Summary
Acyl-CoA synthases are important for lipid synthesis and breakdown, generation of signaling molecules and lipid modification of proteins, highlighting the challenge of understanding metabolic pathways within intact organisms. From a C. elegans mutagenesis screen, we found that loss of ACS-3, a long-chain acyl-CoA synthase, causes enhanced intestinal lipid uptake, de novo fat synthesis, and accumulation of enlarged, neutral lipid rich intestinal depots. Here, we show that ACS-3 functions in seam cells, epidermal cells anatomically distinct from sites of fat uptake and storage, and that acs-3 mutant phenotypes require the nuclear hormone receptor NHR-25, a key regulator of C. elegans molting. Our findings suggest that ACS-3 derived long chain fatty acyl-CoAs, perhaps incorporated into complex ligands such as phosphoinositides, modulate NHR-25 function, which in turn regulates an endocrine program of lipid uptake and synthesis. These results reveal a link between acyl-CoA synthase function and an NR5A family nuclear receptor in C. elegans.
Introduction
The ability to store nutrients, primarily in the form of triacylglycerides, is necessary to provide energy during periods when energy demands exceed caloric intake, however excessive diversion of nutrients for storage can impair growth, reduce reproductive fitness, and is associated with adverse health effects (Guh et al., 2009; Reeves et al., 2007). Numerous studies in organisms ranging from bacteria to humans have identified enzymatic components of metabolism as well as its neural and endocrine regulators. Although many of these components have already been characterized at the biochemical and structural levels, understanding the precise mechanisms by which metabolic activities of various tissues are coordinated and how dysregulation of these processes may result in aberrant metabolic regulation remain significant challenges.
One complexity of metabolism is that many enzyme families are composed of multiple and seemingly redundant members. For instance, at least 26 known or predicted acyl-CoA synthase (ACS) genes are found in the human genome (Watkins et al., 2007). The ACS family of enzymes catalyzes conversation of free fatty acids to acyl-CoA derivates. Although the ACS enzymes have been classified based on their substrate chain length preference into very long chain, long chain, medium and short chain families, the precise physiological roles of most of these enzymes are unknown. Another complexity of metabolism is that the same metabolic intermediates can, depending on the pathway into which they are channeled, have vastly different fates. For instance, acyl-CoAs can be substrates for fat breakdown through mitochondrial or peroxisomal β–oxidation, or alternatively they may be substrates for synthesis pathways, generating triglycerides for storage or structural components of membranes such as phospholipids (Coleman et al., 2002). Additionally, acyl-CoAs serve as substrates for lipid modification of proteins and function as signaling molecules either on their own or as components of more complex molecules, such as phosphoinositols and sphingolipids (Bartke and Hannun, 2009; Black and DiRusso, 2007; Faergeman and Knudsen, 1997; Payrastre et al., 2001). Thus, the consequences of misregulation of an individual ACS are quite difficult to predict even when the biochemical activity of that ACS is known.
C. elegans is an emerging model for the study of metabolism in the context of intact animals (Jones and Ashrafi, 2009; Mullaney and Ashrafi, 2009; Watts, 2009). Like most free-living organisms, C. elegans normally live in environments where food availability is dynamic. Thus, they are able to sense nutrient levels and coordinate their behavioral, physiological, and metabolic responses accordingly. Despite obvious differences, many known mammalian fat regulatory mechanisms are well conserved in C. elegans. These include fat and sugar uptake, transport, breakdown, and synthesis pathways, transcriptional regulators such as nuclear hormone receptors (NHRs) and sterol response element binding protein (SREBP), energy sensing mechanisms such as AMP-activated kinase and target of rapamycin (TOR) kinase, as well as endocrine regulators such as insulin (Ashrafi, 2007). The genetic tractability of C. elegans allows for the study of metabolic regulation through use of suppressor and enhancer screens to identify components of complex pathways (Hodgkin, 2005).
Thus far, of the more than twenty acyl-CoA synthases encoded by the C. elegans genome, phenotypes have been reported for only a few: loss of function mutations in acs-20 and acs-22 cause defective cuticle formation (Kage-Nakadai et al., 2010), while acs-4 and acs-5 are required for serotonin induced fat reduction (Srinivasan et al., 2008). In this study, we show that reduction of function of acs-3, encoding a long-chain acyl-CoA synthase, causes altered patterns of neutral lipid deposition and increased rates of fatty acid uptake and de novo synthesis. We show that acs-3 function in epidermal seam cells is sufficient for restoration of wild type lipid deposition to acs-3 mutants. Since seam cells are anatomically distinct from sites of fat uptake, de novo synthesis, and storage, our findings suggest that acs-3 must exert its effects on whole animal fat metabolism through mediators that can act cell non-autonomously. To uncover these mechanisms, we conducted suppressor screens and found that the acs-3 mutant phenotypes could be reverted to wild type by additional mutations in metabolic enzymes that consume acyl-CoAs and by nhr-25, the only C. elegans member of the NR5A family of nuclear hormone receptors that includes mammalian steroidogenic factor-1 (SF-1) and liver receptor homolog-1 (LRH-1). Our genetic and biochemical analyses suggest a model whereby ACS-3 generated products ultimately modulate the function of NHR-25, which in turn could regulate an endocrine program of fat uptake and synthesis.
Results
ft5 mutants exhibit altered fat storage
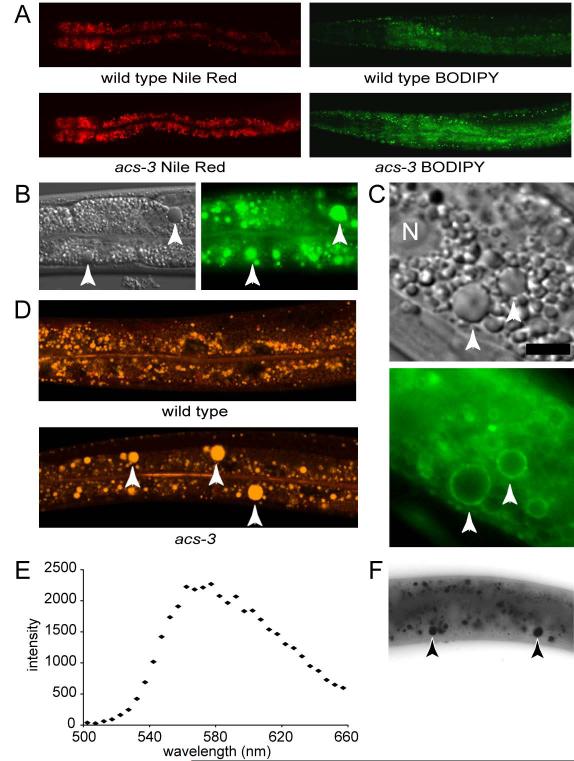
To identify genes important in the regulation of fat storage in C. elegans, we performed an unbiased mutagenesis screen, and isolated mutants with elevated staining of Nile Red, a solvatochromatic vital dye that has been used in mammalian, Drosophila, C. elegans and other experimental systems to identify and characterize metabolic pathways involved in fat metabolism (Chen et al., 2009; Flynn et al., 2009; Fowler and Greenspan, 1985; Jones et al., 2008; McKay et al., 2003; Siloto et al., 2009; Suh et al., 2007; Van Gilst et al., 2005). We found a recessive mutant, ft5, that exhibited dramatically elevated staining, but developed at nearly the same rate as wild-type animals (Figure 1A). To verify that the phenotype was not specific to the Nile Red dye, we grew animals on bacteria containing BODIPY labeled fatty acid, and observed a similar increase in staining intensity (Figure 1A). Additionally, ft5 mutants exhibited large intestinal granules not seen in wild type animals. These enlarged granules were visible with DIC microscopy (Figure 1B), stained with BODIPY-labeled fatty acids (Figure 1B) and stained in fixed animals with Sudan Black (Figure 1F), a diazo dye used for detection of lipids (Greer et al., 2008). They were also encircled by a GFP reporter fused to C. elegans ATGL, a lipase that in mammalian and Drosophila cells is important for hydrolysis of triglycerides from lipid droplets (Grönke et al., 2005; Zimmermann et al., 2004) (Figure 1C). Similar morphological and ATGL localization phenotypes were recently described in C. elegans with increased fat accumulation due to disrupted peroxisomal fat breakdown (Zhang et al., 2010).
Figure 1. Mutation of acs-3, a long chain acyl-CoA synthase, results in altered fat storage.
From a mutagenesis screen we identified a line, acs-3(ft5), that exhibits elevated staining when fed bacteria labeled both with Nile Red (50nM) and BODIPY-labeled fatty acid (A). Further analysis of acs-3(ft5) mutants revealed the presence of large refractile droplets visible using DIC microscopy, which were not seen in wild type animals. These large droplets stain with BODIPY-labeled fatty acid (B). The lipase ATGL, a lipid droplet associated protein, appears as rings associated with the perimeter of these large droplets in acs-3(ft5) mutant animals (C). When acs-3(ft5) mutant animals are labeled with 5μM (100x) Nile Red, these droplets stain yellow-gold, while intestinal cells of wild type animals lack these large droplets (D). To measure emission spectra, we utilized a spectral confocal microscope, exciting with a 488nm laser, and collecting emitted light at 5nm intervals between 500nm and 660nm. The emission peak for an individual droplet is between 470-480nm, indicating that these large droplets contain neutral lipids (E). When fixed and stained with the lipid dye Sudan Black, these large droplets are clearly labeled (F). In all panels, arrows indicate the position of abnormally large lipid droplets. In (C), the scale bar represents 5μM and N denotes the position of an intestinal cell nucleus.
