Summary
The essential proteins DnaB, DnaD, and DnaI of Bacillus subtilis are required for initiation, but not elongation, of DNA replication, and for replication restart at stalled forks. The interactions and functions of these proteins have largely been determined in vitro based on their roles in replication restart. During replication initiation in vivo, it is not known if these proteins, and the replication initiator DnaA, associate with oriC independently of each other by virtue of their DNA binding activities, as a (sub)complex like other loader proteins, or in a particular dependent order. We used temperature sensitive mutants or a conditional degradation system to inactivate each protein and test for association of the other proteins with oriC in vivo. We found that there was a clear order of stable association with oriC; DnaA, DnaD, DnaB, and finally DnaI-mediated loading of helicase. The loading of helicase via stable intermediates resembles that of eukaryotes and the established hierarchy provides several potential regulatory points. The general approach described here can be used to analyze assembly of other complexes.
Keywords: Bacillus subtilis, DnaB, DnaD, helicase, replication initiation
Introduction
Faithful replication and its regulation are crucial for genome stability. In many types of bacteria, replication initiates from a single chromosomal origin of replication, oriC, and proceeds bidirectionally around a circular chromosome. Initiation of replication involves binding of the replication initiator and, in bacteria, local melting of the origin DNA, loading of the replicative helicase at the origin leading to further DNA unwinding, and assembly of the rest of the DNA synthesis machinery to form the replisome.
The replication initiator DnaA is found in virtually all bacteria and is functionally analogous to ORC in eukaryotes and archaea. DnaA is an AAA+ ATPase and also functions as a transcription factor (reviewed in Messer, 2002; Kaguni, 2006). DnaA binds to specific sequences in oriC and needs to be in the ATP-bound form to cause melting of an AT-rich sequence necessary for replication initiation (Bramhill & Kornberg, 1988; Kornberg & Baker, 1992; Messer et al., 2001; Speck & Messer, 2001).
After the action of the initiator protein the replicative helicase is loaded onto oriC. Different organisms use different mechanisms for loading helicase. In yeast, loading of the MCM helicase requires two accessory proteins: CDC6 and CDT1 (Sivaprasad et al., 2006). E. coli uses a single protein, DnaC (reviewed in Kornberg & Baker, 1992; Davey & O'Donnell, 2003), to load helicase at oriC and at stalled replication forks (replication restart). In contrast, B. subtilis uses three proteins, DnaB, DnaD, and DnaI, to load helicase at oriC and at stalled forks during replication restart (Bruand et al., 1995; Bruand et al., 2001; Marsin et al., 2001; Polard et al., 2002; Velten et al., 2003; Rokop et al., 2004; Bruand et al., 2005; Ioannou et al., 2006). DnaB, DnaD, and DnaI are conserved in low G+C Gram-positive bacteria and are required for replication initiation in some of these organisms (Bruck & O'Donnell, 2000; Li et al., 2004; Li et al., 2007), indicating that the mechanism of helicase loading used by B. subtilis is likely conserved. It is not known if the B. subtilis helicase loading proteins DnaD, DnaB, and DnaI form a complex or sub-complexes before association with oriC and assembly of helicase, if they associate with oriC independently of each other, possibly through their DNA binding activity, or if there is ordered dependent association of the individual proteins with oriC.
There are several protein-protein and protein-DNA interactions involving DnaA, DnaB, DnaD, and DnaI in B. subtilis. Some, perhaps all, of these interactions are important for replication initiation. In addition to binding specifically to sequences in oriC, DnaA interacts with DnaD in a yeast two-hybrid assay (Ishigo-Oka et al., 2001). DnaB and DnaD bind to dsDNA and ssDNA, and although specific binding sites have not been identified (Marsin et al., 2001; Turner et al., 2004; Zhang et al., 2005; Zhang et al., 2006), it is possible that they associate with oriC independently of other replication initiation proteins. DnaB and DnaD interact weakly (Marsin et al., 2001; Rokop et al., 2004; Bruand et al., 2005). DnaB is membrane-associated (Hoshino et al., 1987; Sueoka, 1998; Rokop et al., 2004) and interacts with helicase and the DnaI-helicase complex and facilitates helicase loading onto DNA (Velten et al., 2003). In addition, it is generally thought that replication initiation occurs at the inner surface of the cell membrane in both E. coli and B. subtilis (Garner et al., 1998; Sueoka, 1998).
