Abstract
The aim of this study was to investigate whether the VKORC1*3 (rs7294/9041 G > A), VKORC1*4 (rs17708472/6009 C > T), and CYP4F2 (rs2108622/1347 C > T) polymorphisms were associated with elevated warfarin maintenance dose requirements in patients with myocardial infarction (n = 105) from the Warfarin Aspirin Reinfarction Study (WARIS-II). We found significant associations between elevated warfarin dose requirements and VKORC1*3 and VKORC1*4 polymorphisms (P = .001 and P = .004, resp.), whereas CYP4F2 (1347 C > T) showed a weak association on higher warfarin dose requirements (P = .09). However, analysing these variant alleles in a regression analysis together with our previously reported data on VKORC1*2, CYP2C9*2 and CYP2C9*3 polymorphisms, gave no significant associations for neither VKORC1*3, VKORC1*4 nor CYP4F2 (1347 C > T). In conclusion, in patients with myocardial infarction, the individual contribution to warfarin dose requirements from VKORC1*3, VKORC1*4, and CYP4F2 (1347 C > T) polymorphisms was negligible. Our results indicate that pharmacogenetic testing for VKORC1*2, CYP2C9*2 and CYP2C9*3 is more informative regarding warfarin dose requirements than testing for VKORC1*3, VKORC1*4, and CYP4F2 (1347 C > T) polymorphisms.
1. Introduction
Warfarin has an established role in secondary prevention of atherothrombotic disease, reducing new thromboembolic events [1]. However, warfarin dosage is a challenge to clinicians because of its narrow therapeutic range and patient variability in obtained INR levels. An important issue is to improve individual warfarin dose prediction, especially in the initial phase of treatment, in order to avoid thrombosis and treatment-induced bleeding. An increasing number of genetic variations affecting warfarin pharmacokinetics and/or pharmacodynamics have recently been reported to have major impact on dosage requirements, that is, polymorphisms in the CYP2C9, VKORC1, and CYP4F2 genes [2–7]. In Caucasians, the VKORC1 gene is represented by three main haplotypes: VKORC1*2 (rs9934438/6484 C > T), VKORC1*3 (rs7294/9041 G > A), and VKORC1*4 (rs17708472/6009 C > T) [8]. Previously we have found VKORC1*2 to account for 24.5%, and the combination of CYP2C9*2 and CYP2C9*3 (rs1799853 and rs1057910) to account for 7.2% of the warfarin maintenance dose variation in patients from the Warfarin Aspirin Reinfarction Study (WARIS-II) [9]. In the present investigation, we aimed to study whether the high-dose polymorphisms VKORC1*3, VKORC1*4, and CYP4F2 (rs2108622/1347 C > T) were associated with elevation in warfarin maintenance dose requirements in the same patient group.
2. Methods
2.1. Patients
The patients in this study were a subgroup of the WARIS-II trial who had been assigned to three different antithrombotic regimens after acute myocardial infarction [10]. All patients provided written informed consent before participation in the study. The patients were randomly assigned to treatment with either a daily dose of 160 mg aspirin, warfarin (target INR 2.8–4.2), or 75 mg of aspirin combined with warfarin (target INR 2.0–2.5) and followed for 4 years. Blood samples for genotyping were available for 212 patients from Oslo University Hospital, Ullevål. Of the 140 patients on warfarin treatment, from both treatment groups combined, 105 patients were informative for weekly warfarin maintenance dose and used for genetic association analyses (Warfarin total group, Table 1).
Table 1.