Unlike BODIPY-labeled fatty acids or dyes used for detection of lipids in fixed samples, an advantage of Nile Red is that its spectral properties are exquisitely sensitive to the polarity of the environment in which it resides (Fowler and Greenspan, 1985; Greenspan and Fowler, 1985). For instance, in relatively polar phospholipid rich environments, Nile Red’s fluorescence emission spectra exhibits a peak intensity at 629 nm while in relatively neutral lipid rich environments such as triglyceride rich fat droplets, it emits yellow-gold fluorescence characterized by an intensity peak at 576 nm (Greenspan and Fowler, 1985). Because of this property, Nile Red has been used in mammalian systems to distinguish among various particles that stain with lipophilic dyes to specifically define neutral lipid rich deposits (Fowler and Greenspan, 1985). Nevertheless, use of Nile Red as a vital dye to interrogate C. elegans fat storage has recently been criticized, in part, due to the claim that this dye fails to stain lipid depots in tissues such as the skin-like hypodermis or in developing embryos within the hermaphrodite gonad (Brooks et al., 2009; O’Rourke et al., 2009). We found that by simply increasing the concentration of Nile Red fed to living animals it was possible to visualize yellow fluorescence in the enlarged droplets seen in ft5 mutants (Figure 1D) as well as in hypodermis and developing embryos (data not shown). Moreover, the yellow fluorescent granules emitted with peak intensity between 570-580 nm, indicative of an environment rich in neutral lipids (Figure 1E).
The ft5 mutation maps to a seam-cell expressed acyl-CoA synthase
To identify the causative mutation in ft5 we performed positional cloning followed by sequence analysis and found a G to A mutation in the T08B1.6 gene. This mutation results in a glycine to glutamic acid substitution at amino acid 118 of the predicted protein (Table 1). T08B1.6 encodes a predicted long-chain fatty acyl-CoA synthase, which we named acs-3.
Table 1.
Mutations isolated
| Allele | Gene ID | Gene Name | Gene Function | Locus | Mutation |
|---|---|---|---|---|---|
| ft5 | T08B1.6 | acs-3 | Acyl-CoA Synthase | V:-16.58 | Gly118->Glu |
| ft8 | Y45F3A.3 | acdh-11 | Acyl-CoA Dehydrogenase | III:3.73 | Asp309->Asn |
| ft12 | Y45F3A.3 | acdh-11 | Acyl-CoA Dehydrogenase | III:3.73 | Ala244->Thr |
| ft11 | T08G2.3 | acdh-10 | Acyl-CoA Dehydrogenase | X:24.09 | Gly259->Glu |
| ft14 | F41H10.8 | elo-6 | Fatty Acid Elongase | IV:1.64 | Gly205->STOP |
| ft13 | Y46E12BL.4 | spsb-1 | Unknown | II:24.58 | Gly3->Glu |
Molecular identities of mutant alleles identified in this study. acs-3(ft5) was isolated from a screen for animals with altered Nile Red staining, while all other mutants were isolated from an acs-3(ft5) suppressor screen by identifying animals that do not arrest on 110uM LY294002. Each allele identified in this screen also suppressed the fat storage phenotype of acs-3(ft5).
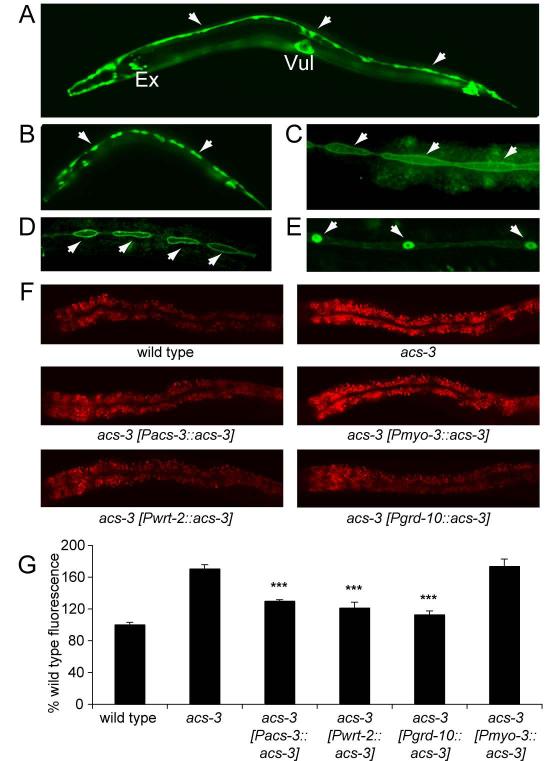
To begin characterization of acs-3, we first generated transgenic animals carrying 2.5kb of the acs-3 upstream regulatory sequence fused to a GFP reporter. GFP expression was observed beginning during embryogenesis and continuing through adulthood. From larval stage L1 to L4, expression appeared restricted to epidermal seam cells, the excretory cell, vulva cells and a subset of unidentified cells in the head and tail (Figure 2A, 2B and S1). In addition to prominent seam cell expression, adult animals also exhibited moderate intestinal GFP expression. Tissue expression patterns observed by the promoter-reporter fusion were recapitulated by transgenic animals in which full length acs-3 cDNA was tagged at the C-terminus with GFP and driven by the same 2.5kb upstream regulatory elements (Figure S1). Nile Red fat staining of these transgenic animals was restored to near wild-type levels indicating that the 2.5kb promoter is sufficient for regulation of lipid storage (Figure 2F and 2G).
Figure 2. acs-3 has a limited expression pattern, and expression of acs-3 in seam cells rescues the fat storage phenotype.
To examine the expression pattern of acs-3, we fused 2.5kb of upstream regulatory sequence to GFP. This construct yields high expression in a limited set of tissues. In L4 animals, strong expression is evident in seam cells, which are beginning to fuse at this stage, the excretory cell (marked Ex) and vulva (marked Vul) as well as cells in the head and tail (A). In an L2 animal, individual seam cells are clearly visible (B). We also generated constructs driving the acs-3 cDNA fused to GFP. We drove this construct with the previously characterized seam cell promoters wrt-2 (C) and grd-10 (D). With both constructs, ACS-3 is clearly localized to the cell membrane (C-D). When we drove the acs-3(ft5) mutant cDNA we observed misslocalization of the protein to seam cell nuclei (E). In panels A-E, arrows mark the position of seam cells. Elevated Nile Red staining of acs-3(ft5) animals can be rescued by expression of acs-3 cDNA with regulatory elements upstream of acs-3 start site, as well as with the wrt-2 and grd-10 seam cell specific promoters (E). Expression in the body-wall muscle, a tissue in which acs-3 is not normally expressed, driven by the myo-3 promoter, fails to rescue the high Nile Red staining of acs-3(ft5) animals (F). We quantified Nile Red intensity in these lines (G). Data are shown as a percentage of the wild-type average +/− SEM. ***p < 0.001 compared to acs-3(ft5).
To determine whether ACS-3 function in a subset of tissues could be sufficient for fat storage regulation, we expressed full length wild-type acs-3 using a variety of tissue specific promoters in acs-3(ft5) mutants. Expression of acs-3 in seam cells, using the previously characterized wrt-2 and grd-10 seam cell specific promoters (Aspock et al., 1999), was sufficient to almost fully rescue the fat storage phenotype of acs-3(ft5) animals (Figure 2F and 2G). We attempted numerous injections of an intestinally driven acs-3 construct but they caused lethality in nearly all instances. In one case, a single line carrying intestinally expressed acs-3 was generated, however, the transgenic animals exhibited numerous phenotypes associated with sickness such as slow growth or growth arrest, small body size and clear intestine (data not shown). Expression of acs-3 specifically in body-wall muscle, a tissue in which we never observed expression with the acs-3 promoter, failed to alter excess Nile Red staining of acs-3(ft5) animals (Figure 2F and 2G). Taken together, these data indicated that reconstitution of acs-3 in seam cells was sufficient to confer wild-type fat staining to acs-3(ft5) mutants but expression at similar or higher level in another tissue did not rescue the altered lipid storage of these mutants.
Within the seam cells, expression of full length ACS-3 fused to GFP at the C-terminus driven by the rescuing 2.5kb promoter resulted in localization of the fusion protein throughout the cytoplasm. Expression of the same reporter fusion driven by the seam cell specific promoters wrt-2, or grd-10, which allowed for nearly full rescue of acs-3(ft5) fat storage phenotype, resulted in clear localization of the reporter protein to the plasma membrane of seam cells (Figure 2C and 2D). Based on its amino acid sequence, the ACS-3 protein is not predicted to have any transmembrane domains. Thus, ACS-3 is likely a cytosolic enzyme that is localized to the inner leaflet of the plasma membrane of seam cells and the difference in subcellular localization using the indicated promoters is likely a consequence of differences in the extent of promoter activity. Consistent with this, there was no obvious overlap between reporter fusions to ACS-3 and CLK-1, a C. elegans mitochondrial protein (Felkai et al., 1999) (Figure S1).