Using assays for the association of DnaA, DnaB, DnaD, and helicase with oriC during replication initiation in vivo, we determined which initiation proteins were required for the association of the others. DnaA associates with oriC independently of any of the other replication initiation proteins. However, we found that association of DnaD with oriC was dependent on DnaA, but not on DnaB or DnaI, and association of DnaB with oriC was dependent on DnaA and DnaD, but not on DnaI. Finally, assembly of the helicase was dependent on DnaA, DnaD, DnaB, and DnaI. These results demonstrate a hierarchical order of assembly of replication initiation proteins at oriC. This ordered assembly provides several potential points for regulation.
Results
Rationale and approaches
We determined the dependence and order of association of replication initiation proteins during the early stages of assembly of the replisome at oriC in vivo. Our approach was to inactivate a specific replication initiation protein, using temperature sensitive replication initiation mutants or conditional degradation (see below), allow ongoing rounds of replication to finish and replication to arrest at the execution point of the inactivated protein, and to use a chromatin immunoprecipitation (ChIP) assay to determine which initiation proteins were associated with oriC.
Temperature sensitive mutations
We used temperature sensitive replication initiation mutants and inactivated the mutant protein by shift to non-permissive temperature. Some of these mutants are able to re-initiate replication in a relatively synchronous manner after shift-down to permissive temperature, allowing for a stronger signal in immunoprecipitations in comparison to an asynchronously growing population of cells. In these mutants, we also measured association of the replication proteins with the oriC region after resumption of replication. The use of temperature sensitive mutants took advantage of previously characterized mutations and allowed for the rapid inactivation and re-activation of some of the mutant proteins. However, each of the proteins participates in multiple interactions and functions and it is generally not known if all or only some of these interactions and functions are defective in the temperature sensitive mutants.
Conditional degradation alleles
In contrast to temperature sensitive mutations, degradation of a protein should eliminate all its interactions and functions. We constructed conditional degradation alleles of dnaD, dnaB, dnaI, and dnaC (helicase) and used these to degrade each protein. We inserted a modified E. coli ssrA (ssrA*) at the 3'-end of the target gene. Degradation of the gene product is induced by expression of the E. coli adaptor protein SspB that is required for efficient ClpXP-mediated degradation of the ssrA*-tagged protein (Griffith & Grossman, 2008). In each case, we tested the resulting strains for degradation of the tagged protein, effects on replication, and association of the other proteins with oriC.
Degradation of helicase or helicase-loader components causes an arrest in replication
Strains in which ssrA* was at the 3' end of genes for helicase (dnaC) and the helicase loader components (dnaD, dnaB, and dnaI) had a conditional lethal phenotype upon expression of sspB. In all cases, the ssrA*-tagged gene was the only copy in the cell, was expressed from the native promoter, and supported cell growth. Typically, by 30 min after SspB induction, the amount of the ssrA*-tagged protein was greatly reduced (Fig. 1A–D). In addition, expression of sspB in strains containing the ssrA*-tagged alleles caused a marked decrease in plating efficiency (<1×10−3), consistent with degradation of an essential protein.
Figure 1. Degradation of replication initiation proteins leads to an arrest in replication.
Cells containing the indicated ssrA* fusion were grown in defined minimal medium and expression of the adaptor SspB was induced with either IPTG (from Pspank-sspB) or xylose (from Pxyl-sspB) at time=0. Samples were taken at the indicated times for determination of levels of the indicated proteins (A–D) or DNA synthesis (E).
A–D. Western blot analysis of cell lysates obtained from strains for conditional degradation. Blots were probed for the indicated replication protein and the adaptor SspB. A. DnaD-ssrA* (WKS265); B. DnaB-ssrA* (WKS649); C. DnaI-ssrA* (WKS738); D. DnaC-ssrA* (WKS66).
