CIM. In order to produce sufficient quantity of DNA⊂SNPs for subsequent transfection studies, 50 cycles were performed continuously in CIM within 90 s, affording eight different DNA⊂SNP solutions (ca. 10 μL each, 100 ng DNA). (See the supporting movie clip and Supporting Information). The resulting DNA⊂SNPs libraries were incubated in the 96-well plates for 20 min at room temperature prior to transferring to the 96-well plates containing NIH 3T3 cells (ca. 8000 cells/well) for transfection studies in parallel. The gene transfection efficiency was evaluated by a plate reader after culturing the cells at 37°C (5% CO2) for 24 h.
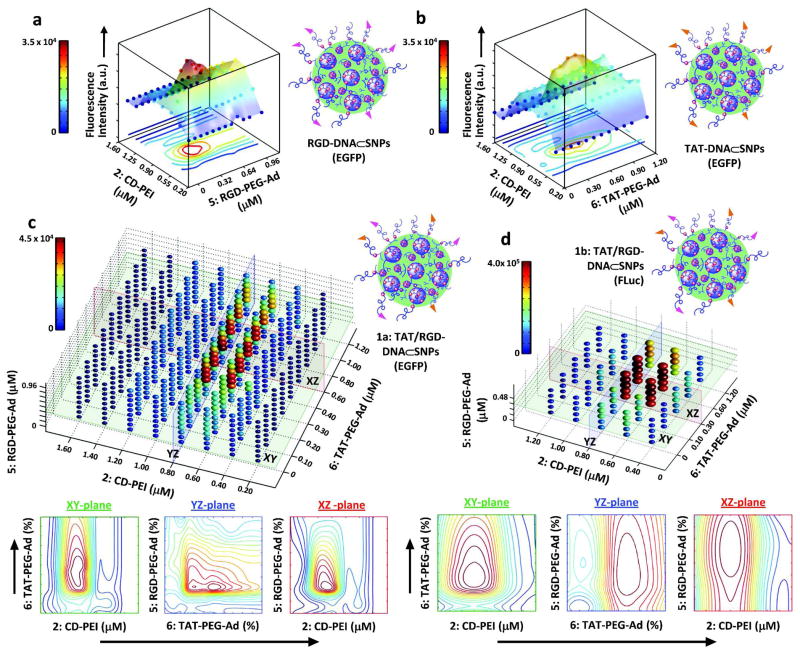
In proof-of-concept trials a CMV promoter-driven EGFP-encoded plasmid DNA was encapsulated into the DNA⊂SNPs. Before a full-scale screening with three variables (i.e., CD-PEI (2), RGD-PEG-Ad (5) and TAT-PEG-Ad (6)), simplified studies on two pairs of variables (i.e., CD-PEI (2) vs. RGD-PEG-Ad (5) and CD-PEI (2) vs. TAT-PEG-Ad (6)) were conducted in search of the optimal transfection performance of RGD-DNA⊂SNPs and TAT-DNA⊂SNPs, respectively. The transfection outcomes (96 data points in each case) were fitted into two 3-dimensional (3D) profiles (Figure 2a and b). The full-scale screening was accomplished by systematically programming the three variables, i.e., 8 different concentrations for CD-PEI (2), 9 for RGD-PEG-Ad (5) and 9 for TAT-PEG-Ad (6). Using the digital DCM, a combinatory library composed of 648 different DNA⊂SNPs was generated within 2.5 h to fill up seven 96-well plates. To ensure the operation fidelity, all the experiments were conducted in triplicate. A 4D gene expression plot was employed to summarize the results from the full-scale screening of the combinatorial library revealing that the optimal transfection performance of DNA⊂SNPs was achieved at CD-PEI (2) concentration of 0.6–0.8 μM, a RGD-PEG-Ad (5) of 0.24–0.48 μM and TAT-PEG-Ad (6) of 0.4–0.8 μM. We also note the transfection performance of the TAT/RGD-DNA⊂SNPs (1a) is significantly improved compared with those observed for RGD-DNA⊂SNPs and TAT-DNA⊂SNPs. To validate the general applicability of this developmental pathway we performed a relatively smaller screening to identify DNA⊂SNPs capable of highly efficient delivery of FLuc-encoded plasmid DNA. We manipulated three variables (5 × 5 × 5 concentrations) resulting in a combinatorial library of 125 different TAT/RGD-DNA⊂SNPs (1b). The consistent results (Figure 2d) were observed for the transfection performance of TAT/RGD-DNA⊂SNPs (1b) for delivering FLuc-DNA plasmid. All four sets of screening studies suggested that the TAT/RGD-DNA⊂SNPs (1) composed of CD-PEI (2, 0.7 ± 0.1 μM), RGD-PEG-Ad (5, 0.28 ± 0.04 μM) and TAT-PEG-Ad (6, 0.60 ± 0.20 μM) show the best transfection performance.
