Abstract
RNA splicing is required to remove introns from pre-mRNA and alternative splicing generates protein diversity. Topoisomerase I (Top1) has been shown to be coupled with splicing by regulating SR splicing proteins. Prior studies on isolated genes also showed that Top1 poisoning by camptothecin (CPT), which traps Top1 cleavage complexes (Top1cc), can alter RNA splicing. Here we tested the impact of Top1 inhibition on splicing at the genome-wide level in human colon carcinoma HCT116 and breast carcinoma MCF7 cells. The RNA of HCT116 cells treated with CPT for various times was analyzed with ExonHit Human Splice Array. Unlike to other exon array platforms, the ExonHit arrays include junction probes that allow the detection of splice variants with high sensitivity and specificity. We report that CPT treatment preferentially affects the splicing of splicing-related factors, such as RBM8A, and generates transcripts coding for inactive proteins lacking key functional domains. The splicing alterations induced by CPT are not observed with cisplatin or vinblastine, and are not simply due to reduced Top1 activity as TOP1 downregulation by siRNA did not alter splicing like CPT treatment. Inhibition of RNA polymerase II (Pol II) hyperphosphorylation by DRB blocked the splicing alteration induced by CPT, which suggests that the rapid Pol II hyperphosphorylation induced by CPT interferes with normal splicing. The preferential effect of CPT on genes encoding splicing factors may explain the abnormal splicing of a large number of genes in response to Top1cc.
Keywords: Topoisomerase I, splicing, camptothecin, transcription, RNA polymerase II
Introduction
Alternative splicing is observed with more than 95 % of the genes (1, 2) and constitutes the main source for protein diversity, allowing the generation of different proteins from a given pre-mRNA by the differential use of splice sites (3). Alternative splicing is produced by several mechanisms: exclusion/inclusion of exons, alternative 3′-splice sites, alternative 5′-splice sites, mutual exclusion of exons, unsplicing of an exon, multiple promoters, multiple poly(A) sites (Fig. 1C & D). Splicing takes place at the pre-mRNA level within spliceosomes; each of them containing around 300 polypeptides and five snRNP (small nuclear ribonucleoproteins) (2). In addition, serine/arginine-rich proteins (SR proteins) and heterogeneous nuclear ribonucleoproteins (hnRNP) are well-known splicing regulators (2). The SR proteins activate splicing by binding exonic splicing enhancers (ESE) whereas hnRNP repress splicing by binding exonic or intronic splicing silencers (ESS, ISS) (1, 2). In addition to the SR proteins and the hnRNP that are ubiquitously expressed, splicing regulators like CELF, Nova, nPTB, FOX1, FOX2, ESRP1, ESRP2 and nSR100 are tissue-specific (2).
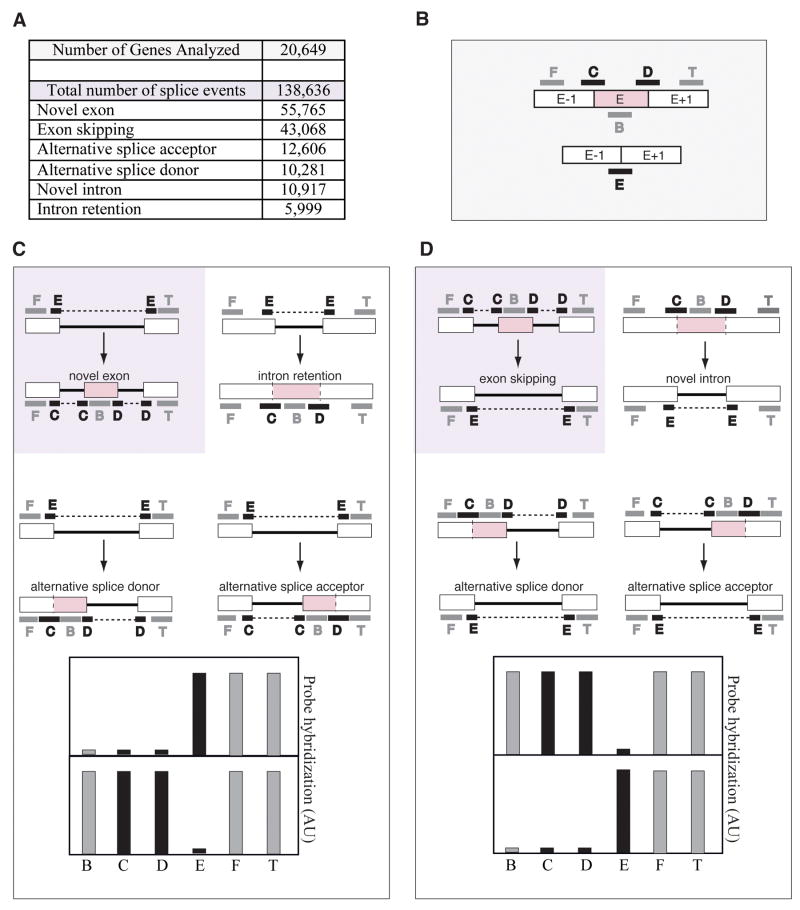
Figure 1.
Description of the ExonHit Array. A, Number of genes and spliced events in ExonHit array. B, Schematic representation of the position of the ExonHit probes for splicing events concerning exon E. C, Probe hybridization profile for novel exon and related events. D, Probe hybridization profile for exon skipping and related events.
Splicing consists of two transesterification reactions. First, the hydroxyl of the branch point site attacks the phosphate of the 5′-splice site, generating a free exon and an intermediate lariat. Second, the hydroxyl of the 5′-splice site attacks the phosphate of the 3′-splice site, generating the mature mRNA and a lariat of introns that will be finally eliminated. Over 70% of the splice events change the protein sequence, and 19% generate truncated proteins (4).