E. Replication rates were set at 100% for each untreated sample. Relative rate of incorporation of 3H-thymidine into TCA-precipitable counts is plotted as a function of time after expression of sspB or addition of HPUra to block replication. Error bars (standard error of the mean) fall within the symbols of the graphs and are omitted for clarity (n=3).
Degradation of helicase (DnaC-ssrA*) caused a rapid decrease in the rate of replication. Shortly after induction of SspB, the rate of DNA synthesis was strongly reduced (Fig. 1E), as measured by incorporation of 3H-thymidine into DNA. This reduction was comparable to that caused by addition of HPUra (Fig. 1E), an inhibitor of the replicative DNA polymerase PolC (Brown, 1970). This decrease is also comparable to that caused by shifting temperature sensitive helicase mutants (dnaCts) to non-permissive temperature (Karamata & Gross, 1970; Sakamoto et al., 1995). These findings indicate that degradation of the replicative helicase causes DNA synthesis to stop quickly, i.e., a “fast-stop” phenotype. They also indicate that even when helicase-ssrA* is part of the replisome, it is sensitive to SspB-mediated ClpXP degradation.
In contrast to the fast stop in DNA synthesis caused by degradation of helicase-ssrA*, degradation of the initiation proteins DnaD-ssrA*, DnaB-ssrA*, and DnaI-ssrA*, caused a slower, more gradual decrease in replication (a “slow stop” phenotype). Incorporation of 3H-thymidine slowly decreased after induction of SspB (Fig. 1E), similar to the effects of shifting temperature sensitive dnaD, dnaB, and dnaI mutants to the non-permissive temperatures (Karamata & Gross, 1970; Li et al., 2004; Li et al., 2007). Together, our results show that expression of SspB causes rapid degradation of the ssrA*-tagged replication proteins, a defect in growth, and corresponding defects in replication. Since each ssrA*-tagged protein is degraded, all interactions and functions of that protein should be compromised shortly after expression of SspB. We have not used a dnaA-ssrA* allele. Instead, we have used the temperature sensitive dnaA1ts (dnaAts) mutation. The DnaA1 mutant protein is rapidly degraded at non-permissive temperature causing a block in replication initiation (Moriya et al., 1990). It is not known if any of the other replication initiation proteins associate with the oriC region after loss of DnaA.
Association of replication initiation proteins DnaD and DnaB and helicase with oriC depends on dnaA
We found that association of helicase and the replication initiation proteins DnaD and DnaB with oriC depends on DnaA. We shifted the dnaAts mutant to non-permissive temperature (52°C) for 60 min and measured association of DnaA, DnaD, DnaB, and helicase (DnaC) with oriC using ChIP. There was no significant association of any of these proteins with oriC under these conditions (Fig. 2). We were not able to reliably detect association of DnaI with oriC under our experimental conditions, likely because this association is transient (Ioannou et al., 2006).
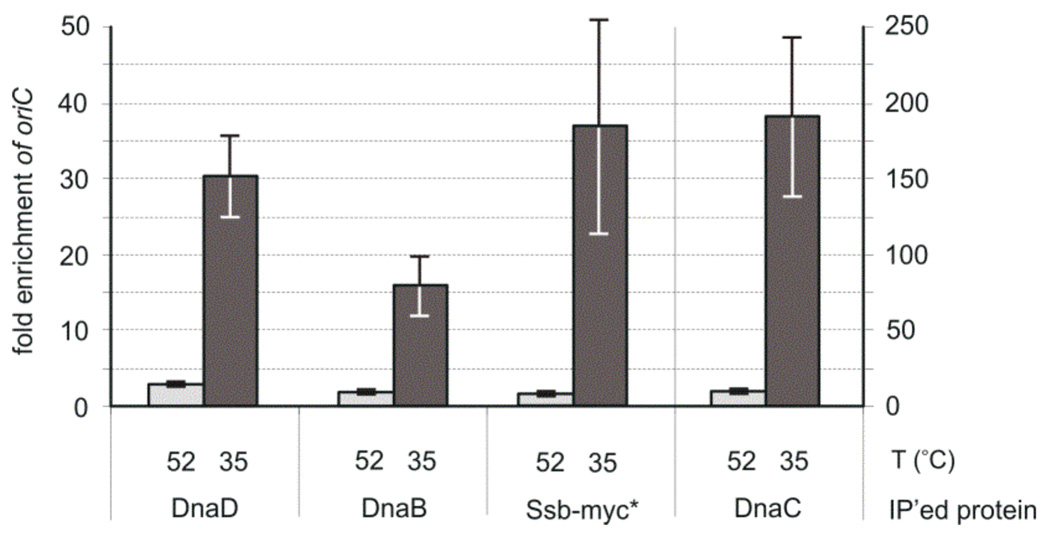
Figure 2. Association of DnaD, DnaB, DnaC (helicase) and Ssb depend on DnaA.