Localization of ACS-3 to seam cells was unexpected, as these cells have not been previously implicated in fat storage or regulation. Seam cells are relatively poorly characterized cells that are part of the skin-like epidermis of C. elegans. During development, seam cells exhibit stem cell characteristics as they go through multiple rounds of division whereby a subset of daughter cells regenerates the seam cell population after each division, while the remainder differentiate into epidermal, glial or neuronal cells (Sulston and Horvitz, 1977).
Recombinant ACS-3 exhibits acyl-CoA synthetase activity in vitro, and prefers long chain fatty acid substrates
To confirm the predicted function of ACS-3, we generated and purified recombinant protein. Using an in vitro colorimetric assay, we tested the activity of this recombinant ACS-3 protein, along with a commercially available Pseudomonas ACS as a positive control. Pseudomonas ACS exhibited strong ACS activity, and could use a broad range of fatty acid chain lengths as substrate, in accordance with previous findings (Hosaka et al., 1981; Knoll et al., 1994) (Figure S2). Recombinant C. elegans ACS-3 also exhibited ACS activity, but was capable of activating a narrower range of fatty acid chain lengths. 18-carbon fatty acid was the preferred substrate chain length, but the enzyme exhibited significant activity on 20- and 22-carbon fatty acids as well (Figure S2). These findings confirm the predicted long-chain fatty acyl-CoA (LCFA-CoA) synthase function of ACS-3.
We next tested the activity of recombinant mutant ACS-3 protein in which we introduced the Glu 118 to Gly mutation identified in acs-3(ft5) animals. Surprisingly, this protein exhibited activity equivalent to wild-type protein (Figure S2). Thus, we asked whether the Glu118 to Gly mutation disrupts other aspects of ACS-3 function such as its localization or expression pattern. As assessed by the full-length GFP reporter, the mutant ACS-3 protein mislocalized to the nucleus (Figure 2E). The incorrect trafficking of the mutant protein raised the possibility that the mutant phenotype may arise from inappropriate ACS activity in the seam cell nucleus. However, the recessive nature of the ft5 mutation, coupled with the fact that acs-3(ft5) mutants can be rescued by expression of the wild-type cDNA, indicated that the mutant phenotype corresponded to acs-3 loss of function rather than neomorphic activity. Thus, the Glu118 to Gly mutation likely disrupts ACS-3 function by mislocalizing this enzyme to the nucleus.
Measures of metabolic parameters demonstrate that acs-3(ft5) animals exhibit elevated rates of fatty acid uptake and increased rates of de novo fatty acid synthesis
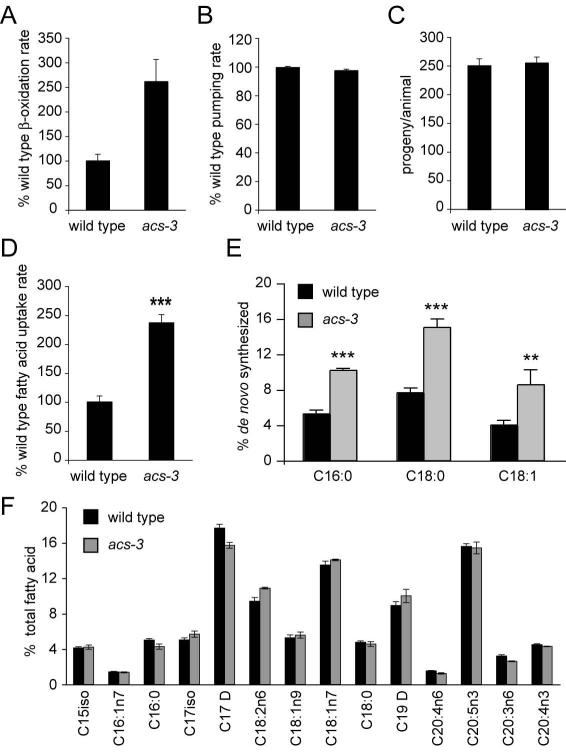
To better understand the altered fat storage phenotype of acs-3(ft5) animals, we assayed various behavioral and metabolic parameters of these mutants. C. elegans intake nutrients through pharyngeal pumping, the rate of which is subject to modulation by food availability and food quality, as well as experience of starvation prior to food exposure (Avery and Horvitz, 1990; You et al., 2008). We found that pumping rates in acs-3(ft5) and wild-type animals were equivalent. Similarly, brood size and rates of movement of acs-3(ft5) were indistinguishable from those of wild-type animals (Figure 3B, 3C and data not shown). Finally, we found that under conditions of full starvation acs-3 mutants tended to survive slightly longer than wild type animals, although the differences in overall survival rate were not statistically significant (Figure S1).
Figure 3. Characterization of metabolic parameters reveals acs-3(ft5) animals have an increased rate of fatty acid uptake and elevated de novo fatty acid synthesis.
We examined a number of physiological parameters that could impact lipid storage. To assess β-oxidation, we incubated animals in media containing tritiated fatty acids, collected that media and measured amount of tritiated water produced (see methods for more details). We found that acs-3(ft5) animals exhibited an increased rate of β-oxidation (A). Measures of pharyngeal pumping rate and progeny production revealed no differences between wild type and acs-3(ft5) mutants (B-C). To examine fatty acid uptake, we incubated animals in media containing BODIPY-labeled fatty acids for 20 minutes, then measured fluorescence intensity. We found that acs-3(ft5) mutants exhibit more than double the rate of fatty acid uptake observed in wild type animals (D). For panels A-D, data are displayed as a percentage of the wild-type average +/− SEM. ***p < 0.001 compared to wild-type. Using 13C labeling, we found that acs-3(ft5) mutants exhibit a higher percentage of de novo synthesized fatty acids (E). **p<0.01, ***p < 0.001 compared to wild type. GC/MS analysis reveals no substantial changes in fatty acid composition between acs-3(ft5) mutants and wild type animals (F). Data are shown as a percentage of total fatty acid +/− SEM.
The requirement of ACS enzymes for consumption of fatty acids suggested defective β-oxidation as a likely explanation for the observed fat storage phenotype. To directly test this hypothesis, we fed animals radiolabeled oleic acid and measured rates of production of labeled water, a byproduct of fat oxidation. Surprisingly, acs-3(ft5) animals showed an increase in β-oxidation rate (Figure 3A). Since this assay is dependent on uptake of radiolabeled oleic acid by whole animals, it raised the possibility that the observed enhanced rate of labeled water production could reflect increased rate of fatty acid absorption rather than increased fat oxidation. To evaluate this possibility, we adapted an assay for fatty acid uptake that has been previously utilized in yeast and mammalian cell culture based experiments (Jia et al., 2007; Li et al., 2005a; Schaffer and Lodish, 1994) as well as recently in C. elegans (Spanier et al., 2009).. We empirically determined conditions whereby feeding animals a short pulse of the fluorescently labeled fatty acid C1-BODIPY 500/512 C12 allowed for comparison of rate of fatty acid uptake. Based on this assay, acs-3(ft5) animals exhibited a striking increase in uptake rate compared to wild-type animals (Figure 3D). After uptake of a pulse of fluorescently-labeled fatty acids, the rate of diminishment of fluorescence was similar in wild-type and mutant animals (data not shown), indicating that the increased accumulation of labeled dietary fatty acids in acs-3(ft5) animals reflected increased rate of fat uptake rather than reduced efflux. Assuming that the assay conditions did not saturate the capacity of β-oxidation enzymes, the increased rate of β-oxidation of acs-3(ft5) mutants likely reflects increased rate of labeled oleate uptake. Thus, when normalized to the rate of fatty acid uptake, the rate of β-oxidation of acs-3(ft5) animals may be similar to that of wild-type animals.
Next, we assessed rates of de novo fatty acid synthesis. acs-3(ft5) animals exhibited approximately 50% increased rates of synthesis relative to wild-type animals (Figure 3E). To determine whether either the increased rate of uptake or synthesis would alter composition of fats, we examined the fatty acid profile of wild-type and acs-3(ft5) mutants. There were no significant differences in fatty acid profiles between these strains (Figure 3F).
Together, these data indicated that the altered fat storage of acs-3(ft5) animals is not a consequence of altered behaviors or defective fat oxidation, but rather acs-3(ft5) mutants exhibit a metabolic shift favoring an increased rate of dietary fatty acid uptake and an elevated rate of de novo fat synthesis, processes consistent with the lipid storage phenotype of these animals.