DNA topoisomerase I (Top1) is required to remove DNA superhelical tensions generated by DNA replication and transcription (5-8). During transcription, positive and negative DNA supercoilings are produced ahead of and behind the elongating RNA polymerase II (Pol II) complex, respectively (9). Top1 relaxes both positive and negative supercoilings by producing transient Top1 cleavage complexes (Top1cc), which are Top1-linked DNA single-strand breaks (7, 10). Several studies have suggested the implication of Top1 in splicing. First, Rossi et al. showed that Top1 can phosphorylate the SR splicing proteins (11, 12), and two domains of Top1 were implicated in this activity: one as an ATP binding site in the carboxy-terminal region of Top1 and the other as a binding site for SF2/ASF and a protein kinase domain in the amino-terminal region of Top1 (12). More recently, this kinase activity of Top1 has been confirmed (13) and Top1 has been proposed to shift from its classical DNA relaxation activity to its kinase activity after binding the SR-splicing factors (14, 15). SF2/ASF can interact by its two RRM (RNA Recognition Motif) domains with Top1, and inhibit DNA relaxation by the enzyme (16). Top1 is also important in the regulation of gene expression by its preferential association with transcriptionally active regions (17-19) and by controlling promoter activity independently from its DNA relaxing activities (20, 21).
Top1 is the target of the plant alkaloid camptothecin (CPT), and its clinical derivatives, topotecan and irinotecan are widely used as anticancer agents (7). CPT and its derivatives are non-competitive, reversible specific Top1 inhibitors that prevent DNA religation due to the trapping (poisoning) of Top1cc (7, 10). Poisoning of Top1 also occurs under normal conditions when the DNA template contains damaged bases including abasic sites, mismatches, oxidized bases or carcinogenic adducts (22, 23). The induction of Top1cc by CPT has been shown to affect transcription in several ways. RNA elongation is rapidly arrested by Top1cc (24) with reduction of Pol II density at promoter pausing sites (19), activation of low abundance antisense RNAs (17) and rapid hyperphosphorylation of Pol II in response to CPT treatment (25). Pol II hyperphosphorylation is rapidly reversible upon CPT removal or CDK inhibition by 5,6-dichloro-1-ß-D-ribobenzimidazole (DRB) (25). The induction of Top1cc by CPT has been shown to impact RNA splicing, but published studies have only been done on some specific genes (26-29).
Here, we tested the implication of Top1cc in splicing at the global genome level in human carcinoma cells to determine whether Top1 inhibition selectively affects particular families of genes. For this purpose, we used ExonHit arrays. Unlike arrays that contain only exon probes, the ExonHit arrays also contain junction probes, which allows the detection and quantitation of novel splice variants (30).
Materials and Methods
Chemicals and cells
Camptothecin, cisplatin and vinblastine were obtained from Sigma-Aldrich (St. Louis, MO). Human HCT116 and MCF7 cell lines were obtained from ATCC (Rockville, MD) and grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini Bio-products, West Sacramento, CA) at 37°C in 95% air and 5% CO2.
Western blotting and antibodies
Western blotting was performed according to standard protocols (31). The C21 Top1 mouse monoclonal antibody was a kind gift from Dr Yung-Chi Cheng (Yale University, New Haven, CT). The other primary antibodies used were anti-Pol II (sc-899; Santa Cruz Biotechnology, Santa Cruz, CA), anti-PS5-Pol II (ab5131; Abcam, Cambridge, MA) and anti-GAPDH (14C10; Cell Signaling, Danvers, MA).
Short interfering RNA (siRNA)
For TOP1 down-regulation, cells were transfected with an siRNA duplex (Qiagen, Valencia, CA) against the sequence AAGGACTCCATCAGATACTAT from the TOP1 mRNA. A negative control siRNA duplex was obtained from Qiagen (target DNA sequence: AATTCTCCGAACGTGTCACGT). Cells were seeded in 6-well plates, at a density of 150,000 cells per well 16 hours before transfection (31).
RT-PCR
Cells were washed in PBS. RNA extraction was performed with the “Nucleospin RNA II” kit (Macherey-Nagel, Bethlehem, PA). The “OneStep RT-PCR” kit (Qiagen) was used under the following conditions: 1× buffer, 400 μM of each dNTP, 0.6 μM of each primer, 2 μL of enzyme mix, and 1 μg of template RNA in a total volume of 50 μL. RNA was reverse-transcribed for 30 min at 50°C, the initial PCR step was activated by heating for 15 min at 95°C before PCR (1 min denaturation at 94°C, 1 min annealing at 60°C, 1 min extension at 72°C, 32 cycles) using a MJ Research PTC-200 Thermo Cycler (MJ Research, Reno, NV). The PCR products were analysed on agarose gels. The amplified DNA fragments were stained with ethidium bromide and fluorescence was detected by video camera imaging using the Quantity One software (BIO-RAD, Hercules, CA). Primer sequences are listed below in Supplemental Table 1.
Probeset Annotations and Data Normalization
ExonHit probe sequences were aligned to known variants in the SpliceMiner database (32). Exons were numbered consecutively according to genomic position, accounting for strand. Background subtraction and quantile normalization of the probes was done using Partek statistical software (Partek Inc., St. Louis, MO). The median polish step was done using R. A probe is designated “body” or “junction” depending on whether it interrogates a known exon or an exon-exon boundary. A junction probe is designated “J1” if it interrogates consecutive exons and “J2” if it interrogates a junction which skips one or more intervening exons.
Principal Component Analysis
The study design consisted of 2 controls (c4h, c20h) collected at 4 and 20 hours, and 5 camptothecin (10 μM) treated samples (1h, 2h, 4h, 15h and 20h) collected at 1, 2, 4 15 and 20 hours, respectively. The RMA intensity values for both the body and junction probes were subjected to Principal Component Analysis. A bi-plot of PC1 vs. PC2, accounting for 69% of the total variability, revealed 3 clusters comprised of control samples (c4h, c20h), early treated (1h, 2h and 4h) and late treated samples (15h, 20h).
Statistical model
The ExonSVD analysis modifies the traditional 3-way ANOVA model with a new parameter for probeset responsiveness. Unlike the ANOVA model which is prone to give false positive signals for alternative splicing in the presence of “dead” or unresponsive probes, the ExonSVD model (y ijk = μ + A′i Dk + Eik + βj(i) + Ck + εijk) includes an explicit parameter, D, for probeset responsiveness. Also, the ExonSVD model can directly handle body and junction probes directly without further elaboration. The body probes primarily determine the differential expression while the junction probes which are sensitive to the varying levels of exon-exon junctions, are the primary source of information regarding alternative splicing. The E effect of the model (alternative splicing effect) is determined as the residual after differential expression effects are accounted for, and is tested for significance to determine whether alternative splicing has occurred. This new model alleviates the need for pre-filtering to eliminate dead and unresponsive probesets. The p-values for E were calculated using the F-distribution where the degrees of freedom, which depend on the number of exons, were determined by fitting rational polynomials to the expected sum-of-squares in a numerical simulation.