Genotype distribution of CYP4F2 and VKORC1 polymorphisms and warfarin maintenance dose according to genotype in the investigated patients.
| Total material | Aspirin | Warfarin | Warfarin + Aspirin | Warfarin total | ||||
|---|---|---|---|---|---|---|---|---|
| N = 212 | N = 72 | N = 58 | N = 47 | N = 105 | ||||
| Genotype N (%) | Genotype N (%) | Genotype N (%) | Dosea | Genotype N (%) | Dosea | Genotype N (%) | Dosea | |
| 41.7 (38.1–45.3) | 34.4 (31.0–37.8) | 38.4 (35.9–41.0) | ||||||
| CYP4F2 | ||||||||
| CC | 123 (58.0) | 42 (58.3) | 32 (55.2) | 39.0 (34.1–43.8) | 26 (55.3) | 33.9 (29.1–38.6) | 58 (55.2) | 36.7 (33.3–40.0) |
| CT | 74 (34.9) | 24 (33.3) | 21 (36.2) | 46.0 (39.9–52.1) | 19 (40.4) | 34.7 (29.1–40.3) | 40 (38.1) | 40.6 (36.2–45.0) |
| TT | 15 (7.1) | 6 (8.3) | 5 (8.6) | 41.3 (25.7–56.9) | 2 (4.3) | 38.8 (-) | 7 (6.7) | 40.5 (28.5–52.6) |
| VKORC1 | ||||||||
| *1/*4 | 1 (0.5) | — | — | — | — | — | — | — |
| *2/*2 | 28 (13.2) | 10 (13.9) | 9 (15.5) | 26.7 (21.2–32.2) | 7 (14.9) | 30.0 (19.9–40.1) | 16 (15.2) | 28.1 (23.4–32.8) |
| *2/*3 | 68 (32.1) | 25 (34.7) | 20 (34.5) | 36.3 (32.0–40.5) | 9 (19.1) | 31.0 (24.6–37.4) | 29 (27.6) | 34.6 (31.2–38.1) |
| *2/*4 | 31 (14.6) | 7 (9.7) | 9 (15.5) | 43.2 (33.1–53.3) | 8 (17.0) | 26.9 (19.9–33.9) | 17 (16.2) | 35.5 (28.5–42.6) |
| *3/*3 | 39 (18.4) | 12 (16.7) | 11 (19.0) | 52.7 (46.5–58.9) | 8 (17.0) | 35.3 (26.9–43.7) | 19 (18.1) | 45.4 (39.2–51.6) |
| *3/*4 | 36 (17.0) | 14 (19.4) | 8 (13.8) | 53.8 (44.0–63.5) | 11 (23.4) | 42.5 (33.1–52.0) | 19 (18.1) | 47.2 (40.5–54.0) |
| *4/*4 | 9 (4.2) | 4 (5.6) | 1 (1.7) | 55.0 | 4 (8.5) | 40.63 (24.6–56.7) | 5 (4.8) | 43.5 (30.0–57.0) |
aMean weekly warfarin maintenance dose (95% CI).
2.2. Genotyping
Genomic DNA was isolated from 200 μL EDTA whole blood as described earlier [9]. The VKORC1*3 (rs7294/9041 G > A) polymorphism was PCR amplified on a LightCycler (Roche.com) real-time instrument under standard conditions essentially as reported [9] with an annealing temperature of 60°C. Forward and reverse primers were 5'-GAGCCTTGCCTAAGGG and 5'-TGGAAAGAGCTTTGGAGAC, respectively, whereas the LC Red640-labelled sensor probe and the fluorescence-labelled anchor probe were 5'-TGTGCGGGTATGGCAGGA and 5'-CGTGTGGCACATTTGGTCCATTGT, respectively. Genotypes were determined by melting point analyses. CYP4F2 (rs2108622/1347 C > T) and VKORC1*4 (rs17708472) were genotyped by TaqMan assays on ABI 7900 HT Fast PCR system (Applied Biosystems, CA, USA). Primers and probes for the CYP4F2 (1347 C > T) assay were commercially available at ABI (cat no C_16179493_40), whereas primers and probes for VKORC1*4 (rs17708472/6009 C > T) were custom made by ABI (forward primer: 5'GCCCGGCCCTTAAGTAATTCTT, reverse primer: 5'TCCCAGTCTCTGATGCAAAACC, wild-type probe: VIC-TGAACCGTTATACTAGCCTTG, mutant probe: Fam-CCGTTATACCAGCCTTG). Genotyping of VKORC1*2 (rs9934438/6484 C > T), CYP2C9*2 (rs1799853), and CYP2C9*3 (rs1057910) was done previously [9].