Loss of function of acyl-CoA consuming enzymes suppress the effects of acs-3(ft5) mutation
Seam cells are anatomically distant from the intestinal lumen where uptake of dietary fatty acid occurs. Moreover, many genes that encode components of lipid metabolism are expressed in intestinal and epidermal cells, while few have thus far been reported to be expressed in seam cells (Hunt-Newbury et al., 2007; Srinivasan et al., 2008). To elucidate how loss of ACS-3 activity in seam cells elicits the observed whole animal metabolic changes, we sought to identify additional genes required for the observed phenotypes of acs-3(ft5) mutants. To do so, we took advantage of the fortuitous observation that treatment of animals with 110 μM of LY294002, a broad spectrum inhibitor of PI-3 kinases, results in developmental arrest in acs-3(ft5) yet causes only minor developmental delay of wild-type animals (Figure 4A). LY294002 has been reported to functionally inhibit AGE-1, the sole C. elegans class-I PI-3 kinase (Babar et al., 1999). However, we found that growth arrest of drug treated acs-3 mutants was unlikely to be only due to inhibition of AGE-1 since acs-3(ft5); age-1(hx546) double mutants did not arrest in the absence of drug treatment, nor were they sensitive to treatment with LY294002 (Figure S3). Despite the fact that we did not know the targets through which LY294002 caused growth arrest in acs-3(ft5) mutants, the striking arrest phenotype provided a facile screening strategy to search for potential suppressors of acs-3(ft5). We anticipated that such an approach would be more specific that screening for suppression of the acs-3(ft5) Nile Red staining phenotype, as inactivation of many genes is known to alter Nile Red staining (Ashrafi et al., 2003).
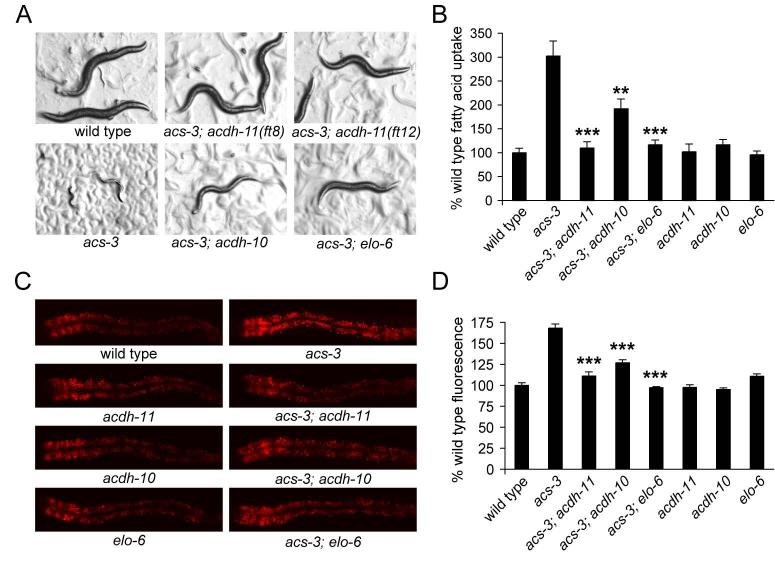
Figure 4. An unbiased mutagenesis screen identifies two acyl-CoA dehydrogenases and a fatty acyl-CoA elongase as suppressors of acs-3(ft5) growth and fat storage phenotypes.
acs-3(ft5) animals exhibit a developmental arrest when treated with the PI-3 kinase inhibitor LY294002. We performed an unbiased mutagenesis screen utilizing this phenotype, and identified two alleles of acdh-11, one allele of acdh-10 and one allele of elo-6 as suppressors of this developmental phenotype (A). These mutations also suppressed the high fatty acid uptake of acs-3(ft5) mutants, but do not affect uptake in an otherwise wild type background (B). The elevated Nile Red staining phenotype of acs-3(ft5) mutants was also suppressed by these mutations, while each of these suppressors, when outcrossed to an otherwise wild type background exhibited normal Nile Red staining phenotype (C-D). Data are displayed as a percentage of the wild-type average +/− SEM. **p < 0.01, ***p < 0.001 compared to acs-3(ft5).
F2 progeny of mutagenized acs-3(ft5) animals were grown on 110 μM LY294002, and animals that developed to adult stage in 72 hours, a time point at which acs-3(ft5) animals are 100% arrested in the L2 or L3 larval stages, were selected (Figure 4A). Newly identified acs-3(ft5) suppressors were assayed for Nile Red staining independent of LY294002 treatment. In all cases, high Nile Red staining of acs-3(ft5) animals was reduced to nearly wild-type levels (Figure 4C and 4D) suggesting that the identified mutations indeed suppressed the effects of acs-3(ft5), at least on Nile Red staining, rather than simply preventing LY294002 action.
We identified the molecular lesions in ft8, ft11, ft12, and ft14 suppressor lines (Table 1). In ft8 and ft11, we found missense mutations in the Y45F3A.3 gene, encoding a predicted very long-chain acyl-CoA dehydrogenase, which we named acdh-11. In ft11 we found a missense mutation in the T08G2.3 gene, encoding a predicted medium chain acyl-CoA dehydrogenase, which we named acdh-10. Finally, in ft14, we identified a nonsense mutation in the F41H10.8 gene, encoding the fatty acid elongase elo-6 (Table 1). Identified mutations in acdh-10, acdh-11, and elo-6 suppressed acs-3(ft5) in a recessive manner, indicating they were loss of function mutations. The early STOP in elo-6(ft14) was consistent with a loss of function, and treatment of acs-3(ft5) mutants with two-generation RNAi of either acdh-10 or acdh-11 suppressed LY294002-induced growth arrest of these animals in a similar manner to the mutations identified (data not shown).
A number of observations indicated that losses of each acdh-10, acdh-11, and elo-6 specifically counteracted the effects acs-3(ft5) reduction of function. First, although the LY294002 induced growth arrest, high Nile Red staining, enlarged droplet size, and the high fatty acid uptake rate of acs-3(ft5) animals were either partially or fully reverted to wild type levels by each of elo-6(ft14), acdh-10(ft11) and acdh-11(ft8), none of these mutations on their own caused a detectable change in these parameters when compared to wild type animals (Figure 4A, 4B, 4C, 4D and S4). Second, acdh-10(ft11), acdh-11(ft8), and elo-6(ft14) mutants all developed and grew at or near wild-type rates and exhibited normal pharyngeal pumping rates further indicating that these mutations do not correct the altered fat storage of acs-3(ft5) mutants merely due to detrimental effects on animal viability or changes in food intake (Figure S5 and data not shown).
To begin elucidating how the newly identified suppressors counter the effects of loss of acs-3, we first investigated their expression patterns. In each case, expression of GFP reporter fusion was first detected during embryonic development and persisted through larval and adult stages (Figure S5 and data not shown). As previously reported (Pauli et al., 2006), we found that elo-6 expression was largely confined to the intestinal cells and undetectable in the seam cells (Figure S5). Similarly, there was no detectable acdh-11 expression in the seam cells although expression was clearly visible in many other cell types including the fat storing hypodermal and intestinal cells (Figure S5). Reporter fusion of acdh-10 exhibited a broad expression in numerous tissues (Figure S5). Thus, reduction of function of ACS-3 can be ameliorated by deficiencies in metabolic enzymes in tissues other than the seam cells.
Since elo-6 regulates production of mono-methyl branched fatty acids (mmBCFA) (Kniazeva et al., 2004; Seamen et al., 2009), we tested whether elo-6(ft14) suppression of acs-3(ft5) is due to diminished mmBCFA levels. As expected, acs-3(ft5); elo-6(ft14) double mutants had reduced levels of mmBCFA (Figure S6). However, dietary supplementation with these fatty acids failed to reverse the elo-6(ft14) suppression of acs-3(ft5) (Figure S7), suggesting low mmBCFA levels do not suppress acs-3(ft5). Additionally, there were no obvious changes in mmBCFA caused by losses of acdh-10 and acdh-11 (Figure S6).
A unifying explanation for how mutations in each of acdh-10, acdh-11, and elo-6 restore wild type growth and lipid metabolic phenotypes to acs-3(ft5) mutants is that each of these enzymes modulates fatty acyl-CoA turnover. As an elongase enzyme, ELO-6 acts on fatty acyl-CoAs, effectively depleting the fatty acyl-CoA pool of shorter chain lengths through production of longer chain fatty acyl-CoAs, while ACDH enzymes are well known to function downstream of ACS enzymes in β-oxidation of fatty acyl-CoAs (Eaton et al., 1996). As such, the deficiency in generation of acyl-CoA products of ACS-3 could be countered by simultaneous deficiencies in enzymes that consume fatty acyl-CoAs. Our findings additionally indicate that these enzymes need not function in the very same tissue.
acs-3 regulation of fat storage and growth is dependant on the nuclear hormone receptor nhr-25, a member of the SF-1 and LRH-1 family
How might changes in ACS-3 generated acyl-CoA products in the seam cells cause changes in fatty acid uptake and lipid storage phenotypes in intestinal cells? In addressing this question we were guided by the molecular identity of yet another suppressor of acs-3(ft5). In the suppressor line ft13 we identified a missense mutation in Y46E12BL.4, a previously uncharacterized gene (Table 1). As in the acdh and elo-6 suppressors, the ft13 mutation suppressed both the LY294002 induced growth arrest and Nile Red staining phenotypes of acs-3(ft5) mutants, without causing obvious changes in growth and staining patterns of otherwise wild type animals (Figure S8). We generated RNAi constructs targeting Y46E12BL.4, and found that knockdown of this gene in acs-3(ft5) mutants recapitulated the suppression observed in the mutant background, confirming the identity of the suppressor mutation (data not shown).