Significance Index and Event Type
We developed a “significance index” calculated as the negative log10(p-value) for alternative splicing plus two times the maximum absolute value of the E term, for each gene. Using this significance index, one can order the entire set of genes according to evidence for alternative splicing.
For the genes with evidence of alternative splicing, we define events of four types: Exon gained early, Exon lost early, Exon gained late, Exon lost late, in comparison to the control condition. For each interior exon, we interrogate the junctions between it and its preceding (J1-1st) or its following exon (J1-2nd), and the junction joining the preceding to the following exon (J2). In some cases, the J2 probe was not available. In others, other J1 probes were not available, so a determination of the event could not be made. An event of certain type was declared if corresponding residuals from the ExonSVD model satisfied a set of conditions involving the signs of differences of Eik. E.g. for “exon loss early”, [E(early,J2) - E(c, J2)] > 0, [E(early,J1-1st) - E(c, J1-1st)] <0 and [E(early,J1-2nd) - E(c, J1-2nd)] < 0. We also required that the magnitude of one of the differences be greater than 2-fold. Not all of the significant genes contained one of these 4 types of events. Some had multiple events, some had both gain and loss of different exons.
High-Throughput GoMiner (HTGM)
HTGM leverages the Gene Ontology (GO) to identify “biological processes” represented in a list of genes. High-Throughput GoMiner (HTGM) (33) was used here, is an enhancement of GoMiner that efficiently processes an arbitrary number of such gene lists. The gene lists obtained from AnovaSVD were sorted in decreasing order with respect to the significance index. We submitted the top 998 genes for HTGM analysis. The HTGM parameters are listed in Supplemental Table 2.
A GO category is considered to be enriched if the number of changed genes that HTGM assigned to it is greater than the number expected by chance. The enrichment of a category is considered to be statistically significant if its false discovery rate (FDR) is less than or equal to a given threshold (typically 0.10). See (33) for details.
Genesis Clustering Program
Clustered image maps (CIMs) were produced in our studies by the Genesis program (33). We chose the Euclidean distance metric and average linkage for hierarchical clustering. Large generic categories were removed from all CIMs to facilitate visualization.
Results
Genome-wide analysis of splicing alterations induced by Top1 poisoning
The ExonHit array allows the analysis of 138,636 splice events among 20,649 genes. The probes are designed to mostly recognize two kind of splicing events: exon skipping and novel exon (Fig. 1A). The array uses a combination of exon and junction probes that are targeted inside exon sequences (probes F, T, B) and at junctions between two neighboring exons (probes C, D, E), respectively (Fig. 1B). A novel exon (exon gain) is characterized by an increase of the B, C, D probes and a decrease of the E probe (Fig. 1C). A skipped exon (exon loss) produces an increase of the E probe and a decrease of the B, C, D probes (Fig. 1D).
To study the impact of Top1 poisoning on splicing, we purified total RNA from human colon carcinoma HCT116 cells treated with 10 μM CPT for 1, 2, 4, 15 and 20 hours (Fig. 2A), and performed ExonHit array analysis for each sample. Controls samples were analyzed at 4 and 20 h following DMSO treatment (0.1%; the solvent used to dissolve CPT).
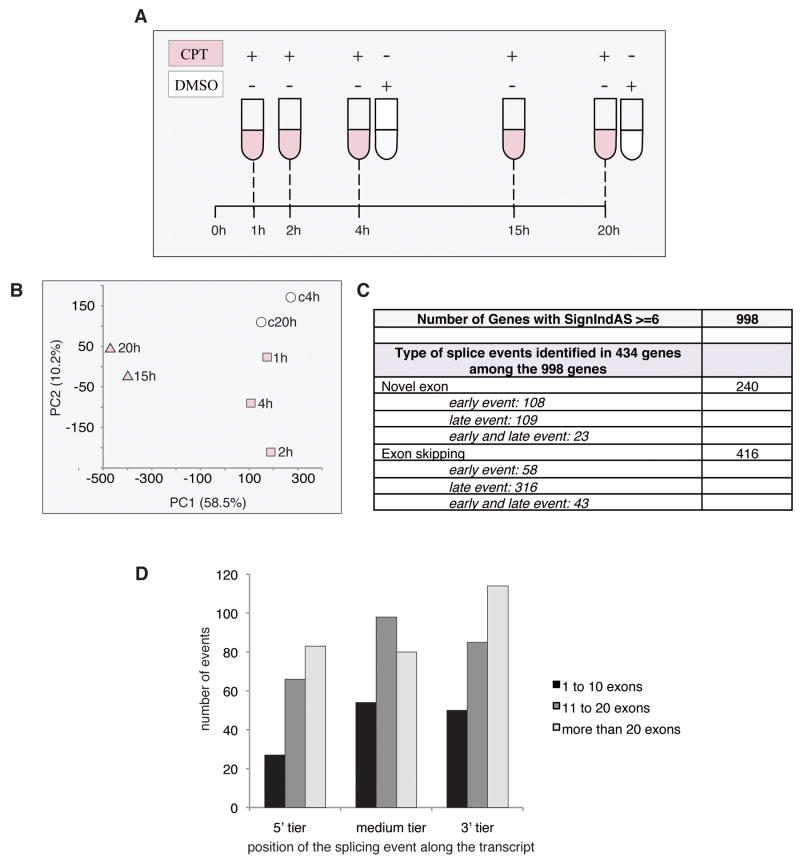
Figure 2.
Overall analysis of the splice events induced by CPT. A, Experimental design. Human colon carcinoma HCT116 cells were treated with camptothecin (CPT) at 10 μM during 1, 2, 4, 15 and 20 h. Controls samples received vehicle alone (0.1 % DMSO, 4 h and 20 h). B, PCA organization of the results, shown in principal component space, shows grouping: (control: c4h, c20h, white circle), (early: 1h, 2h, 4h, gray square), (late: 15h, 20h, gray triangle). C, Organization of splice events by types and as early or late events. D, Position of the splicing event along transcripts depending of the length of the transcripts. The black bars represent transcripts containing 1 to 10 exons, the dark gray bars the transcripts containing 11 to 20 exons and the light gray bars the transcripts containing more than 20 exons. The data presented on panels C and D have been obtained using the genes having a significance index (SI) of 6 or more, i.e. 998 genes.