2.3. Statistical Analysis
Statistical analysis was performed using SPSS 16.0 statistic software (SPSS Inc., Chicago, IL, USA) and PLINK (http://pngu.mgh.harvard.edu/purcell/plink/). SNPs were tested for deviation from Hardy-Weinberg equilibrium using exact tests [11]. The Mann-Whitney U-test was used to test the difference in warfarin dosage between two groups. Multivariate regression analyses were performed to evaluate the individual impact of VKORC1*3, *4, and CYP4F2 (1347 C > T) polymorphisms on mean warfarin maintenance dose, after adjustment for the covariates age and treatment group. The significance level of P ≤ .05 was set for entry into the final model.
3. Results
Genotype distributions of the VKORC1*2, VKORC1*3, VKORC1*4, and CYP4F2 (1347 C > T) polymorphisms in 212 patients from the Oslo subset of the WARIS-II trial are summarized in Table 1. None of the Single Nucleotide Polymorphisms (SNPs) deviated from the Hardy-Weinberg equilibrium. Combined with our previous WARIS-II data [9], three polymorphisms in the VKORC1 gene have been analysed representing three distinct haplotypes: VKORC1*2, VKORC1*3, and VKORC1*4. Lack of any of these haplotypes was considered as VKORC1*1. The VKORC1*1 haplotype was found in one patient only (Table 1).
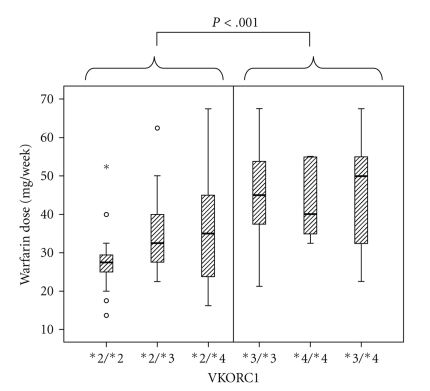
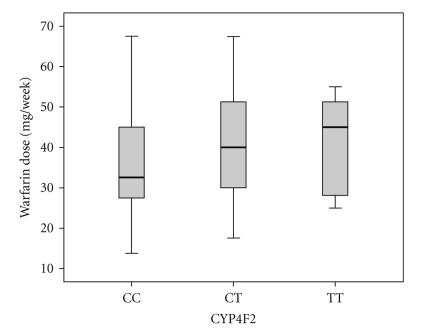
Boxplot of mean weekly warfarin dose for combinations of different VKORC1 haplotypes is shown (Figure 1). Patients homozygous for the VKORC1*2 haplotype had significantly lower mean warfarin maintenance dose compared to patients homozygous for either VKORC1*3 or *4 haplotypes (P < .001 and P = .003, resp., Mann-Whitney), whereas there was no significant difference in warfarin dosage between homozygotes of VKORC1*3 and *4 (P = .80, Mann-Whitney). Heterozygotes with combinations of VKORC1*2 and VKORC1*3 or *4 haplotypes had significantly lowered maintenance dose compared to heterozygotes of VKORC1*3 and *4 (P = .002 and P = .023, resp., Mann-Whitney). There was a slight increase in mean weekly warfarin dose in patients who were heterozygous (CT), but not for patients who were homozygous (TT) for the CYP4F2 (1347 C > T) T allele compared to patients with the normal CC-genotype (P = .14 and P = .51, resp., Mann-Whitney), indicating a lack of trend between the three CYP4F2 genotypes (Figure 2). In three separate multivariate regression analyses, VKORC1*3 and VKORC1*4 were associated with higher dose requirements when adjusted for age and treatment group only (P = .001 and P = .004, resp.), while CYP4F2 (1347 C > T) showed a weak association on higher warfarin dose requirement (P = .09). However, a stepwise multivariate regression analysis of VKORC1*3, *4, and CYP4F2 (1347 C > T) together with our earlier reported data on VKORC1*2, CYP2C9*2, and CYP2C9*3, age and treatment group gave no significant associations for neither VKORC1*3, *4 nor CYP4F2 (1347 C > T).