Figure 2.
a) A 3D profile of gene transfection performance of RGD-DNA⊂SNPs (EGFP) with variation of CD-PEI (2) and RGD coverage (96 data points). b) A 3D profile of gene transfection performance of TAT-DNA⊂SNPs (EGFP) with variation of CD-PEI (2) and TAT coverage (96 data points). c) A 4D profile of gene transfection performance of TAT/RGD-DNA⊂SNPs (EGFP) with variation of CD-PEI (2), TAT and RGD coverages (648 data points). The XY, YZ and XZ plates across the best performance were simplified and denoted 2D contour images. d) A 4D profile of gene transfection performance of TAT/RGD-DNA⊂SNPs (FLuc) with variation of CD-PEI (2), TAT and RGD coverages (125 data points). The XY, YZ and XZ plates across the best performance were simplified and denoted 2D contour images.
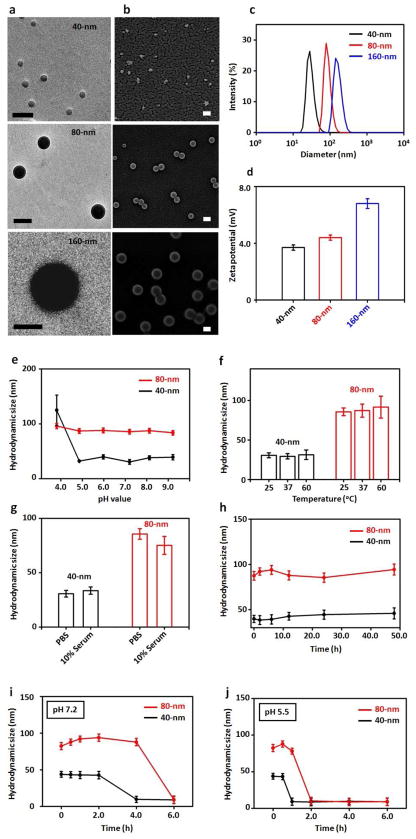
To understand how the identified synthetic variables contribute to the optimal transfection performance we scaled up the production of TAT/RGD-DNA⊂SNPs (1a) under three sets of synthetic variables for characterization at three CD-PEI (2) concentrations (0.6, 0.8 and 1.2 μM) while keeping concentrations of the RGD-PEG-Ad (0.28 μM) and TAT-PEG-Ad (0.60 μM) constant. We realize that CD-PEI (2) concentration affects the cross-linking degrees of the CD-PEI/Ad-PAMAM hydrogel cores in TAT/RGD-DNA⊂SNPs (1), leading to the size variation. Both transmission electron microscopy (TEM, Figure 3a) and scanning electron microscopy (SEM, Figure 3b) were employed to examine the morphology and sizes of the resulting TAT/RGD-DNA⊂SNPs (1). Three distinct sizes of 42 ± 4, 86 ± 9 and 160 ± 13 nm were observed for those produced at CD-PEI (2) concentrations of 0.6, 0.8 and 1.2 μM, respectively. Interestingly, we also noticed that the size distributions of the DCM-produced DNA⊂SNPs are much narrower than those prepared manually (Tabe 1), and these results were confirmed by dynamic light scattering (Zetasizer Nano, Malvern Instruments Ltd) measurements (Figure 3c). We attribute the greater control upon DNA⊂SNP size distribution to the precision and reproducibility of the sampling, metering and mixing processes in the digital DCM. In addition, no significant size variations were observed by introducing different concentrations ranging from 0 to 1.2 μM of ligands (5 and/or 6). We also characterized the surface potentials of these TAT/RGD-DNA⊂SNPs (Figure 3d), which spanned a range of 3.7±0.2–6.8±0.3 mV. In addition, the actual surface coverages of RGD-PEG-Ad (5) and TAT-PEG-Ad (6) were estimated by measuring absorption intensity of a FITC-labeled analog (i.e., FITC-PEG-Ad)22 on the DNA⊂SNPs prepared by the respective synthetic parameters (See Supporting Information). The results indicated that 0.28 and 0.60 μM of RGD-PEG-Ad (5) and TAT-PEG-Ad (6) reflect 5 and 9 % of surface coverage on the resulting DNA⊂SNPs, respectively. In short, TAT/RGD-DNA⊂SNPs (1) with sizes of 40 and 80 nm, as well as 5% RGD-PEG-Ad (5) and 9% TAT-PEG-Ad (6) coverages22 exhibited optimal cell transfection performance. Finally, in order to understand the dynamic stability of 40- and 80-nm TAT/RGD-DNA⊂SNPs (1) we employed real-time DLS measurements to monitor their size variation (i) at different pH values (pH 3.8–9.2) and temperatures (25, 37 and 60°C), (ii) in the presence of 10% serum, and (iii) at different storage times in presence of physiological salt concentrations (Figure 3e–h). The stability of the TAT/RGD-DNA⊂SNPs (1) can be attributed to the multivalent CD/Ad recognition and electrostatic