The data were analyzed using a new ExonSVD model (see Materials and Methods) (all the data are in GEO; accession number “GSE23677”; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23677). Figure 2B shows the principal component analysis (PCA) organization of the results, with the controls, the early CPT treatment time points (1, 2 and 4 h) and the later time points (15 and 20 h) grouping separately. This demonstrates a time-dependent effect on splicing and a correlation between splicing alterations and CPT treatment. Nine hundred ninety eight genes among all the genes analyzed (around 5%) had a significance index of 6 or greater, indicative of a statistically significant and/or large changes in splicing in response to CPT (Supplemental Table 3). The type of alternative splicing event could be determined for 50% of the genes (434 genes) and corresponded to novel exon or exon skipping (Fig. 2C). For the other 50% of the genes, determination was not possible due to the complexity of the apparent event or because the array did not probe the relevant splice junctions. The appearance of novel exon was both an early as well as a late event (Fig. 2C). On the other hand, exon skipping, in most cases, was only detectable at the late time treatments with CPT (15 or 20 h) (Fig. 2C). Next, we wanted to know whether the splice events occurred in specific regions of the transcript. We divided the 434 genes with splicing alterations into three groups according to the number of exons contained in each gene (based on the publicly available GenBank information) [1 to 10 (103 genes), 11 to 20 (164 genes), more than 20 exons (167 genes)]. Splice events occurred all along the transcript and tended to increase with the length of the transcripts (Fig. 2D). Multiple events on a same transcript concerned 22% of the short transcripts, 35% of the medium transcripts and 45% of the long transcripts. Together these results suggest that splicing alteration occurred both on short and long transcripts but tended to increase with the length of the transcript.
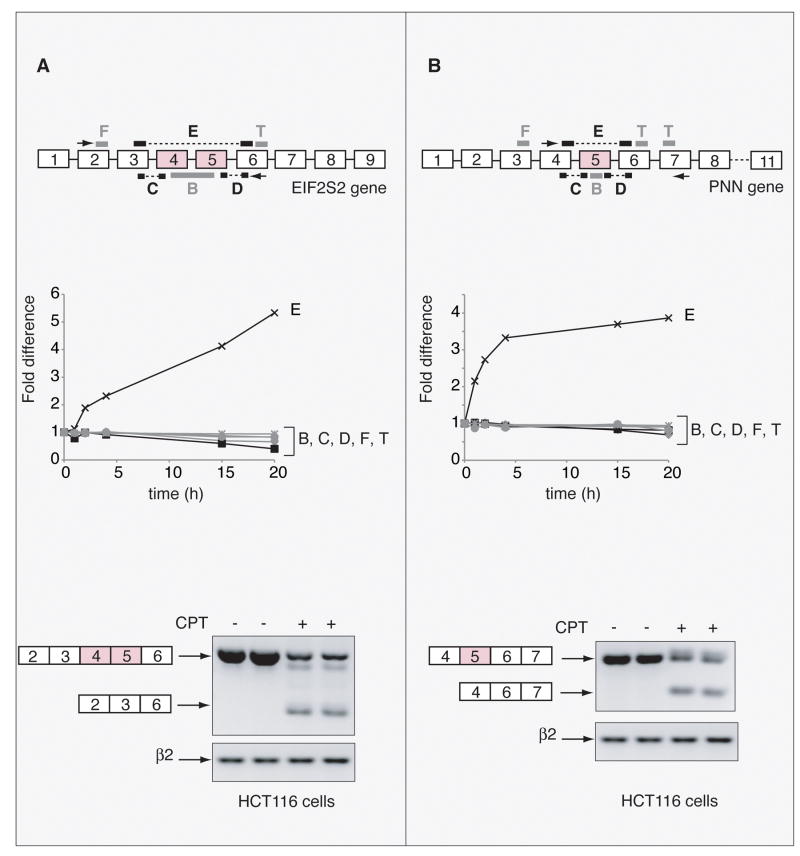
To validate the ExonHit genome-wide results, we took 12 genes which ranked near the top of the list of the 998 genes, according to significance index (Supplemental Table 4) and performed RT-PCR with gene-specific primers. The RT-PCR results were in agreement with the ExonHit results for more than 90% of the splice alterations analyzed (see Supplemental Table 4). Figure 3 showed two examples of exon skipping for the EIF2S2 and PNN genes. CPT induced the skipping of exons 4 and 5 in the EIF2S2 transcript. The ExonHit data showed an increase of the E probe (that links exon 3 to exon 6), and the RT-PCR showed the decrease of the long transcript (containing exons 4 and 5) following CPT treatment and the appearance of the short transcript (without exons 4 and 5). For the PNN transcript, CPT induced the skipping of exon 5 with an increase of the E probe (that links exon 4 to exon 6) in the ExonHit analysis. Accordingly, RT-PCR showed a decrease of the long transcript (containing exon 5) and the appearance of the short transcript (without exon 5) following CPT treatment. Exon gain was also validated in response to CPT for the caspase-2 gene (Supplemental Fig.1).
Figure 3.
Validation of the ExonHit results in HCT116 cells treated with CPT (10 μM, 20 h). A, Example of the EIF2S2 gene. Upper panel: schematic gene representation and position of the ExonHit probes (B, C, D, E, F, T) and of the primers used for the RT-PCR (black arrows). Middle panel: fold difference for each of the probes depending on CPT treatment normalized to the untreated controls. Lower panel: RT-PCR showing the effect of CPT on exons 4 and 5 exclusion. EIF2S2 mRNA was analyzed by RT-PCR using EIF2S2 E2s and EIF2S2 E6as primers. B, Example of the PNN gene. Upper panel: schematic gene representation and position of the ExonHit probes (B, C, D, E, F, T) and of the primers used for the RT-PCR (black arrows). Middle panel: fold difference for each of the probes depending on CPT treatment normalized to the untreated controls. Lower panel: RT-PCR showing the effect of CPT on exon 5 exclusion. PNN mRNA was analyzed by RT-PCR using PNN E4s and PNN E7as primers. Controls cells received vehicle alone. β2 microglobulin (β2) mRNA was used as a standardizing control.