Figure 1.
Box plot of mean weekly warfarin maintenance dose for combinations of different VKORC1 haplotypes. The boxes represent the values from the 25th to 75th percentile. The middle lines represent the median. The vertical lines extend from the minimum values to the maximam values, excluding outliers and extreme values marked with open circles and asterisk, respectively. The P-value indicates the statistical significance of the difference in mean weekly warfarin dose between patients with or without the VKORC1*2 haplotype (Mann-Whitney test).
Figure 2.
Box plot of mean weekly warfarin maintenance dose for combinations of different CYP4F2 genotypes. The boxes represent the values from the 25th to 75th percentile. The middle lines represent the median. The vertical lines extend from the minimum values to the maximum values.
4. Discussion
In our earlier study of 212 patients with myocardial infarction (the WARIS-II Study), up to 32% of the variability in warfarin dose requirements could be explained by the VKORC1*2, CYP2C9*2, and CYP2C9*3 polymorphisms [9]. This is in accordance with results by Wadelius et al. [6], where 201 Swedish patients with mixed indications (mainly atrial fibrillation and heart valve prosthesis) were studied. Whereas all these three SNPs contributed to reduced warfarin dosage requirements, the VKORC1*3 and *4 haplotypes and recently the CYP4F2 (rs2108622/1347 C > T)-polymorphism have been associated with increased dose requirement [2–6]. Carriers of any of these six genetic variants may be at increased risk of being outside the narrow therapeutic range for INR, especially during the initial phase of the anticoagulation treatment. The combined effect of different genetic variants upon warfarin dose requirement may therefore be difficult to foresee and needed to be clarified by further studies.
The frequencies of the three VKORC1 haplotypes (*2, *3, and *4) in this study were in concordance with previous reports [8, 12–14]. In line with Osman et al. [13] and Spreafico et al. [14], we found that all carriers of the VKORC1*2 haplotype needed lower doses of warfarin compared to VKORC1*3 and *4 carriers, irrespective of homozygosity or heterozygosity of the *2 allele. In separate multivariate regression analysis, VKORC1*3 and VKORC1*4 were associated with higher dose requirements, however, inclusion of both VKORC1*3 and *4 into our previously described regression model [9], did not explain more of the variability in warfarin dose requirements, possibly because the VKORC1 activities associated with VKORC1*3 and VKORC1*4 were similar. Since the contribution of differentiating between *3 and *4 (high dose variants) to warfarin dosage was negligible compared to the considerable contribution of *2 (low dose variant), genotyping for VKORC1*2 instead of VKORC1*2, *3, and *4 may be sufficient for routine purposes in Caucasians.
The genotype frequencies of the CYP4F2 (1347 C > T) polymorphism in our study were similar to results from others [3, 15]. This CYP4F2 variant, first discovered by Caldwell and coauthors [3], has been found to explain from 1.5%–7% of warfarin dosing variability [3, 15–17]. In our study of 105 patients we had 80% power to detect significance for CYP4F2 explaining 7% of the dose variance and 24% power to detect significance explaining a modest dosing variability of 1.5%. The lack of association between the CYP4F2 (1347 C > T) polymorphism and warfarin dosage in this study indicates that the impact of this polymorphism on warfarin dosage is likely to be small in Caucasians. However, four of the seven patients who were homozygous for the CYP4F2 TT-genotype were carriers of either VKORC1*2 or CYP2C9*3 (2 were homozygous for VKORC1*2 and 2 were heterozygous for both VKORC1*2 and CYP2C9*3) which may have confounded the CYP4F2 effect.