Y46E12BL.4 encodes an uncharacterized protein predicted to contain a conserved SPRY domain, most closely related to the SPRY domain of D. melanogaster GUSTAVUS, an RNA binding protein with roles in early embryonic development, and the mouse Ssb-1, -2, -3 and -4, proteins whose physiological functions remain poorly understood. Each of these homologous proteins contains both a central SPRY domain as well as a C-terminally encoded SOCS box domain. This latter domain was lacking from the wormbase prediction of Y46E12BL.4. We performed 3′ RACE to identify the 3′ end of the Y46E12BL.4 cDNA and found that this gene does indeed encode a protein with both an SPRY and a C-terminal SOCS box domain. Therefore, we named this gene spsb-1 (SPry and Socs Box gene).
Although C. elegans spsb-1 is uncharacterized, and our attempts at obtaining transgenic animals expression GFP fusions to spsb-1 were unsuccessful, its homology to GUSTAVUS provided a potential clue to mechanisms through which acs-3 may exert its effects on metabolism. In a yeast two hybrid screen, Drosophila GUSTAVUS was found to interact with FTZ-F1 (Giot et al., 2003), an NR5A NHR family member (Asahina et al., 2000). Similar to acs-3, nhr-25, the sole NR5A family member in C. elegans, is expressed in seam cells. Moreover, in response to as-yet-unidentified signals, nhr-25 regulates molting, an endocrine program that is initiated in the seam cells but exerts effects throughout C. elegans tissues (Asahina et al., 2000; Frand et al., 2005; Kostrouchova et al., 1998). The fact that some NHRs are regulated by lipophilic hormones, coupled with the identities of suppressors of acs-3, suggested that fatty acyl-CoA product of ACS-3 may directly or indirectly regulate function of NHR-25 to in turn regulate an endocrine program of fat uptake and de novo fat synthesis.
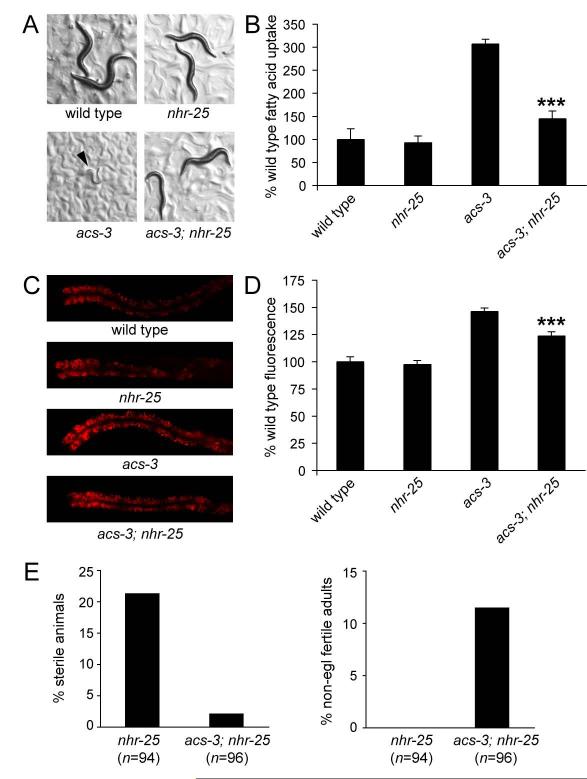
At least 27 C. elegans nhrs, including nhr-25, are reported to be expressed in the seam cells (identified in a search of www.wormbase.org). We tested whether RNAi mediated inactivation of any of these genes could either recapitulate the acs-3(ft5) phenotype, or suppress it. Only RNAi knockdown of nhr-25, but none of other NHRs tested, suppressed the high Nile Red staining of acs-3(ft5) mutants without altering staining in wild-type animals (data not shown). Unambiguous interpretation of this finding was not possible because of pleiotropic phenotypes such as sterility, partially penetrant lethality and reduced body size (Asahina et al., 2000; Chen et al., 2004), associated with nhr-25 RNAi treatment. Therefore, we generated acs-3(ft5); nhr-25(ku217) double mutants. ku217 is a hypomorphic allele of nhr-25 that exhibits reduced DNA binding activity, yet retains enough activity to allow for relatively normal rates and patterns of development up until the adult stage in a majority of animals, at which point all animals exhibit a severe defect in egg-laying (Chen et al., 2004). Experiments with this allele were performed before the onset of the egg-laying deficient phenotype. Nile Red staining, large droplet formation and fatty acid uptake were significantly reduced in acs-3(ft5); nhr-25(ku217) double mutants compared to acs-3(ft5), while nhr-25(ku217) alone exhibited wild-type fatty acid uptake, Nile Red staining levels and lipid droplet size (Figure 5B, 5C, 5D and S4). Moreover, nhr-25(ku217) partially suppressed the growth arrest phenotype of LY294002 treated acs-3(ft5) animals (Figure 5A). Together, these data suggested that phenotypes causes by acs-3 reduction of function were dependent on nhr-25. Consistent with this interpretation, nhr-25 is expressed in seam cells, the excretory cell and cells of the developing vulva (Asahina et al., 2000; Chen et al., 2004; Gissendanner and Sluder, 2000) a pattern very similar to that of acs-3 (Figure 2A, 2B and S1).
Figure 5. Mutation of nhr-25 suppresses the high Nile Red staining and fatty acid uptake phenotypes of acs-3(ft5) animals.
The hypomorphic nhr-25(ku217) allele partially suppresses the developmental arrest of acs-3(ft5) mutant animals grown on LY294002 (A). Arrow indicates position of an arrested acs-3(ft5) animal. nhr-25(ku217) also suppresses the high fatty acid uptake phenotype and high Nile Red staining phenotype of acs-3(ft5) animals, but nhr-25(ku217) in an otherwise wild type background exhibits wild type fatty acid uptake and Nile Red staining (B-D). Data are shown as a percentage of the wild-type average +/− SEM. ***p < 0.001 compared to acs-3(ft5). We also tested the affect of acs-3(ft5) on previously described nhr-25(ku217) phenotypes. We found that loss of acs-3 function can partially suppress both the sterility and egg-laying phenotypes of nhr-25(ku217) (E-F). Data are shown as a percent of all animals assayed. Because the severity of the egg-laying and sterility phenotypes in nhr-25(ku217) is temperature dependant, we performed these assays at 24 degrees, a temperature at which these phenotypes are strong, but most animals develop to adulthood.
To further investigate the genetic interaction between acs-3 and nhr-25, we examined a number of nhr-25 dependant phenotype. As previously reported, we found that nhr-25(ku217) animals exhibit partially penetrant sterility, while fertile animals that develop to adulthood exhibit a strong egg-laying defective phenotype caused by improper vulval development (Chen et al., 2004). The vulval development defect was partially corrected by the acs-3(ft5) mutation as was the sterility associated with loss of nhr-25 function (Figure 5E). nhr-25(ku217) animals also exhibit extranumerary seam cells, however this phenotype was neither suppressed nor enhanced in acs-3(ft5); nhr-25(ku217) double mutants (data not shown). Together, these findings provide additional evidence supporting a genetic regulatory relationship between acs-3 and nhr-25 consistent with the possibility that loss of acs-3(ft5) is associated with enhanced activity of NHR-25.
The nhr-25 ligand-binding domain accommodates phosphoinositide lipids containing long chain fatty acids
nhr-25 is the single C. elegans member of the NR5A family of NHRs. This family includes D. melanogaster FTZ-F1, and mammalian LRH-1 and SF-1. One possible mechanism linking acs-3 and nhr-25 was suggested by biochemical studies of SF-1 and LRH-1. While the existence and identity of potential ligands of these mammalian receptors have been debated, these receptors accommodate phospholipids containing long chain fatty acids in their ligand-binding pockets (Krylova et al., 2005; Li et al., 2005b). In vitro experiments with mammalian SF-1 have shown that phosphoinositides are efficiently exchanged into the ligand-binding domain, raising the possibility that these lipids may act as ligands for these receptors (Krylova et al., 2005; Sablin et al., 2008).