interactions between Ad-PAMAM/CD-PEI hydrogel and DNA. In order to study the role of Ad-PEG (4) for dynamic stability of DNA⊂SNPs we monitored the size variation of DNA⊂SNPs (Figure 3i and j) by removing the excess amount of Ad-PEG (4) in the mixture through membrane dialysis (MWCO 10kD) under pH 5.5 and 7.2. It was found that the 40- and 80-nm TAT/RGD-DNA⊂SNPs (1) were disassembled gradually within 6 h at both pH conditions. In addition, the TAT/RGD-DNA⊂SNPs (1) exhibited faster disassembly behavior at pH 5.5 (< 2 h) than at pH 7.2, which allows the TAT/RGD-DNA⊂SNPs (1) efficiently degraded inside the endosomes and released the DNA intracelluarly.36
Figure 3.
a) and b) TEM and SEM images of the resulting TAT/RGD-DNA⊂SNPs with different sizes of 42 ± 4, 86 ± 9 and 160 ± 13 nm. Scale bars: 100 nm. c) Dynamic light scattering (DLS) was employed to measure DNA⊂SNPs hydrodynamic sizes in PBS buffer. d) Zeta potentials variations of DNA⊂SNPs in PBS buffer. Stability studies of 40- and 80-nm DNA⊂SNPs under different conditions: e) pH-dependent size variations of DNA⊂SNPs in the respective buffer solutions with pH values ranging from 3.8 to 9.2. Error bars are obtained from three measurements. f) Temperature-dependent size variation of DNA⊂SNPs in PBS buffer (pH = 7.2) with different temperatures at 25, 37 and 60 °C. g) Size variations of DNA⊂SNPs in presence and absence of 10% serum containing PBS buffer after 4 h incubation. Error bars are obtained from three measurements. h) Time-dependent size variation of DNA⊂SNPs from 0–48 h in PBS (pH 7.2). i and j) Time-dependent dynamic stability of DNA⊂SNPs under dialysis at pH 5.5 and 7.2, respectively.
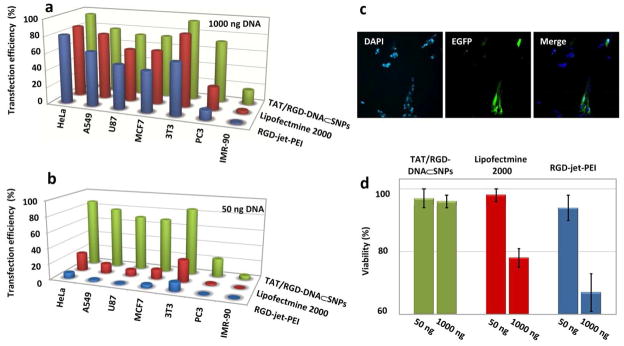
To compare the transfection performance of the optimal 40-nm TAT/RGD-DNA⊂SNPs (1) with the leading transfection reagents (i.e., RGD-jet-PEI and lipofectamine 2000) we carried out the dose-dependent gene transfection studies in 24-well plates with encapsulated-DNA (50 and 1000 ng per well) by using a collection of cells, including NIH 3T3 (Mouse fibroblast cell line), HeLa (human cervix epithelial carcinoma cell line), A549 (human lung cancer cell line), U87 (human glioblastoma cell line), MCF7 (human breast adenocarcinoma cell line), PC3 (human prostate cancer cell line), and IMR-90 (human fibroblast cell line). The gene transfection studies results (Figure 4a and b) indicated that the 40-nm TAT/RGD-DNA⊂SNPs (1) exhibited significantly improved transfection performance compared to those observed for RGD-jet-PEI and lipofectamine 2000 across the different cancer and fibroblast cell lines at high dosage of DNA (1000 ng DNA per well). Higher than 70% transfection efficiencies were observed across various cancer cell lines we tested, even for the PC3 cell line that is difficult to be transfected by those two commercial reagents. IMR-90, one of the hard-to-transfect human fibroblast cell lines, was tested and the transfection efficiency of DNA⊂SNPs toward IMR-90 cells reached to 17±7 %, which can be visualized by the fluorescence micrographs (Figure 4c). The transfection efficiency of 40-nm TAT/RGD-DNA⊂SNPs (1) for IMR-90 cells is 11-fold and 5-fold higher than that of RGD-jet-PEI and lipofectamine 2000, respectively. It is important to note that 40-nm TAT/RGD-DNA⊂SNPs (1) still achieved high transfection efficiency at low dosage of DNA (50 ng per well), indicating the outstanding gene delivery ability of DNA⊂SNPs. Moreover, the cell viability assay results indicated that the 40-nm TAT/RGD-DNA⊂SNPs (1) exhibited negligible cytotoxicity. More than 96 ± 3 % of DNA⊂SNPs-transfected cells showed normal viability even in the presence of high dosage of reagent (1000 ng DNA per well). In contrast, at high DNA dose more than 34% and 22% NIH 3T3 cells died after treatment by RGD-jet-PEI and lipofectamine 2000 for 48 h (Figure 4d).