dnaAts mutant cells (WKS588) were grown at permissive temperature (30°C) and shifted to non-permissive temperature (52°C) for 1hr to inactivate DnaA and allow most ongoing rounds of replication to finish (light grey bars). Cells were then shifted to permissive temperature (35°C) in the presence of HPUra (dark grey bars) to allow replication to re-initiate and to trap initiation complexes at oriC. Indicated proteins were immunoprecipitated (IP’ed) from the cell lysates after crosslinking with formaldehyde. Error bars indicate the standard error of the mean (n=3). Note the different scale for the helicase IP on the right axis.
We also tested association of the replication proteins with oriC after release of the replication block, that is, after return of the dnaAts mutant to permissive temperature. Since the DnaA1 mutant protein is unstable (Moriya et al., 1990), re-initiation of replication after shift-down to permissive temperature is asynchronous due to the differences in accumulation of DnaA and concomitant replication initiation in individual cells. This asynchrony makes it difficult to detect association of specific replication proteins at oriC. To restrict movement of the replisome away from the origin and trap the replication complex near oriC after return to the permissive temperature, we simultaneously added the replication inhibitor HPUra (Brown, 1970). We found that there was significant association of DnaD, DnaB, and helicase with oriC by 15 minutes after shift-down to permissive temperature in the presence of HPUra (Fig. 2). We also observed origin-association of the single stranded DNA binding protein Ssb, using an Ssb-myc fusion (Fig. 2), indicative of DNA unwinding at oriC. Together, the results indicate that association of helicase and the replication initiation proteins DnaD and DnaB with oriC depends on dnaA. Association of DnaA with oriC in vivo is independent of each of the other replication initiation proteins (Goranov et al., 2005; Breier & Grossman, 2009).
Association of DnaD with oriC does not require dnaB or dnaI
DnaD binds dsDNA and ssDNA in vitro in the absence of other proteins (Marsin et al., 2001; Turner et al., 2004; Zhang et al., 2005; Zhang et al., 2006). We found that in vivo, specific association of DnaD with oriC was dependent on DnaA (Fig. 2), but not on DnaB or DnaI (Fig. 3A, B). DnaD was associated with oriC in the dnaB134ts (dnaBts) and dnaI2ts (dnaIts) mutants after 60 min at the non-permissive temperature (Fig. 3A). We also monitored association of DnaD with oriC in the dnaB-ssrA* and dnaI-ssrA* degradation mutants. Before induction of SspB, there was some association of DnaD with oriC (Fig. 3B). This level is indicative of the amount of DnaD at oriC in a population of asynchronously growing cells. Degradation of DnaB-ssrA* or DnaI-ssrA* prevented initiation of replication while ongoing replication finished (Fig. 1B), essentially causing all cells to arrest at the start of replication. Under these conditions, when most cells are poised to initiate a new round of replication, association of DnaD with oriC increased relative to that in the asynchronous population (Fig. 3B), indicating that association of DnaD with oriC was independent of DnaB and DnaI. The enrichment of oriC in the ChIP signal was not due to a cross-reacting protein since there was no significant enrichment after degradation of DnaD-ssrA* (Fig. 3B) or in the dnaD23ts (dnaDts) mutant at non-permissive temperature (Fig. 3A).