Splicing alterations are enriched in genes coding for splicing factors
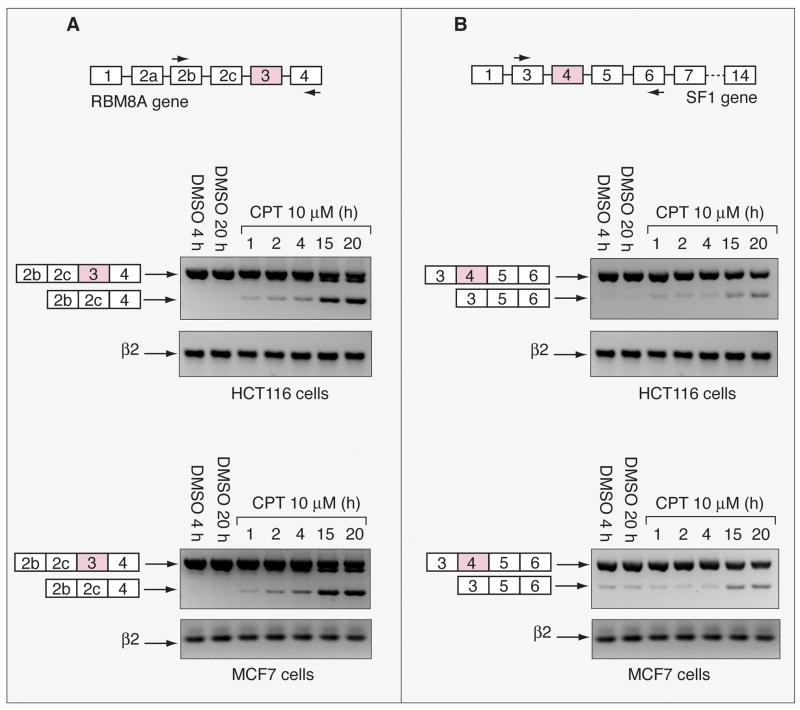
Next, we tested whether CPT affected the alternative splicing of specific gene families. The 998 differentially spliced genes were analyzed by GoMiner software (http://discover.nci.nih.gov/gominer/index.jsp). The categories of genes that tended to be preferentially affected are listed in Figure 4 and Table 1; and include those related to splicing, mitosis and methylation. Splicing categories were the most affected by CPT treatment. Individual splicing events were then further analyzed for the genes encoding splicing-related factors. The splicing alterations induced by CPT on the RBM8A, ZRANB2, BAT1 and SF1 genes were further examined (Supplemental Table 4 and Fig. 5). Kinetic experiments were performed for the RBM8A and SF1 genes not only in HCT116 cells but also in human breast carcinoma MCF7 cells treated with CPT. By RT-PCR the effect on splicing for RBM8A, which is the skipping of exon 3, was detected early, beginning 1 h after CPT treatment in both cell lines (Fig. 5A). Splicing of SF1, which corresponds to the skipping of exon 4, was detectable later, at 15 h CPT treatment (Fig. 5B). Together these results demonstrate preferential splicing alterations in the genes encoding splicing factors by Top1 poisoning.
Figure 4.

GoMiner categories demonstrate enrichment for the genes encoding splicing factors. HTGM categories versus genes CIM. The genes mapping to significantly enriched GO categories are indicated in red.
Table 1.
List of the best GoMiner categories. The table displays the name of the categories with lowest false discovery rate (FDR) concerning the genes having a significance index (SI) of 6 or more, i.e. 998 genes.
| Go categories | FDR | Number of spliced genes |
|---|---|---|
| GO:0006397_mRNA_processing | 0 | 30 |
| GO:0000398_nuclear_mRNA_splicing__via_spliceosome | 0 | 25 |
| GO:0000375_RNA_splicing__via_transesterification_reactions | 0 | 25 |
| GO:0016071_mRNA_metabolic_process | 0 | 30 |
| GO:0008380_RNA_splicing | 0 | 27 |
| GO:0022613_ribonucleoprotein_complex_biogenesis | 0 | 17 |
| GO:0007052_mitotic_spindle_organization | 0 | 6 |
| GO:0000278_mitotic_cell_cycle | 0 | 34 |
| GO:0000245_spliceosome_assembly | 0.01 | 8 |
| GO:0015850_organic_alcohol_transport | 0.01 | 4 |
| GO:0000279_M_phase | 0.01 | 23 |
| GO:0007051_spindle_organization | 0.01 | 7 |
| GO:0000280_nuclear_division | 0.01 | 17 |
| GO:0007067_mitosis | 0.01 | 17 |
| GO:0048285_organelle_fission | 0.02 | 17 |
| GO:0000087_M_phase_of_mitotic_cell_cycle | 0.02 | 17 |
| GO:0022618_ribonucleoprotein_complex_assembly | 0.03 | 9 |
| GO:0007049_cell_cycle | 0.05 | 46 |
| GO:0006281_DNA_repair | 0.05 | 21 |
| GO:0032259_methylation | 0.08 | 8 |
| GO:0043414_biopolymer_methylation | 0.08 | 8 |
| GO:0022402_cell_cycle_process | 0.08 | 36 |
| GO:0006730_one-carbon_metabolic_process | 0.09 | 8 |
| GO:0022403_cell_cycle_phase | 0.1 | 26 |
Figure 5.
Impacts of CPT on the splicing of splicing-related proteins in HCT116 and MCF7 cells. A, Alternative splicing of RBM8A pre-mRNA in response to CPT. Upper panel: schematic gene representation and position of the primers used for the RT-PCR (black arrows). Middle and lower panels: RT-PCR showing the effect of CPT on exon 3 exclusion. RBM8a mRNA was analyzed by RT-PCR using RBM8A E2s and RBM8A E4as primers. B, Alternative splicing of SF1 pre-mRNA in response to CPT. Upper panel: schematic gene representation and position of the primers used for the RT-PCR (black arrows). Middle and lower panels: RT-PCR showing the effect of CPT on exon 4 exclusion. SF1 mRNA was analyzed by RT-PCR using SF1 E3s and SF1 E6as primers. Controls cells received vehicle (DMSO) alone. β2 microglobulin (β2) mRNA was used as a standardizing control.