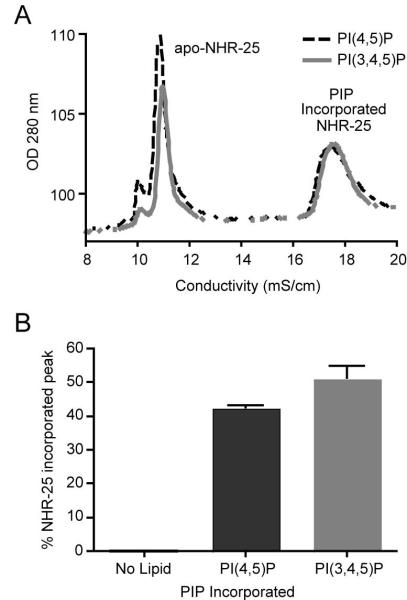
To investigate potential biochemical similarities between mammalian and C. elegans NR5A class of receptors, we generated purified recombinant NHR-25 ligand-binding domain protein. Utilizing a previously described assay (Sablin et al., 2008), we tested the ability of this purified protein to incorporate phospholipids in vitro. We found that, as in the mammalian NR5A ligand-binding domains (Krylova et al., 2005), NHR-25 ligand-binding domain could indeed accommodate PI(3,4,5)P3 and PI(4,5)P2 (Figure 6A and 6B).
Figure 6. NHR-25 ligand-binding domain binds phosphoinositides.
Mammalian SF-1, an nhr-25 homolog, has been shown to bind phosphoinositide species. To test the ability of NHR-25 to bind these lipids, we generated and purified recombinant NHR-25 ligand binding domain. Incubation of this protein with PI(4,5)P and PI(3,4,5)P alters retention of the protein in ion exchange chromatography (A) indicating the protein has bound the lipid. Incubation of each phohsphoinositide species with NHR-25 ligand binding domain protein induced ~50% of the protein to bind lipid (B).
The ability of purified NHR-25 to bind PI(3,4,5)P3 and PI(4,5)P2 along with the observation that acs-3(ft5) mutants exhibit a synthetic growth arrested when treated with the PI-3 kinase inhibitor LY294002, suggest the possibility that phosphoinositides may act as endogenous regulators of nhr-25. Since attachment of fatty acids to the inositol sugar is dependent on availability of fatty acyl-CoAs, our findings are consistent with a model whereby ACS-3 exerts control over NHR-25 activity by determining the chain lengths of fatty acids present on these rings.
Discussion
We found that animals deficient in acs-3, encoding an acyl-CoA synthase that preferentially produces LCFA-CoA species, exhibited increased fat uptake, increased de novo fatty acid synthesis and altered fat storage, including formation of large lipid droplets. Reconstitution of acs-3 in the seam cells alone was sufficient to correct the fat storage phenotypes associated with loss of acs-3. Since seam cells are a cellular component of C. elegans epidermis that are not directly involved in dietary fat uptake and de novo fat synthesis, our findings indicated that ACS-3 activity modulates organismal fat metabolism through signals that initiate in the seam cells but ultimately act in other tissues. Consistent with this hypothesis, we found that the metabolic consequences of acs-3 loss require nhr-25, a seam cell expressed NHR previously implicated in the regulation of molting, an endocrine process that links nutrient availability and developmental timing. Our genetic suppressor analyses and biochemical characterization of purified NHR-25 are consistent with a model whereby loss of long chain acyl-CoAs generated by ACS-3 results in aberrant function of NHR-25, which in turn, regulates an endocrine signaling cascade that ultimately modulates dietary fat uptake and de novo fat synthesis.
The abnormally large lipid droplets in acs-3(ft5) intestinal cells, which can be suppressed by mutations in acdh-10, acdh-11, elo-6 or nhr-25, are reminiscent of those recently described in animals deficient in peroxisomal β-oxidation (Zhang et al., 2010). As has been shown for the peroxisomal mutants, the acs-3(ft5) mutant large droplets stain with BODIPY labeled fatty acids and are ringed by ATGL::GFP transgenically expressed in intestinal cells. We found that these large droplets stain with Nile Red with an emission peak intensity of 570-580 nm, indicating that these large droplets are primarily comprised of neutral lipids. These findings underscore the utility of Nile Red staining as an assay to investigate properties of different lipid storage compartments in intact animals.
It was already known that SF-1 and nhr-25 are both required for gonadal development and fertility, while both nhr-25 and LRH-1 are necessary for embryonic development (Asahina et al., 2000; Fayard et al., 2004; Hammer and Ingraham, 1999). Our findings indicate that the elevated fatty acid uptake of acs-3 mutants is nhr-25 dependent. Similarly, LRH-1 regulates bile acid production, and is required for efficient intestinal uptake of dietary lipids (Mataki et al., 2007). Finally, beyond its well-recognized role in sex determination, mammalian SF-1 functions in energy balance as a key transcriptional regulator of glucocorticoid synthesis in the adrenal glands (Bakke et al., 2001). Thus, in C. elegans as in mammals, an NR5A family receptor modulates metabolic related pathways.
One model consistent with our findings is that ACS-3 functions to generate PI(3,4,5)P3 and PI(4,5)P2 species that contain long chain fatty acids, which in turn, act as inhibitory ligands for NHR-25. Crystallographic studies with recombinant mammalian SF-1 and LRH-1 have identified bacterial phospholipids as fortuitous ligands bound within the ligand pocket of these receptors (Krylova et al., 2005; Li et al., 2005b). A number of other phospholipids can also be accommodated by the ligand pocket of these receptors (Krylova et al., 2005; Li et al., 2005b; Sablin et al., 2008). Among these, phosphoinositides PI(3,4,5)P3 and PI(3,5)P2 readily bound the ligand-binding domain and mutations interfering with phosphoinositide binding were found to reduce transcriptional activity of SF-1 (Krylova et al., 2005; Sablin et al., 2008). We found that NHR-25 could bind PI(3,4,5)P3 and PI(4,5)P2 under the same conditions reported in experiments with mammalian SF-1 and LRH-1 (Sablin et al., 2008). Moreover, sensitivity of acs-3 mutants to the PI-3kinase inhibitor LY294002 indicated an in vivo regulatory relationship between phosphoinositides and nhr-25.
Since mammalian NR5A receptors exhibit constitutive activity in many cell types (Fayard et al., 2004), physiological ligands of these receptors may function to inhibit activity or modulate activity specifically toward subsets of transcriptional targets. Given the preference of ACS-3 for long chain fatty acids, it is possible that its reduction of function may preferentially diminish production of PI(3,4,5)P3 and PI(4,5)P2 species containing long chain fatty acids. Our genetic data suggest that such species normally act to inhibit activity of NHR-25 toward mechanisms that promote dietary fat uptake and de novo fat synthesis. Consistent with the notion that chain lengths of fatty acids present on various lipid species could modulate activity of NR5A class of receptors, there is evidence that phospholipids with medium chain fatty acids enhance in vitro binding of purified SF-1 to a short peptide motif derived from the TIF-2 co-activactor, while phospholipids containing long chain fatty acids competitively disrupt this binding (Li et al., 2005b). However, since acyl-CoAs are substrates for generation of a variety of complex lipid species, we cannot rule out the possibility that ACS-3 derived long chain fatty acyl-CoAs could exert their effects on NHR-25 through mechanisms other than phosphoinositides. Similarly, ACS-3 derived fatty acyl-CoAs may exert their effects on NHR-25 through modulation of potential co-activators of NHR-25 or upstream regulatory proteins.
Identification of in vivo physiological lipid ligands of NHRs has been a fundamental challenge in understanding these complex transcriptional master-regulators. Such ligands may be present at very low levels, and may only associate with NHRs under specific physiological conditions, making ligand identification very challenging. Our studies point to ACS enzymes as a previously unappreciated in vivo mechanism for regulating the function of the NR5A family of NHRs. Though in vitro experiments have demonstrated phospholipids can bind and modulate activity of the NR5A family nuclear receptor SF-1, our data represent the first in vivo demonstration that genes acting in lipid metabolism function upstream of this family of receptors.
Methods
Vital dye lipid staining and quantification
Synchronized animals were plated on NGM plates containing OP50 and 50nM Nile Red dye. All lipid staining experiments were conducted at 25°C, as acs-3(ft5) animals exhibited the strongest phenotype at this temperature. Animals were grown for 40 hours to the young adult stage. Images were acquired on a Zeiss Axioplan II microscope outfitted with a digital CCD camera. Images shown were obtained using a 16x objective. For quantification, images were collected using a 5x objective. The same circular region of interest (ROI) that encompasses one entire animal was applied to each of these images. The minimum fluorescence value was subtracted from the mean of the ROI to correct for background signal. For each experiment, 8-12 animals of each strain were imaged and analyzed.
For BODIPY staining, a 1mg/ml stock of C1-BODIPY 500/512 C12 (Invitrogen) was added to NGM plates seeded with OP50 at a final dilution of 1:50,000. Images were acquired with a 16x objective as described for Nile Red.
For high Nile Red staining, synchronized L4 animals were plated on seeded NGM plates containing 5uM Nile Red (100x) and allowed to stain for 2-4 hours. Spectral data were collected on a Nikon C1si spectral confocal scope utilizing a 488nm laser for excitation and collecting emitted light at 5nm intervals from 500nm to 660nm.