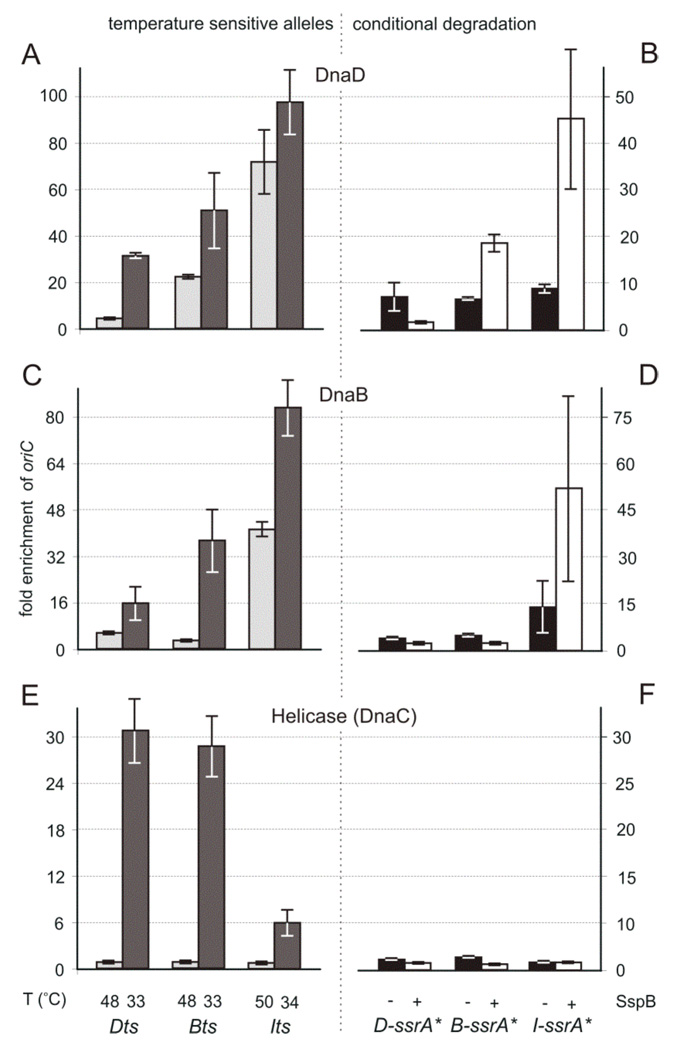
Figure 3. Order of dependence of DnaD, DnaB, and DnaC ( helicase) for association with oriC.
The relative enrichment of oriC after crosslinking and immunoprecipitation of the indicated proteins, DnaD (A, B), DnaB (C, D), and DnaC (E, F), was determined. Note the different scales on the y-axes. The indicated protein (x-axis) was inactivated by shift to non-permissive temperature (A, C, E) or expression of SspB to induce degradation of ssrA*-tagged proteins (B, D, F).
A, C, E. Temperature sensitive mutants included: dnaDts (KPL73), dnaBts (KPL69), and dnaIts (KPL147). Cells were grown at the permissive temperature, shifted to non-permissive (high) temperature (indicated) to inactive the mutant protein and allow ongoing rounds of replication to finish, and then shifted back to low temperature to allow replication to re-initiate. Samples were taken one hour after shift to high temperature (light gray bars) and 2 minutes after shift-down to permissive temperature (dark gray bars).
B, D, F. Conditional degradation mutants included: DnaD-ssrA* (WKS265), DnaB-ssrA* (WKS649), and DnaI-ssrA* (WKS738). Samples were taken for ChIP analyses during asynchronous exponential growth under permissive conditions, i.e., in the absence of inducer and little or no expression of SspB (black bars) and 1 hour after addition of inducer (IPTG or xylose) to induce expression of SspB and cause degradation of the indicated ssrA*-tagged protein. Error bars indicate the standard error of the mean (n=3).