Pol II hyperphosphorylation is associated with Top1-induced splicing alterations
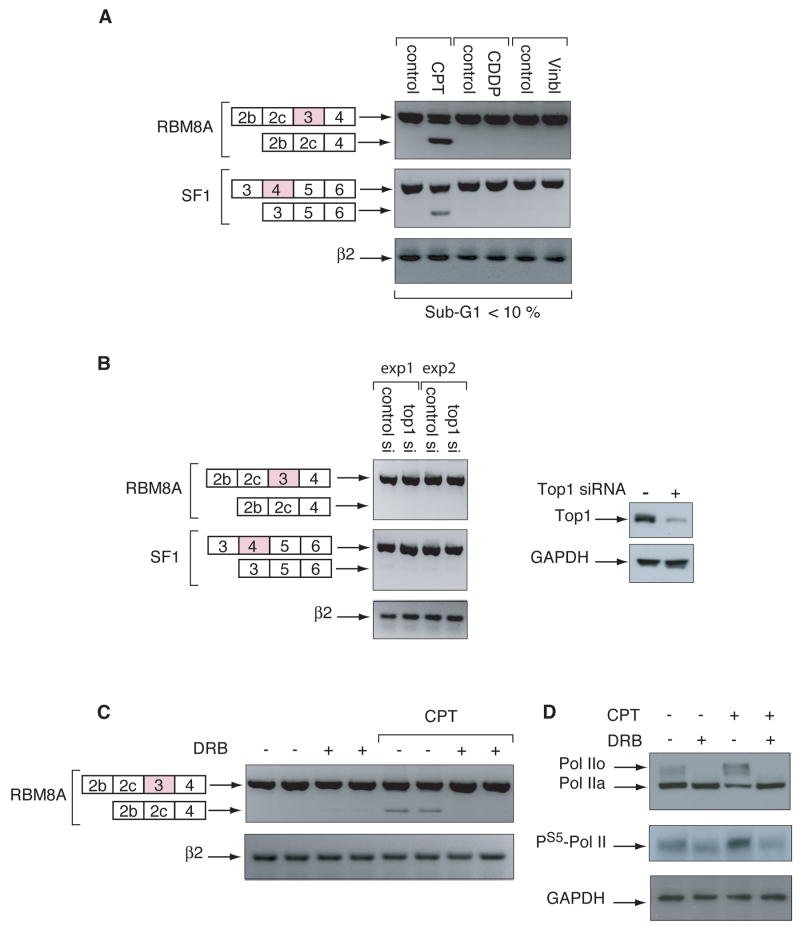
To determine the specificity of CPT in inducing altered splicing, other anticancer agents that do not target Top1 were tested. Neither cisplatin, a DNA alkylating drug, nor vinblastine, a mitotic spindle poison affected the splicing of RBM8A or SF1 (Fig. 6A). To test whether the effect of CPT was related to Top1cc or due to Top1 depletion by its sequestering in Top1cc, we down-regulated Top1 using an siRNA (Fig. 6B, right panel). Down-regulation of Top1 had no effect on alternative splicing of the RBM8A or SF1 genes (Fig. 6B, left panel), indicating that the generation of Top1cc rather than Top1 inactivation is necessary to induce splicing.
Figure 6.
CPT-induced alternative splicing is Top1cc-dependent and linked to Pol II hyperphosphorylation. A, HCT116 cells were treated with camptothecin (CPT, 10 μM, 20 h), cisplatin (CDDP, 50 μM, 20 h), or vinblastine (0.1 μM, 20 h), and RBM8A and SF1 were analyzed by RT-PCR using RBM8A E2s / RBM8A E4as and SF1 E3s / SF1 E6as, respectively. Controls cells received vehicle (DMSO) alone. β2 microglobulin (β2) mRNA was used as control. The percentage of cells in sub-G1 (37) is indicated at the bottom of the panel. B, Downregulation of Top1 does not induce the alternative splicing of RBM8A or SF1. HCT116 cells were transfected with a negative control siRNA or a siRNA against Top1. Left panel: RBM8A or SF1 mRNA were analyzed by RT-PCR using RBM8A E2s / RBM8A E4as and SF1 E3s / SF1 E6as respectively. β2 microglobulin (β2) mRNA was used as a standardizing control. exp: experiment. Right panel: Western blotting showing the efficiency of Top1 downregulation by siRNA. Top1 was analyzed by Western blotting. GAPDH was used as a loading control. C, The CDK inhibitor, 5, 6-Dichloro-β-D ribofuranosyl benzimidazole (DRB) suppresses the effect of CPT on alternative splicing. HCT116 cells were pretreated with DRB (100 μM, 1 h) prior treatment by CPT (10 μM, 2 h). RBM8A splicing was analyzed by RT-PCR using RBM8A E2s and RBM8A E4as primers. Controls cells received vehicle (DMSO) alone. β2 microglobulin (β2) mRNA was used as a standardizing control. D, DRB prevents the Pol II hyperphosphorylation induced by CPT. HCT116 cells were pretreated with DRB as in panel C. Pol II was analyzed by Western blotting. Pol IIa (hypophosphorylated) and Pol IIo (hyperphosphorylated) forms are indicated. GAPDH was used as a loading control.
Because Pol II hyperphosphorylation has recently been reported in response to CPT-induced Top1cc (25) and Pol II hyperphosphorylation has been implicated in alternative splicing (34, 35), we tested the relationship between CPT-induced Pol II hyperphosphorylation and splicing alterations. HCT116 cells were pretreated with 5,6-dichloro-β-D ribofuranosyl benzimidazole (DRB), an inhibitor of CDK (36), before exposing the cells to CPT. DRB pretreatment completely abrogated the CPT-induced splicing of RBM8A (Fig. 6C) and caspase-2 (Supplemental Fig. 2). Under these conditions, DRB abrogated CPT-induced Pol II hyperphosphorylation (Fig. 6D) (37). These results suggest a relationship between CPT-induced Pol II hyperphosphorylation and splicing alterations.
Discussion
This genome-wide analysis study shows that Top1cc alters the splicing of a large number of genes with a preference for the genes encoding splicing or splicing-related factors. Previous studies on the splicing effects of Top1 inhibitors focused on single genes (26-29, 38) and did not reveal the important contribution of the splicing factors genes as preferential splicing targets for Top1 poisons.