Figure 4.
a) and b) Transfection efficiencies of the optimal 40-nm TAT/RGD-DNA⊂SNPs at high (1000 ng DNA per well) and low (50 ng DNA per well) DNA dosage along with control delivery reagents (lipofectamine 2000 and RGD-jet-PEI) for a collection of cell lines, including NIH 3T3 (Mouse fibroblast cell line), HeLa (human cervix epithelial carcinoma cell line), A549 (human lung cancer cell line), U87 (human glioblastoma cell line), MCF7 (human breast adenocarcinoma cell line), PC3 (human prostate cancer cell line) and IMR-90 (human fibroblast cell line) in 24-well plates. c) The representative fluorescence micrographs of IMR-90 cells transfected by 40-nm TAT/RGD-DNA⊂SNPs at high dosage of DNA (1000 ng DNA/well). d) Cytotoxicity of RGD-jet-PEI, lipofectamine 2000 and 40-nm TAT/RGD-DNA⊂SNPs at high and low dosage of DNA transfected NIH 3T3 cells determined by cell viability assay after 48 h of transfection.
Experimental Section
General
Reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO) and used as received without further purification or otherwise noted. Branched polyethylenimine (PEI, MW = 10 kD) was purchased from Polysciences Inc (Washington, PA). Polymers contain primary, secondary and tertiary amine groups in approximately 25/50/25 ratio. 1st-generation polyamidoamine dendrimer (PAMAM) with 1, 4-diaminobutane core and amine terminals in 20% wt methanol solution was purchased from Dendritic Nanotechnologies, Inc (Mount pleasant, MI). 1-Adamantanamine (Ad) hydrochloride and β-cyclodextrin (β-CD) were purchased from TCI America (San Francisco, CA). N-hydroxysuccinimide (SCM) and maleimido (MAL) hetero-functionalized polyethylene glycol (SCM-PEG-MAL, MW = 5 kD) was obtained from NANOCS Inc (New York, NY). Phosphate-buffered saline (PBS, 1X, pH 7.2 ± 0.05) for sample preparation. 6-Mono-tosyl-β-cyclodextrin (6-OTs-β-CD) was prepared following the literature reported method.37 Octa-Ad-grafted polyamidoamine dendrimer (Ad-PAMAM), CD-grafted branched polyethylenimine (CD-PEI) and Ad-grafted polyethylene glycol (Ad-PEG) were prepared as the method we reported before.17 Dry CH2Cl2 was obtained by refluxing over CaH2 and freshly distilled before use. NIH 3T3 (mouse embryonic fibroblast cell line), HeLa (human cervix epithelial carcinoma cell line), A549 (human lung cancer cell line), U87 (human glioblastoma cell line), MCF7 (human breast adenocarcinoma cell line), PC3 (human prostate cancer cell line) and IMR-90 (human primary fibroblasts cell line) were purchased from American Type Culture Collection (ATCC). The Dulbecco’s Modified Eagle Medium (DMEM), Earl’s Modified Eagle’s Medium (EMEM) growth medium, RPMI-1640, Opti-MEM reduced serum medium and Penicillin/streptomycin were obtained from Invitrogen (Carlsbad, CA). Fetal Bovine Serum (FBS) and EGFP-encoded plasmid DNA (pMAX EGFP®, 4.3 kb) and luciferase-encoded plasmid DNA (FLuc-DNA, 6.6 kb) were obtained from Lonza Walkerrsville Inc (Walkerrsville, MD). 4′, 6-Diamidino-2-phenylindole (DAPI) was purchased from Invitrogen (Carlsbad, CA). RGD (RGDC) and TAT peptides (CGRKKRRQRRR) were purchased from GenScript Corp. (Piscataway, NJ).