In vitro assay of ACS function
The acs-3 cDNA was cloned into the bacterial expression vector pBAD-DEST49 (Invitrogen) allowing for L-arabinose inducible expression of the ACS-3 protein with an N-terminal thioredoxin tag to improve solubility, and a C-terminal 6x His tag. This construct was expressed from E. coli, and purified with nickel resin. ACS activity was assayed using an enzyme coupled colorimetric assay, modified for use in 96-well format (Knoll et al., 1994). ACS activity generates acyl-CoA species from free fatty acid and CoA. Bacterial acyl-CoA oxidase in the reaction mix acts on acyl-CoA, generating H2O2. Catalase in the assay solution produces formaldehyde from H2O2 and methanol. Formaldehyde reacts with 4-amino-3-hydrazino-5-mercapto-1,2,4-triazole and IO4− to generate a purple dye that can be detected at 550 nm. We empirically determined the absorption coefficient of the resulting dye to be 3×107 M−1 0.5cm−1 and used this to calculate the amount of fatty acyl-CoA produced in each experiment. We generated a standard curve and verified that absorption at 550 nm is linear with acyl-CoA concentration within the range tested. Commercially available Pseudomonas ACS protein (Sigma) was used as a positive control. All control assays were performed three times, while experimental assays using ACS-3 protein were performed five times.
Generation of transgenic animals
To assess the expression pattern of acs-3, 2.5kb of the upstream regulatory sequence of T08B1.6 amplified by PCR and fused to GFP unc-54 3′ UTR. This product was injected into N2 adults at 5 ng/μl with a rol-6 coinjection marker to generate strain KQ341. To examine localization of the ATGL protein, we cloned the C05D11.7a cDNA into a Gateway donor vector (Invitrogen). This was recombined with the vha-6 promoter into a Gateway destination vector containing GFP and the unc-54 3′ UTR for intestinal expression. This construct was injected into acs-3(ft5) mutant animals at 5ng/ul with Ptph-1::GFP as a coinjection marker to generate strain KQ1982. To rescue acs-3(ft5), and determine subcellular localization of the ACS-3 protein, 2.5kb of the upstream regulatory sequence was cloned into a Gateway Donor vector, recombined with the T08B1.6 cDNA (Open Biosystems) into a Gateway destination vector containing GFP and the unc-54 3′ UTR, and injected into acs-3(ft5) adults at a concentration of 3 ng/μl with Podr-1::RFP (gift of C. Bargmann) as a coinjection marker to generate strain KQ519. To determine subcellular localization of the mutant ACS-3 protein, the ft5 mutation was introduced by site directed mutagenesis into the T08B1.6 cDNA Gateway donor vector, which was recombined with the donor vector containing 2.5kb of T08B1.6 upstream regulatory sequence into a GFP destination and injected into N2 animals at a concentration of 2 ng/μl with a Pmyo-2::mCherry coinjection marker to generate strain KQ1332. For seam cell rescue, 1.4kb of upstream regulatory sequence from wrt-2 (Open Biosystems), or 0.9kb upstream regulatory sequence of grd-10, was recombined with the T08B1.6 cDNA into a GFP destination vector, and injected at 5 ng/μl with Podr-1::RFP coinjection marker into acs-3(ft5) generate line KQ579 (wrt-2) and KQ577 (grd-10). For body-wall specific expression, 2kb of myo-3 upstream regulatory sequence (Open Biosystems) was recombined with T08B1.6 cDNA into a GFP destination vector and injected at 5 ng/μl with a Pmyo-2::mCherry coinjection marker into acs-3(ft5) animals to generate line KQ1260. For intestine specific expression, 2kb of elt-2 upstream regulatory sequence (Open Biosystems) was recombined with the T08B1.6 cDNA into a GFP destination vector and injected at 5 ng/μl with a Pmyo-2::mCherry coinjection marker into acs-3(ft5) animals to generate line KQ1335. To examine potential mitochondrial localization of ACS-3, we generated a construct driving the mitochondrial protein clk-1 (Open Biosystems) with the wrt-2 seam cell promoter, and fused to mCherry. This construct was injected into acs-3(ft5) mutant animals at 5ng/ul with Ptph-1::GFP as a coinjection marker to generate line KQ795. To examine potential colocalization, this line was crossed with line KQ579, carrying Pwrt-2::acs-3::GFP. To confirm acs-3 is expressed in seam cells, we generated a Pacs-3::mCherry promoter fusion construct utilizing 2.5kb of acs-3 5′ regulatory sequence, and injected this into strain JR667, which carries an integrated SCM::GFP marker.
Fatty acid uptake assay
We developed an assay similar to that reported by Spanier et al, 2009. Synchronized L1 animals were plated on NGM plates seeded with OP50 and grown at 25°C to the young adult stage. The pellet from a saturated OP50 culture was resuspended in an equal volume of S-Basal medium to generate assay solution. To this solution 0.75 μl of a 1mg/ml stock solution of C1-BODIPY 500/512 C12 in DMSO was added per ml. Animals were washed off the plates and rinsed 2x in S-Basal medium. After the final rinse, animals were allowed to settle to the bottom of a 15ml conical tube, then a 30 μl aliquot was added 0.5 ml of assay solution in a foil wrapped 1.5ml tube. The animals were incubated at room temperature for 20 minutes while rotating. Following incubation, animals were allowed to settle to the bottom of the tube for 1 minute, then pipeted onto a slide containing an agar pad and pre-chilled on a cold aluminum block. For quantification of uptake, images were collected using a 5x objective. The same circular region of interest (ROI) that encompasses one entire animal was applied to each of these images. The minimum fluorescence value was subtracted from the mean of the ROI to correct for background signal. For each experiment, 8-12 animals of each strain were imaged and analyzed.
Measurement of de novo fatty acid synthesis and fatty acid composition
Metabolic labeling was performed essentially as described (Perez and Van Gilst, 2008). Briefly, equal amounts of bacteria grown in either LB (12C media) or isogro (98.5% 13C enriched Sigma) were mixed and plated onto agarose plates. For each sample, 30,000 synchronized L1 animals prepared from bleaching gravid adults were added to these plates and grown for 44-48 at 20° C. Lipids were extracted and fatty acids analyzed as described (Perez and Van Gilst, 2008).
Fatty acid oxidation assay
Synchronized L4 animals were washed off plates with 0.9% NaCl, then rinsed three times in 0.9% NaCl. Following washes, animals were incubated for 20 minutes in 0.9% NaCl to allow animals to empty their intestines and then resuspended in S-basal in a final volume of 540 μl. An aliquot (20 μl) was stored at −80° C for subsequent protein determination. [9,10(n)-3H] Oleic acid (GE Healthcare) was added to worms to a final concentration of 20 μM (specific activity 67-93 Ci/mol), complexed to fatty acid free BSA in a 2:1 molar ratio. Samples were incubated on an orbital shaker for one hour at 20° C. Subsequently, TCA was added to a final concentration of 5% w/v. After mixing, samples were centrifuged (14000xg, 10min) and the supernatant (1ml) was transferred to a new microcentrifuge tubes. Phosphate buffered saline (250 μl) and 5M NaOH (100 μl) were added prior to ion exchange chromatography on Dowex 1×8 (200-400 MESH) ion exchange columns (1ml). Samples were eluted using water (1ml) and the amount of 3H2O in the eluate was determined by scintillation counting. Samples without animals were included as background controls. Addition of NaN3 (10mM) to wild-type animals inhibited fatty acid oxidation to approximately 10%. In addition, feeding animals with fadR bacteria, a strain incapable of taking up fatty acids, did not affect labeled water production, demonstrating that bacterial fatty acid oxidation does not contribute significantly to measured rates (data not shown).
acs-3(ft5) suppressor screen
Synchronized L4 acs-3(ft5) animals were mutagenized with 47mM EMS (ethyl methanesulfonate) essentially as described (Brenner, 1974). Mutagenized animals were rinsed and plated on NGM plates containing OP50. After 24 hours, animals were collected and bleached to obtain F1 embryos. These animals were incubated overnight in S-Basal medium to synchronize the population, split into 10 pools and plated on NGM plates with OP50. 40,000 F1 animals were collected 72 hours later, and their embryos were collected by bleach treatment. F2 animals were synchronized overnight in S-Basal, then plated at a high density on 10cm plates containing 110uM LY294002 and OP50. Plates were screened 72-96 hours later, and animals that had reached the gravid adult stage were picked. These lines were rescreened, yielding seven lines exhibiting moderate to strong suppression.
NHR-25 LBD protein purification and ligand exchange
The 6XHIS-NHR-25 LBD (amino acids 309-541 of the NHR-25a spliceform) was cloned into pBH4 bacterial expression vector and expressed in DE3 pLysS e. coli and purified by Ni-Trap affinity purification followed by Q-ion exchange chromatography and size excluded into 20 mM Hepes (8.0) with 1 mM EDTA, essentially as previously described for NR5A receptors (Krylova et al., 2005; Sablin et al., 2008). Di-palmitoyl (C16) PI(4,5)P2 and PI(3,4,5)P3 (Cayman Chemical Co, Ann Arbor, MI) were solubilized in water, then immediately added from a Hamilton syringe to a solution of purified NHR-25 LBD (10 μM final) at a five-fold molar excess of PIP (50 μM final) in borosilicate glass tubes. Incorporation of PIPs into NHR-25 LBD was allowed to reach equilibrium while nutating at room temperature for 16 hrs under nitrogen, essentially as previously described for mSF-1 LBD (Sablin et al., 2008). Following determination of the elution profile of NHR-25 LBD alone, the apo-NHR-25 LBD and NHR-25 LBD:PIP species were then separated by MonoQ ion exchange chromatography. Peaks were integrated by AKTA FPLC Unicorn 5.1 software, and the percentage of PIP incorporation calculated by dividing the integrated PIP-incorporated peak by the total area under all NHR-25 LBD peaks.