Our findings based on myocardial infarction patients confirm that among the hitherto known genetic variants suspected to influence warfarin maintenance dosage, VKORC1*2, VKORC1*3, and VKORC1*4 are of major importance in addition to CYP2C9 variants, whereas the CYP4F2 (1347 C > T) polymorphism in Caucasians is likely to be of relatively little importance. Since the low dose variant VKORC1*2 in particular, as well as the CYP2C9*2 and CYP2C9*3 variants explain up to 32% of the variability, these variant alleles would probably be sufficient comprehensive genetic markers in Caucasians for individual warfarin dose prediction in clinical routine. In order to make a more complete survey of the genetic background for enzymes affecting individual warfarin pharmacokinetics and/or pharmacodynamics, a full genotyping of the six current SNPs is required. Populations with higher carrier frequency of the VKORC*1 haplotype (ethnic Africans) would probably also benefit from a more detailed genotyping program to elucidate individual warfarin dosage requirements.
Acknowledgment
This paper has been financed with aid from the South-Eastern Norway Regional Health Authority and EXTRA funds from the Norwegian Foundation for Health and Rehabilitation.
References
- 1.Drapkin A, Merskey C. Anticoagulant therapy after acute myocardial infarction. Relation of therapeutic benefit to patient’s age, sex, and severity of infarction. Journal of the American Medical Association. 1972;222(5):541–548. doi: 10.1001/jama.222.5.541. [DOI] [PubMed] [Google Scholar]
- 2.Aithal GP, Day CP, Kesteven PJL, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. The Lancet. 1999;353(9154):717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell MD, Awad T, Johnson JA, et al. CYK4F-2 genetic variant alters required warfarin dose. Blood. 2008;111(8):4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Andrea G, D’Ambrosio RL, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105(2):645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 5.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. The New England Journal of Medicine. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 6.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics Journal. 2005;5(4):262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Chen H, Momary KM, Cavallari LH, Johnson JA, Sadée W. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 ( VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008;112(4):1013–1021. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisen C, Watzka M, Sittinger K, et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thrombosis and Haemostasis. 2005;94(4):773–779. doi: 10.1160/TH05-04-0290. [DOI] [PubMed] [Google Scholar]
- 9.Haug KBF, Sharikabad MN, Kringen MK, et al. Warfarin dose and INR related to genotypes of CYP2C9 and VKORC1 in patients with myocardial infarction. Thrombosis Journal. 2008;6, article 7 doi: 10.1186/1477-9560-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurlen M, Abdelnoor M, Smith P, Erikssen J, Arnesen H. Warfarin, aspirin, or both after myocardial infarction. The New England Journal of Medicine. 2002;347(13):969–974. doi: 10.1056/NEJMoa020496. [DOI] [PubMed] [Google Scholar]
- 11.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. American Journal of Human Genetics. 2005;76(5):887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oldenburg J, Bevans CG, Fregin A, Geisen C, Müller-Reible C, Watzka M. Current pharmacogenetic developments in oral anticoagulation therapy: the influence of variant VKORC1 and CYP2C9 alleles. Thrombosis and Haemostasis. 2007;98(3):570–578. [PubMed] [Google Scholar]
- 13.Osman A, Enström C, Arbring K, Söderkvist P, Lindahl TL. Main haplotypes and mutational analysis of vitamin K epoxide reductase (VKORC1) in a Swedish population: a retrospective analysis of case records. Journal of Thrombosis and Haemostasis. 2006;4(8):1723–1729. doi: 10.1111/j.1538-7836.2006.02039.x. [DOI] [PubMed] [Google Scholar]
- 14.Spreafico M, Lodigiani C, van Leeuwen Y, et al. Effects of CYP2C9 and VKORC1 on INR variations and dose requirements during initial phase of anticoagulant therapy. Pharmacogenomics. 2008;9(9):1237–1250. doi: 10.2217/14622416.9.9.1237. [DOI] [PubMed] [Google Scholar]
- 15.Sagrieya H, Berube C, Wen A, et al. Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenetics and Genomics. 2010;20(7):407–413. doi: 10.1097/FPC.0b013e328338bac2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgiani P, Ciccacci C, Forte V, et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10(2):261–266. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genetics. 2009;5(3) doi: 10.1371/journal.pgen.1000433. Article ID e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]