1H NMR spectra were recorded on a Bruker Avance 400 spectrometer in deuterated solvents. Mass spectra were acquired using an Applied Biosystems Voyager DE-STR MALDI-TOF mass spectrometer (Framingham, MA). Dynamic light scattering and zeta potentials of DNA⊂SNPs (1) were measured on Zetasizer Nano instrument (Malvern Instruments Ltd., United Kingdom). Transmission electron microscope (TEM) images were measured on Philips CM 120 electron microscope operating with an acceleration voltage of 120 kV. SEM images of DNA⊂SNPs (1) were obtained with a JEOL JSM-6700F SEM. Freeze-dried samples on a silicon surface were sputter-coated with gold before measurement. Cell imaging and gene transfection studies were performed on a Nikon TE2000S inverted fluorescent microscope with a CCD camera (Photomatrix, Cascade II), X-Cite 120 Mercury lamp, automatic stage, and filters for three fluorescent channels (W1 (DAPI), W2 (EGFP and AO) and W3 (PI)).
The fabrication and operations of Digital Dural Core Microreactor (DCM)
The fabrication and operations of Digital Dural Core Microreactor (DCM) are described in the Supporting Information.
Programming supramolecular nanoparticles in a digital dual-core microreactor
By using the digital DCM, two 3D and two 4D screenings were performed sequentially. In Figure 2a, 96 different RGD-DNA⊂SNPs were generated by altering concentrations of CD-PEI (0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4 and 1.6 μM) and RGD-Ad-PEG (0, 0.08, 0.16, 0.24, 0.32, 0.40, 0.48, 0.56, 0.64, 0.72, 0.80, 0.88 and 0.96 μM). In Figure 2b, 96 different TAT-DNA⊂SNPs were generated by altering concentrations of CD-PEI (0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4 and 1.6 μM) and TAT-Ad-PEG (0, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 1.00, 1.10 and 1.20 μM). In Figure 2c, 648 different TAT/RGD-DNA⊂SNPs (1a) were generated by altering concentrations of CD-PEI (0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4 and 1.6 μM), TAT-Ad-PEG (0, 0.10, 0.20, 0.30, 0.40, 0.60, 0.80, 1.00, and 1.20 μM) and RGD-Ad-PEG (0, 0.08, 0.16, 0.24, 0.32, 0.48, 0.64, 0.80, and 0.96 μM). In Figure 2d, 125 different TAT/RGD-DNA⊂SNPs (1b) were generated by altering concentrations of CD-PEI (0.4, 0.6, 0.8, 1.0 and 1.2 μM), TAT-Ad-PEG (0, 0.10, 0.30, 0.60 and 1.20 μM) and RGD-Ad-PEG (0, 0.16, 0.24, 0.32 and 0.48 μM).
Dynamic light scattering (DLS)
DLS experiments were performed with a Zetasizer Nano instrument (Malvern Instruments Ltd., United Kingdom) equipped with a 10-mW helium-neon laser (λ = 632.8 nm) and thermoelectric temperature controller. Measurements were taken at a 90° scattering angle. The sizes and the standard derivations of assembled TAT/RGD-DNA⊂SNPs (1) were calculated by averaging the values of at least three measurements.
Transmission electron microscope (TEM)
The morphology and sizes of TAT/RGD-DNA⊂SNPs were directly examined using transmission electron microscope. The studies were carried out on a Philips CM 120 electron microscope, operating at an acceleration voltage of 120 kV. The TEM samples were prepared by drop-coating 2 μL of TAT/RGD-DNA⊂SNPs solutions onto carbon-coated copper grids. Excess amounts of slugs were removed by filter papers after 45 s. Subsequently, the surface-deposited TAT/RGD-DNA⊂SNPs were negatively stained with 2% uranyl acetate for 45 s before the TEM studies.