Supplementary Material
Acknowledgements
B.C.M was supported by a Genentech Fellowship and an NSF Graduate Research Fellowship. This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (DK070149) and a Burroughs Wellcome Career Award to K.A. We thank the Bargmann and Vidal labs for plasmids and Matthew Good for assistance with protein purification. We are grateful to members of the Ashrafi Lab as well as Jordan Ward for helpful discussions and critical reading of the manuscript. In addition, we wish to thank Kurt Thorn and the UCSF Nikon imaging center for assistance with and access to a Spectral Confocal microscope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asahina M, Ishihara T, Jindra M, Kohara Y, Katsura I, Hirose S. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells. 2000;5:711–723. doi: 10.1046/j.1365-2443.2000.00361.x. [DOI] [PubMed] [Google Scholar]
- Ashrafi K. Obesity and the regulation of fat metabolism. WormBook. 2007:1–20. doi: 10.1895/wormbook.1.130.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Aspock G, Kagoshima H, Niklaus G, Burglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–923. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J Exp Zool. 1990;253:263–270. doi: 10.1002/jez.1402530305. [DOI] [PubMed] [Google Scholar]
- Babar P, Adamson C, Walker GA, Walker DW, Lithgow GJ. P13-kinase inhibition induces dauer formation, thermotolerance and longevity in C. elegans. Neurobiol Aging. 1999;20:513–519. doi: 10.1016/s0197-4580(99)00094-9. [DOI] [PubMed] [Google Scholar]
- Bakke M, Zhao L, Hanley NA, Parker KL. SF-1: a critical mediator of steroidogenesis. Mol Cell Endocrinol. 2001;171:5–7. doi: 10.1016/s0303-7207(00)00384-1. [DOI] [PubMed] [Google Scholar]
- Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res. 2009;50(Suppl):S91–96. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN, DiRusso CC. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim Biophys Acta. 2007;1771:286–298. doi: 10.1016/j.bbalip.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks KK, Liang B, Watts JL. The influence of bacterial diet on fat storage in C. elegans. PLoS ONE. 2009;4:e7545. doi: 10.1371/journal.pone.0007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang C, Song L, Sommerfeld M, Hu Q. A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. Journal of Microbiological Methods. 2009;77:41–47. doi: 10.1016/j.mimet.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Chen Z, Eastburn DJ, Han M. The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol Cell Biol. 2004;24:7345–7358. doi: 10.1128/MCB.24.17.7345-7358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lewin TM, Van Horn CG, Gonzalez-Baró MR. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J Nutr. 2002;132:2123–2126. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- Eaton S, Bartlett K, Pourfarzam M. Mammalian mitochondrial beta-oxidation. Biochem J. 1996;320(Pt 2):345–357. doi: 10.1042/bj3200345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323(Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG, Hekimi S. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn EJ, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) The Journal of Lipid Research. 2009;50:1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SD, Greenspan P. Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem. 1985;33:833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- Frand A, Russel S, Ruvkun G. Functional genomic analysis of C elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, nile red. The Journal of Lipid Research. 1985;26:781–789. [PubMed] [Google Scholar]
- Greer ER, Pérez CL, Van Gilst MR, Lee B, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, Jäckle H, Kühnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Guh DP, Zhang W, Bansback N, Amarsi Z, Birmingham CL, Anis AH. The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC public health. 2009;9:88. doi: 10.1186/1471-2458-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GD, Ingraham HA. Steroidogenic factor-1: its role in endocrine organ development and differentiation. Frontiers in neuroendocrinology. 1999;20:199–223. doi: 10.1006/frne.1999.0182. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Genetic suppression. WormBook : the online review of C elegans biology. 2005:1–13. doi: 10.1895/wormbook.1.59.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K, Kikuchi T, Mitsuhida N, Kawaguchi A. A new colorimetric method for the determination of free fatty acids with acyl-CoA synthetase and acyl-CoA oxidase. J Biochem. 1981;89:1799–1803. doi: 10.1093/oxfordjournals.jbchem.a133380. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Moulson CL, Pei Z, Miner JH, Watkins PA. Fatty acid transport protein 4 is the principal very long chain fatty acyl-CoA synthetase in skin fibroblasts. J Biol Chem. 2007;282:20573–20583. doi: 10.1074/jbc.M700568200. [DOI] [PubMed] [Google Scholar]
- Jones K, Ashrafi K. Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Disease Models & Mechanisms. 2009;2:224–229. doi: 10.1242/dmm.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, Alimov AP, Rilo HL, Jandacek RJ, Woollett LA, Penberthy WT. A high throughput live transparent animal bioassay to identify non-toxic small molecules or genes that regulate vertebrate fat metabolism for obesity drug development. Nutrition & metabolism. 2008;5:23. doi: 10.1186/1743-7075-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage-Nakadai E, Kobuna H, Kimura M, Gengyo-Ando K, Inoue T, Arai H, Mitani S. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS ONE. 2010;5:e8857. doi: 10.1371/journal.pone.0008857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M, Crawford QT, Seiber M, Wang CY, Han M. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2004;2:E257. doi: 10.1371/journal.pbio.0020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll LJ, Johnson DR, Gordon JI. Biochemical studies of three Saccharomyces cerevisiae acyl-CoA synthetases, Faa1p, Faa2p, and Faa3p. J Biol Chem. 1994;269:16348–16356. [PubMed] [Google Scholar]
- Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Li H, Black PN, DiRusso CC. A live-cell high-throughput screening assay for identification of fatty acid uptake inhibitors. Anal Biochem. 2005a;336:11–19. doi: 10.1016/j.ab.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005b;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Mataki C, Magnier BC, Houten SM, Annicotte JS, Argmann C, Thomas C, Overmars H, Kulik W, Metzger D, Auwerx J, Schoonjans K. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol Cell Biol. 2007;27:8330–8339. doi: 10.1128/MCB.00852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM. C elegans: a model for exploring the genetics of fat storage. Dev Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney BC, Ashrafi K. C. elegans fat storage and metabolic regulation. BBA - Molecular and Cell Biology of Lipids. 2009;1791:474–478. doi: 10.1016/j.bbalip.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2006;133:287–295. doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payrastre B, Missy K, Giuriato S, Bodin S, Plantavid M, Gratacap M. Phosphoinositides: key players in cell signalling, in time and space. Cell Signal. 2001;13:377–387. doi: 10.1016/s0898-6568(01)00158-9. [DOI] [PubMed] [Google Scholar]
- Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Reeves GK, Pirie K, Beral V, Green J, Spencer E, Bull D, Collaboration MWS. Cancer incidence and mortality in relation to body mass index in the Million Women Study: cohort study. BMJ. 2007;335:1134. doi: 10.1136/bmj.39367.495995.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin E, Blind R, Krylova I, Ingraham J, Cai F, Williams J, Fletterick R, Ingraham H. Structure of SF-1 Bound by Different Phospholipids: Evidence for Regulatory Ligands. Molecular Endocrinology. 2008;23:25–34. doi: 10.1210/me.2007-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer JE, Lodish HF. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell. 1994;79:427–436. doi: 10.1016/0092-8674(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Seamen E, Blanchette J, Han M. P-type ATPase TAT-2 negatively regulates monomethyl branched-chain fatty acid mediated function in post-embryonic growth and development in C. elegans. PLoS Genet. 2009;5:e1000589. doi: 10.1371/journal.pgen.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RM, Truksa M, He X, Mckeon T, Weselake R. Simple methods to detect triacylglycerol biosynthesis in a yeast-based recombinant system. Lipids. 2009;44:963–973. doi: 10.1007/s11745-009-3336-0. [DOI] [PubMed] [Google Scholar]
- Spanier B, Lasch K, Marsch S, Benner J, Liao W, Hu H, Kienberger H, Eisenreich W, Daniel H. How the intestinal peptide transporter PEPT-1 contributes to an obesity phenotype in Caenorhabditits elegans. PLoS ONE. 2009;4:e6279. doi: 10.1371/journal.pone.0006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Sadegh L, Elle IC, Christensen AG, Faergeman NJ, Ashrafi K. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 2008;7:533–544. doi: 10.1016/j.cmet.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JM, Zeve D, McKay R, Seo J, Salo Z, Li R, Wang M, Graff JM. Adipose is a conserved dosage-sensitive antiobesity gene. Cell Metab. 2007;6:195–207. doi: 10.1016/j.cmet.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. The Journal of Lipid Research. 2007;48:2736–2750. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol Metab. 2009;20:58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0912308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.