The lack of association of DnaD with oriC in the dnaDts mutant was rapidly reversible. Two minutes after shift-down of the dnaDts mutant cells to permissive temperature, there was association of DnaD with oriC (Fig. 3A). Two minutes after shift-down of the dnaBts and dnaIts mutants to permissive temperature, DnaD remained associated with oriC, at a level similar to that after shift-down in the dnaDts mutant. Together, these results indicate that association of DnaD with oriC requires DnaA but not DnaB or DnaI.
Association of DnaB with oriC requires dnaD, but not dnaI
DnaB is part of the helicase loader and interacts with DnaD (Velten et al., 2003; Rokop et al., 2004; Bruand et al., 2005). DnaB also binds DNA in the absence of the other initiation proteins (Marsin et al., 2001; Zhang et al., 2005; Zhang et al., 2006), and is required for the enrichment of the origin region in membrane fractions (Hoshino et al., 1987; Sueoka, 1998). We found that association of DnaB with oriC was dependent on DnaA (Fig. 2) and DnaD, but not DnaI (Fig. 3C, D). There was little or no association of DnaB with oriC after 60 min at non-permissive temperature in the dnaDts mutant (Fig. 3C). In contrast, there was significant association of DnaB with the oriC region in the dnaIts mutant (Fig. 3C). Similar results were obtained with the dnaD-ssrA* and dnaI-ssrA* mutants. During exponential growth in the asynchronous population, before induction of SspB, there was low but detectable association of DnaB with oriC (Fig. 3D). Sixty minutes after induction of SspB to induce degradation of the DnaD-ssrA*, there was little or no association of DnaB with oriC (Fig. 3D). In contrast, DnaB was associated with oriC following degradation of DnaI-ssrA* (Fig. 3D). The enrichment of oriC in the ChIP signal was most likely due to immunoprecipitation of DnaB and not a cross-reacting protein since the ChIP signal was greatly reduced after degradation of DnaB-ssrA* (Fig. 3D) and in the dnaBts mutant at non-permissive temperature (Fig. 3C).
The lack of association of DnaB with oriC at the non-permissive temperature in the dnaDts and dnaBts mutants was rapidly reversible. Two minutes after shift-down of the dnaDts or dnaBts mutant to permissive temperature, there was significant association of DnaB with oriC (Fig. 3C). Two minutes after shift-down of the dnaIts to permissive temperature, DnaB remained associated with oriC (Fig. 3C). Together, these results indicate that association of DnaB with oriC requires DnaA and DnaD, but not DnaI.
Requirements for association of helicase with oriC
We determined which proteins were required for the association of helicase with oriC. We found that helicase was not detectably associated with oriC in the dnaAts (Fig. 2), dnaBts, dnaDts or dnaIts mutants after incubation at the non-permissive temperature (Fig. 3E), consistent with previous studies of assembly of helicase onto DNA (Bruand et al., 1995; Imai et al., 2000; Bruand et al., 2001; Velten et al., 2003; Rokop et al., 2004; Bruand et al., 2005; Ioannou et al., 2006). During exponential growth in an asynchronous population, we observed little if any association of helicase with oriC (Fig. 3F), as expected for a population of cells at different stages of the replication cycle. Sixty minutes after induction of SspB to induce degradation of the DnaD-ssrA*, DnaB-ssrA*, or DnaI-ssrA*, and to synchronize replication at the initiation stage, there was little or no detectable association of helicase with oriC (Fig. 3F). Together, these results indicate that DnaA, DnaD, DnaB, and DnaI are needed for loading of the replicative helicase at oriC and that the DnaIts and DnaI-ssrA* proteins are inactivated under non-permissive conditions.
In contrast to the lack of helicase association with oriC at non-permissive temperature, there was significant association after release of the replication initiation block. Two minutes after shift-down of the dnaDts and dnaBts mutants to permissive temperature, there was >20-fold enrichment of the oriC region in the immunoprecipitates of helicase (Fig. 3E). Enrichment was lower in the dnaIts mutant after shift-down, probably because of the longer time for replication to initiate and lack of synchrony in this mutant.