The ExonHit Human Splice Array with its junction probes enabled the detection of early splice alterations with higher sensitivity than arrays that display only exons probes (for instance the Affymetrix GeneChip Human Exon 1.0 ST array). To detect a new splice event, we find the junction probes to be more sensitive than the exon probes, allowing the detection of events with high sensitivity and selectivity (30). The use of exon probes is sufficient to compare expression levels of alternative transcripts in stable cell lines where transcripts are stably produced; however to study the changes in splicing following acute treatments, it is mostly the junction probes that allowed the detection of new variants because of the long half-life of most basal transcripts (median half-life ≈ 10 h) (39). Effectively, in the analysis of our ExonHit data, the junction probes were critical to identify the appearance of new splice events by the detection of change (mostly an increase) in E or C and D probes (see Figure 1).
Among the splicing factors that were abnormally spliced in response to CPT, three of them exemplify the functional impact of CPT treatment. RBM8A is a RNA binding motif (RBM) protein present in intermediate-containing spliceosomes. RBM8A is a component of a splicing-dependent multiprotein exon junction complex (EJC) deposited at splice junction on mRNA (40, 41). It influences downstream processes including nuclear mRNA export, subcellular mRNA localization, translation efficiency and nonsense mediated mRNA decay. The RBM8A polypeptide contains a 90-amino-acid RNA recognition motif (RRM) that binds single-stranded RNA and contains two highly conserved short sequences, RNP1 and RNP2 that are crucial for RNA binding (42). In response to CPT, the skipping of exon 3 removes the RNP2 domain resulting in a truncated RRM (without RNP2) that is predicted to inactivate RBM8A. Similarly, ZRANB2 (43), which interacts with the U2 small nuclear RNA auxiliary factor and SNRP70, has its zf-RanBP (zinc-finger Ran Binding Protein) domain missing due to the skipping of exon 4 after CPT treatment. We also found that BAT1 (44), a splicing factor required for the association of U2snRNP with pre-mRNA and mRNA export from the nucleus to the cytoplasm, has its DEAD domain truncated due to the skipping of exons 3 and 5 after CPT treatment. These results illustrate how CPT, by impacting on key functional domains of splicing factors that are important for their binding with RNA or other splicing-related factors, can inactivate these splicing factors and consequently affect in “trans” the splicing of other genes.
The exposure of HCT116 cells to 10 μM camptothecin during 20 hours does not generate significant amount of apoptosis. The level of apoptosis measured by Hoechst staining was below 10%. Moreover, we previously observed that pre-treatment, by the broad caspase inhibitor, Z-VAD-fmk, does not suppress the effect of camptothecin on the splicing of caspase-2 (29). Also, many of the changes observed at 20 hours were already readily detectable at early time (1 hour). Consequently, the changes observed concerning alternative splicing do not mainly reflect the cells going through the process of dying.
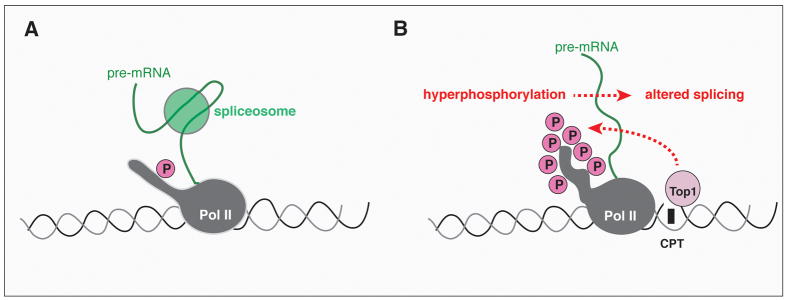
Top1 poisoning by CPT and anticancer drugs, abasic sites, base mismatches, oxidized bases, carcinogenic adducts and strand breaks can deplete Top1 activity by sequestering Top1 in the cleavage complexes (7, 22, 23). However, it is well-established that the anticancer cytotoxic activity of CPT is not due to depletion of Top1 activity but rather to the trapping of Top1cc and subsequent effects of DNA replication and transcription (8, 45). Similarly, our Top1 depletion experiments with siRNA show that the splicing effects of CPT are Top1cc-dependent but are probably unrelated to depletion of Top1 catalytic activity. We recently reported the rapid induction of Pol II hyperphosphorylation in response to CPT-induced Top1cc (25). In this study, we confirmed Pol II hyperphosphorylation after CPT treatment. Pretreatment with DRB, which inhibits CDK (25) and suppresses Pol II hyperphosphorylation (see Fig. 6) (25) abrogated the splicing effects of CPT. It is plausible that CPT alters splicing in response to Pol II hyperphosphorylation (see Fig. 7), which has been proposed to affect Pol II pausing and the selection of splicing sites (34). Similarly, Munoz et al. showed that UVC affects splicing by inducing Pol II hyperphosphorylation and slowing down Pol II elongation (35), which can favor the use a weak splice sites (34, 46). The poisoning of Top1cc by CPT can affect splicing by such a “kinetic coupling model” (47). However, it is also possible that hyperphosphorylation of the carboxy terminal domain (CTD) of Pol II affects its interaction with and recruitment of splicing factors as proposed in the “recruitment coupling model” (48, 49). In summary, transcription could regulate alternative splicing by modulation of Pol II elongation rates (kinetic coupling) (47) and by the association of splicing factors to the transcribing polymerase (recruitment coupling) (50). Our finding that the genes encoding splicing factors are preferentially altered can explain the effect of Top1cc on the splicing of a large variety of genes.
Figure 7.
Schematic representation of the Pol II and splicing interactions during normal conditions (A) or after Top1 poisoning (B).
Supplementary Material
Acknowledgments
This work was supported by the NIH Intramural Program, Center for Cancer Research, National Cancer Institute. We wish to thank Richard Einstein, ExonHit, Gaithersburg, Maryland for contributing the arrays and data analyses. We also wish to thank Dr. David Goldstein, Office and Science Technology Partnership, Center for Cancer Research, National Cancer Institute for support.