Scanning electron microscope (SEM)
We prepared the TAT/RGD-DNA⊂SNPs samples for SEM observation by a standard procedure: Briefly, 2 μL of TAT/RGD-DNA⊂SNPs solution was drop-coated onto silica substrate and freeze-dry by dry ice. Then, the samples were sputter coated with gold before examination with a Hitachi S800 field emission SEM at an accelerating voltage of 10 keV.
Zeta potential measurements
Zeta potentials of TAT/RGD-DNA⊂SNPs (1) were determined by photon correlation spectroscopy using a Zetasizer Nano instrument, (Malvern Instruments, Malvern, Worcestershire, UK). The measurements were performed at 25°C with a detection angle of 90°, and the raw data were subsequently correlated to Z average mean size using a cumulative analysis by the Zetasizer software package.
Stability studies of 40 and 80-nm TAT/RGD-DNA⊂SNPs
Stability of 40 and 80-nm TAT/RGD-DNA⊂SNPs in different pH values
Information on the stability of TAT/RGD-DNA⊂SNPs in variable pH environments is needed to understand its characteristics under physiological conditions and to provide a reasonable pH range for chemical modification of TAT/RGD-DNA⊂SNPs for further experiments. The pH-dependent stability of TAT/RGD-DNA⊂SNPs as a function of pH was tested at a range from 3.8 to 9.2 by DLS analysis. The stock solutions of 40- and 80-nm TAT/RGD-DNA⊂SNPs were prepared in PBS buffer containing monobasic potassium phosphate (I = 1.5 mM), sodium chloride (I = 155 mM), and dibasic sodium phosphate (I = 2.7 mM). The following buffers were used to adjust the pH values of the TAT/RGD-DNA⊂SNPs samples in the stock solution: 100 mM HCl-KCl buffer (pH 0–2.0), 100 mM Glycine-HCl buffer (pH 2.2–3.6), 100 mM CH3COOH-CH3COONa buffer (pH 3.7–5.6), 100 mM Na2HPO4-NaH2PO4 buffer (pH 5.8–8.0), 100 mM Tris-HCl buffer (pH 7.0–9.0) and 100 mM Na2CO3-NaHCO3 buffer (pH 9.2–10.8). Typically, a 100-μL portion of TAT/RGD-DNA⊂SNPs stock solution was mixed with a 900-μL portion of different pH value buffer solutions (I = 100 mM). The resulting solutions were shaken and equilibrated until the pH stabilized. The final pH values were determined by pH meter.
Stability of 40 and 80-nm TAT/RGD-DNA⊂SNPs under different temperatures
To understand the thermal stability of the TAT/RGD-DNA⊂SNPs we employed real-time DLS measurements to monitor the size variation of both of the 40- and 80-nm TAT/RGD-DNA⊂SNPs in PBS at 25, 37 and 60 °C. In each case the samples were equilibrated under a specific temperature for 20 min prior to data acquisitions.
Stability of 40 and 80-nm TAT/RGD-DNA⊂SNPs in 10% Serum
To understand the stability of the TAT/RGD-DNA⊂SNPs in presence of serum we employed DLS measurements to observe the size variation of both of the 40- and 80-nm TAT/RGD-DNA⊂SNPs in the mixture of serum and PBS (1:9, V/V). The samples were kept at room temperature for 4h prior to data acquisitions.
Stability of 40 and 80-nm TAT/RGD-DNA⊂SNPs under a physiological ionic strength
To ensure the in vivo stability of the TAT/RGD-DNA⊂SNPs it is critical to examine the size variation of them under a physiological ionic strength. The 40- and 80-nm TAT/RGD-DNA⊂SNPs were prepared in PBS solutions (pH = 7.2). We employed real-time DLS measurements to monitor the size variation of both of the 40- and 80-nm TAT/RGD-DNA⊂SNPs at different times. The TAT/RGD-DNA⊂SNPs sizes were recorded for 48 h.