Discussion
Ordered assembly of a helicase-loading complex at oriC
B. subtilis has four essential gene products that are needed for replication initiation but not replication elongation: DnaA, DnaB, DnaD and DnaI. Other low-GC Gram-positive organisms appear to use the same four essential products (Bruck & O'Donnell, 2000; Li et al., 2004; Li et al., 2007), and it is therefore believed that the mechanism of replication initiation is conserved. Our work describes a general approach to determine ordered dependence of assembly of a complex in vivo. Applying this approach, we found that there is an ordered hierarchy in the assembly of the replication initiation proteins. DnaA binds to oriC independently of the other initiation proteins (Goranov et al., 2005; Breier & Grossman, 2009). DnaD association with oriC depends on DnaA, but does not require DnaB or DnaI. DnaB association with oriC requires DnaA and DnaD, but not DnaI. Finally, assembly of the helicase depends on all four replication initiation proteins DnaA, DnaD, DnaB, and DnaI. Thus, the order of assembly is DnaA, DnaD, DnaB, then DnaI-helicase.
In addition to their roles in replication initiation at oriC, DnaD, DnaB, and DnaI are needed for replication restart at stalled forks. Much of what is known about DnaD, DnaB, and DnaI is from elegant analyses of replication restart in vitro (Bruand et al., 2001; Marsin et al., 2001; Polard et al., 2002). However, in contrast to replication initiation, replication restart requires PriA and not DnaA, and can occur wherever a replication fork stalls instead of just at oriC. Despite these differences, our findings show that the hierarchal assembly in vivo at oriC is largely similar to the in vitro findings for replication restart.
Conditional degradation system
We used a conditional degradation system that relies on the highly conserved protease ClpXP and its ability to interact with the adaptor protein SspB that facilitates degradation of certain substrates (McGinness et al., 2006; Griffith & Grossman, 2008). The degradation tag that is useful in B. subtilis, ssrA*, was designed to be relatively stable and to require the adaptor protein SspB from E. coli to stimulate rapid degradation (Griffith & Grossman, 2008). Most proteins we have tagged with ssrA* have been functional and are efficiently degraded {e.g., Fig. 1 and (Griffith & Grossman, 2008)}. We anticipate that this system will be broadly applicable for the analysis of many different biological processes and especially for dissecting assembly pathways involving essential proteins. ClpXP orthologs are found in most bacteria, mitochondria and chloroplasts and because of this conservation, the system has the potential to be extended to other organisms.
Model for sequential assembly of a helicase- loading complex
Based on the data presented here, in combination with previous work, we propose the following model for the assembly of the helicase-loading complex in vivo at oriC in B. subtilis, and other Gram-positives (Fig. 4). First, DnaA binds to oriC. Based on in vitro work, this binding event induces local melting of an AT-rich sequence in oriC and melting depends on the presence of DnaA-ATP (Bramhill & Kornberg, 1988; Kornberg & Baker, 1992). Next, DnaD associates with oriC. This association could be by virtue of direct interaction with DnaA, via interaction with the ssDNA or other non-B-type DNA generated by DnaA, or both. Next, DnaB associates with oriC, probably through a direct interaction with DnaD. Because DnaB is associated with the membrane and is required for enrichment of oriC in membrane fractions of cells (Hoshino et al., 1987; Sueoka, 1998), this interaction probably brings the origin region and its associated proteins to the inner surface of the cell membrane. Once this complex is assembled, we propose that a complex of DnaI-helicase interacts with DnaB and the ssDNA in the melted AT-rich region and helicase is assembled into a hexamer encircling the ssDNA.
Figure 4. Model for the association of helicase and helicase loader with oriC in vivo.
The inner surface of the cell membrane is depicted as a gray bar across the top of each panel. DNA is depicted as a double helix with oriC bound to DnaA. Shapes with letters represent the proteins: A=DnaA (initiator), B=DnaB, D=DnaD, I=DnaI (loader ATPase), C=DnaC (helicase). Established protein-protein and protein-DNA interactions (see text) are also shown, except for self-interactions.