References
- 1.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–54. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corcos L, Solier S. Alternative mRNA splicing, pathology and molecular therapeutics. Med Sci (Paris) 2005;21:253–60. doi: 10.1051/medsci/2005213253. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–9. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 5.Champoux JJ. DNA topoisomerases: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 8.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–8. [PubMed] [Google Scholar]
- 11.Rossi F, Labourier E, Forne T, et al. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–2. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- 12.Tazi J, Rossi F, Labourier E, Gallouzi I, Brunel C, Antoine E. DNA topoisomerase I: customs officer at the border between DNA and RNA worlds? J Mol Med. 1997;75:786–800. doi: 10.1007/s001090050168. [DOI] [PubMed] [Google Scholar]
- 13.Soret J, Gabut M, Dupon C, et al. Altered serine/arginine-rich protein phosphorylation and exonic enhancer-dependent splicing in Mammalian cells lacking topoisomerase I. Cancer Res. 2003;63:8203–11. [PubMed] [Google Scholar]
- 14.Andersen FF, Tange TO, Sinnathamby T, et al. The RNA splicing factor ASF/SF2 inhibits human topoisomerase I mediated DNA relaxation. J Mol Biol. 2002;322:677–86. doi: 10.1016/s0022-2836(02)00815-x. [DOI] [PubMed] [Google Scholar]
- 15.Malanga M, Czubaty A, Girstun A, Staron K, Althaus FR. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J Biol Chem. 2008;283:19991–8. doi: 10.1074/jbc.M709495200. [DOI] [PubMed] [Google Scholar]
- 16.Kowalska-Loth B, Girstun A, Trzcinska AM, Piekielko-Witkowska A, Staron K. SF2/ASF protein binds to the cap region of human topoisomerase I through two RRM domains. Biochem Biophys Res Commun. 2005;331:398–403. doi: 10.1016/j.bbrc.2005.03.180. [DOI] [PubMed] [Google Scholar]
- 17.Baranello L, Bertozzi D, Fogli MV, Pommier Y, Capranico G. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1alpha gene locus. Nucleic Acids Res. 2010;38:159–71. doi: 10.1093/nar/gkp817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010 doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khobta A, Ferri F, Lotito L, Montecucco A, Rossi R, Capranico G. Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. J Mol Biol. 2006;357:127–38. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- 20.Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci U S A. 1993;90:11508–12. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–32. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 22.Pommier Y, Barcelo JM, Rao VA, et al. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dexheimer TS, Kozekova A, Rizzo CJ, Stone MP, Pommier Y. The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 2008;36:4128–36. doi: 10.1093/nar/gkn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai SD, Zhang H, Rodriguez-Bauman A, et al. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–50. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sordet O, Larochelle S, Nicolas E, et al. Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J Mol Biol. 2008;381:540–9. doi: 10.1016/j.jmb.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shkreta L, Froehlich U, Paquet ER, Toutant J, Elela SA, Chabot B. Anticancer drugs affect the alternative splicing of Bcl-x and other human apoptotic genes. Mol Cancer Ther. 2008;7:1398–409. doi: 10.1158/1535-7163.MCT-08-0192. [DOI] [PubMed] [Google Scholar]
- 27.Eisenreich A, Bogdanov VY, Zakrzewicz A, et al. Cdc2-like kinases and DNA topoisomerase I regulate alternative splicing of tissue factor in human endothelial cells. Circ Res. 2009;104:589–99. doi: 10.1161/CIRCRESAHA.108.183905. [DOI] [PubMed] [Google Scholar]
- 28.Solier S, De Cian MC, Bettaieb A, Desoche L, Solary E, Corcos L. PKC zeta controls DNA topoisomerase-dependent human caspase-2 pre-mRNA splicing. FEBS Lett. 2008;582:372–8. doi: 10.1016/j.febslet.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 29.Solier S, Lansiaux A, Logette E, et al. Topoisomerase I and II inhibitors control caspase-2 pre-messenger RNA splicing in human cells. Mol Cancer Res. 2004;2:53–61. [PubMed] [Google Scholar]
- 30.Fehlbaum P, Guihal C, Bracco L, Cochet O. A microarray configuration to quantify expression levels and relative abundance of splice variants. Nucleic Acids Res. 2005;33:e47. doi: 10.1093/nar/gni047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solier S, Sordet O, Kohn KW, Pommier Y. Death receptor-induced activation of the Chk2- and histone H2AX-associated DNA damage response pathways. Mol Cell Biol. 2009;29:68–82. doi: 10.1128/MCB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahn AB, Ryan MC, Liu H, Zeeberg BR, Jamison DC, Weinstein JN. SpliceMiner: a high-throughput database implementation of the NCBI Evidence Viewer for microarray splice variant analysis. BMC Bioinformatics. 2007;8:75. doi: 10.1186/1471-2105-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeeberg BR, Qin H, Narasimhan S, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marengo MS, Garcia-Blanco MA. Shedding UV light on alternative splicing. Cell. 2009;137:600–2. doi: 10.1016/j.cell.2009.04.054. [DOI] [PubMed] [Google Scholar]
- 35.Munoz MJ, Perez Santangelo MS, Paronetto MP, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–20. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–36. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 37.Sordet O, Goldman A, Redon C, Solier S, Rao VA, Pommier Y. Topoisomerase I requirement for death receptor-induced apoptotic nuclear fission. J Biol Chem. 2008;283:23200–8. doi: 10.1074/jbc.M801146200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilch B, Allemand E, Facompre M, et al. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001;61:6876–84. [PubMed] [Google Scholar]
- 39.Yang E, van Nimwegen E, Zavolan M, et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–72. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conklin DC, Rixon MW, Kuestner RE, Maurer MF, Whitmore TE, Millar RP. Cloning and gene expression of a novel human ribonucleoprotein. Biochim Biophys Acta. 2000;1492:465–9. doi: 10.1016/s0167-4781(00)00090-7. [DOI] [PubMed] [Google Scholar]
- 41.Salicioni AM, Xi M, Vanderveer LA, et al. Identification and structural analysis of human RBM8A and RBM8B: two highly conserved RNA-binding motif proteins that interact with OVCA1, a candidate tumor suppressor. Genomics. 2000;69:54–62. doi: 10.1006/geno.2000.6315. [DOI] [PubMed] [Google Scholar]
- 42.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 43.Adams DJ, van der Weyden L, Mayeda A, Stamm S, Morris BJ, Rasko JE. ZNF265--a novel spliceosomal protein able to induce alternative splicing. J Cell Biol. 2001;154:25–32. doi: 10.1083/jcb.200010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen J, Zhang L, Zhao R. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem. 2007;282:22544–50. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- 45.Capranico G, Marinello J, Baranello L. Dissecting the transcriptional functions of human DNA topoisomerase I by selective inhibitors: Implications for physiological and therapeutic modulation of enzyme activity. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbcan.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Munoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 47.de la Mata M, Alonso CR, Kadener S, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 49.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–22. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 50.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–80. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.