Cell culture
NIH 3T3, HeLa, A549, U87 and IMR-90 cell lines were routinely maintained in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). MCF7 was cultured in EMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. PC3 was cultured in RPMI-1640 containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
Gene transfection studies
Cells (5×104 cells/well) were plated in 24-well plates and allowed to adhere overnight. EGFP-encoded DNA was diluted in 1× TE buffer. The 40-nm TAT/RGD-DNA⊂SNPs were prepared on DCM and incubated for 20 min at room temperature before transfection experiments. The 40-nm TAT/RGD-DNA⊂SNPs in PBS (10 μL) was diluted with 100 μL Opti-MEM medium and subsequently transferred to each well. For the control groups, RGD-jet-PEI and lipofectamine 2000 were used as a standard transfection reagent and operated according to the protocol provided by the manufacturers. TAT/RGD-DNA⊂SNPs along with controls were incubated with the cells for 4 hours then removed by aspirating, and replaced with 500 μL/well of fresh culture media. Cells were allowed to grow for 24 h at 37°C and 5% CO2 and then fixed (4% paraformaldehyde for 15 min at room temperature), then washed with PBS three times, stained with DAPI, and finally rinsed with PBS prior to EGFP expression analysis by fluorescence microscope.
Conclusion
In conclusion, we have demonstrated a rapid developmental pathway toward generation of a highly efficient gene delivery system by leveraging the powers of a supramolecular synthetic approach and a custom-designed digital microreactor. This pathway can be adopted for the development of nanoparticle-based vectors capable of delivering a variety of loads, such as gene, drugs, proteins and their mixtures. We are currently exploring the use of the DNA⊂SNP-based transfection reagents for reprogramming of human primary fibroblast cells in order to generate induced pluripotent stem cells that is crucial in the field of regulative medicine.38
Supplementary Material
Table 1.
Comparison of TAT/RGD-DNA⊂SNPs synthesized by DCM and conventional pipetting
| DCM | Pipetting22 | |
|---|---|---|
| Throughput | 250 conditions/h | <20 conditions/h |
| Size distribution | Excellent (PDIa: <0.05) | Good (PDIa: 0.10~0.20) |
| Operation error | No | Possible |
| Reproducibility | High | Modest |
PDI: The polydispersity index obtained from DLS measurements.
Acknowledgments
This research was supported by NIH-NCI NanoSystems Biology Cancer Center (U54CA119347), R21 grant (EB008419–01) and California Institute of Regenerative Medicine (RT1-01022-1).
Footnotes
Supporting Information Available: Synthesis of TAT-PEG-Ad, DCM setup and operation, microscope settings, imaging processing and data analysis and cell viability assay. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Clifton K.-F. Shen, Email: kshen@mednet.ucla.edu.
Hsian-Rong Tseng, Email: hrtseng@mednet.ucla.edu.
References
- 1.Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–84. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–82. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 4.Niemeyer CM. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew Chem Int Edit. 2001;40:4128–58. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 5.Ferrari M. Cancer nanotechnology: Opportunities and challenges. Nat Rev Cancer. 2005;5:161–71. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 6.De M, Ghosh PS, Rotello VM. Applications of Nanoparticles in Biology. Adv Mater. 2008;20:4225–41. [Google Scholar]
- 7.Nie SM, Xing Y, Kim GJ, Simons JW. Nanotechnology applications in cancer. Annu Rev Biomed Eng. 2007;9:257–88. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 8.Torney F, Trewyn BG, Lin VSY, Wang K. Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat Nanotech. 2007;2:295–300. doi: 10.1038/nnano.2007.108. [DOI] [PubMed] [Google Scholar]
- 9.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–30. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Cai WB, He LN, Nakayama N, Chen K, Sun XM, Chen XY, Dai HJ. In vivo biodistribution and highly efficient tumour targeting of carbon nanotubes in mice. Nat Nanotech. 2007;2:47–52. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 11.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Del Rev. 2009;61:721–31. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu HJ, Wagner E. Bioresponsive polymers for nonviral gene delivery. Curr Opin in Mol Therapeutics. 2009;11:165–78. [PubMed] [Google Scholar]
- 13.Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8:526–33. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang WD, Selim KMK, Lee CH, Kang IK. Bioinspired application of dendrimers: From bio-mimicry to biomedical applications. Prog Polym Sci. 2009;34:1–23. [Google Scholar]
- 15.Mitragotri S, Lahann J. Physical approaches to biomaterial design. Nat Mater. 2009;8:15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Kim BYS, Rutka JT, Chan WCW. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotech. 2008;3:145–50. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Wang ST, Su H, Chen KJ, Armijo AL, Lin WY, Wang YJ, Sun J, Kamei K, Czernin J, et al. A Supramolecular Approach for Preparation of Size-Controlled Nanoparticles. Angew Chem Int Edit. 2009;48:4344–48. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303:1352–55. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 19.Klajn R, Olson MA, Wesson PJ, Fang L, Coskun A, Trabolsi A, Soh S, Stoddart JF, Grzybowski BA. Dynamic hook-and-eye nanoparticle sponges. Nat Chem. 2009;1:733–38. doi: 10.1038/nchem.432. [DOI] [PubMed] [Google Scholar]
- 20.Lee CC, Sui GD, Elizarov A, Shu CYJ, Shin YS, Dooley AN, Huang J, Daridon A, Wyatt P, Stout D, et al. Multistep synthesis of a radiolabeled imaging probe using integrated microfluidics. Science. 2005;310:1793–96. doi: 10.1126/science.1118919. [DOI] [PubMed] [Google Scholar]
- 21.Lin WY, Wang Y, Wang S, Tseng HR. Integrated microfluidic reactors. Nanotoday. 2009;4:470–81. doi: 10.1016/j.nantod.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Chen KJ, Wang S, Ohashi M, Kamei K-i, Sun J, Ha JH, Liu K, Tseng HR. A small library of DNA-encapsulated supramolecular nanoparticles for targeted gene delivery. Chem Commun. 2010;46:1851–53. doi: 10.1039/b923711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai WB, Chen XY. Nanoplatforms for targeted molecular imaging in living subjects. Small. 2007;3:1840–54. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa A, Zhou YM, Kambe N, Nakayama Y. Enhancement of Star Vector-Based Gene Delivery to Endothelial Cells by Addition of RGD-Peptide. Bioconj Chem. 2008;19:558–61. doi: 10.1021/bc700385r. [DOI] [PubMed] [Google Scholar]
- 25.Merkel OM, Germershaus O, Wada CK, Tarcha PJ, Merdan T, Kissel T. Integrin αvβ3 Targeted Gene Delivery Using RGD Peptidomimetic Conjugates with Copolymers of PEGylated Poly(ethylene imine) Bioconj Chem. 2009;20:1270–80. doi: 10.1021/bc9001695. [DOI] [PubMed] [Google Scholar]
- 26.Zuber G, Dontenwill M, Behr JP. Synthetic Viruslike Particles for Targeted Gene Delivery to αvβ3 Integrin-Presenting Endothelial Cells. Mol Pharm. 2009;6:1544–52. doi: 10.1021/mp900105q. [DOI] [PubMed] [Google Scholar]
- 27.Jung J, Solanki A, Memoli KA, Kamei K-i, Kim H, Drahl MA, Williams LJ, Tseng HR, Lee K. Selective Inhibition of Human Brain Tumor Cells through Multifunctional Quantum-Dot-Based siRNA Delivery 13. Angew Chem Int Ed. 2010;49:103–07. doi: 10.1002/anie.200905126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torchilin VP. Cell penetrating peptide-modified pharmaceutical nanocarriers for intracellular drug and gene delivery. Peptide Sci. 2008;90:604–10. doi: 10.1002/bip.20989. [DOI] [PubMed] [Google Scholar]
- 29.Roy R, Jerry DJ, Thayumanavan S. Virus-Inspired Approach to Nonviral Gene Delivery Vehicles. Biomacromolecules. 2009;10:2189–93. doi: 10.1021/bm900370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–59. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–69. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–73. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 33.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–84. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 34.Vincent ME, Liu W, Haney EB, Ismagilov RF. Microfluidic stochastic confinement enhances analysis of rare cells by isolating cells and creating high density environments for control of diffusible signals. Chem Soc Rev. 2010;39:974–84. doi: 10.1039/b917851a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marre S, Jensen KF. Synthesis of micro and nanostructures in microfluidic systems. Chem Soc Rev. 2010;39:1183–202. doi: 10.1039/b821324k. [DOI] [PubMed] [Google Scholar]
- 36.Overly CC, Lee KD, Berthiaume E, Hollenbeck PJ. Quantitative Measurement of Intraorganelle Ph in the Endosomal Lysosomal Pathway in Neurons by Using Ratiometric Imaging with Pyranine. Proc Natl Acad Sci U S A. 1995;92:3156–60. doi: 10.1073/pnas.92.8.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petter RC, Salek JS, Sikorski CT, Kumaravel G, Lin FT. Cooperative Binding by Aggregated Mono-6-(Alkylamino)-Beta-Cyclodextrins. J Am Chem Soc. 1990;112:3860–68. [Google Scholar]
- 38.Maherali N, Hochedlinger K. Guidelines and Techniques for the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.