Loading of helicase via hierarchical and stable association of proteins is also observed in yeast (Sivaprasad et al., 2006). Loading of the MCM/helicae complex requires two accessory proteins, Cdc6 and Cdt1, in addition to the origin recognition complex ORC. Association of Cdc6 with the origin depends on ORC, but does not require Cdt1 (Bowers et al., 2004; Speck et al., 2005; Randell et al., 2006). Though Cdt1 interacts with ORC directly, it requires Cdc6 for its recruitment to the origin in vivo (Chen et al., 2007). Thus, stable hierarchical assembly during helicase loading seems to be conserved in prokaryotes and eukaryotes.
Temporal and spatial regulation of replication initiation
Replication initiation is highly regulated. In contrast to B. subtilis with four essential replication initiation proteins, E. coli has only two, the initiator DnaA (Marszalek & Kaguni, 1994; Mott et al., 2008) and the E. coli helicase loader DnaC (Baker et al., 1986; Baker et al., 1987; Kornberg & Baker, 1992). Most of the known mechanisms for controlling replication initiation in E. coli affect DnaA and its interactions with oriC. For example, there are mechanisms for sequestration of the oriC region for a period of time after replication initiates (Lu et al., 1994; von Freiesleben et al., 1994), mechanisms for modulating interaction of DnaA with the oriC (Ishida et al., 2004; Keyamura et al., 2007), and mechanisms that couple nucleotide hydrolysis with ongoing replication elongation (Kato & Katayama, 2001). However, the proteins known to modulate replication initiation in E. coli are not widely conserved and are not found in Gram-positive bacteria.
In B. subtilis, DnaA and its interactions with oriC are also likely targets for controlling replication initiation. The level of DnaA is regulated by transcriptional autoregulation (Ogura et al., 2001), like in E. coli (Atlung et al., 1985; Braun et al., 1985). In addition, two different non-essential proteins, Soj and YabA, modulate replication initiation and interact with DnaA (Noirot-Gros et al., 2002; Noirot-Gros et al., 2006; Murray & Errington, 2008), although the mechanisms by which these regulators function are not known.
The existence of three additional replication initiation proteins that assemble in a hierarchical manner at oriC provides additional potential regulatory points. Although the mechanisms of action of regulators of replication initiation in B. subtilis and other Gram-positives are not known, we anticipate that one or more will affect specific steps in the hierarchical assembly of the initiation complex. Consistent with this hypothesis, we have found that a mutation in dnaB (dnaBS371P, also known as dnaB75) affects the frequency of replication initiation in vivo (Rokop et al., 2004). This mutation was isolated as a suppressor of a dnaDts mutation (Rokop et al., 2004; Bruand et al., 2005), and also suppresses the need for priA in replication restart (Bruand et al., 2001). The mutant DnaB protein has increased interaction with DnaD and DnaD is enriched in membrane fractions of cells (Rokop et al., 2004). Moreover, interaction between DnaB and the DnaI-helicase complex stimulates translocase and helicase activities (Velten et al., 2003), whereas DnaD destabilizes the complex of DnaI and helicase in vitro (Turner et al., 2004). Although not yet known, we suspect that there are regulators that normally modulate the interaction between DnaD and DnaB affecting the helicase loading process.
One important consequence of the ordered association of helicase loading proteins described here is to ensure spatial regulation of replication initiation. Replication initiation is thought to take place at the inner face of the membrane. In E. coli, DnaA interacts with the membrane directly (Garner et al., 1998). In addition, E. coli DnaA recruits helicase directly to the origin (Marszalek & Kaguni, 1994; Mott et al., 2008), where it is loaded by a single loader protein. In B. subtilis no evidence exists for a direct interaction between DnaA and helicase or DnaA and the membrane. Yet, like in E. coli, replication initiation is thought to occur at the inner face of the membrane (Winston & Sueoka, 1980; Watabe & Forough, 1987). Enrichment of B. subtilis oriC in membrane fractions depends on DnaB (Hoshino et al., 1987; Sueoka, 1998). DnaD bridges the oriC-DnaA complex and the DnaB-membrane complex, which in turn is responsible for bringing in the DnaI–helicase complex. The use of multiple proteins for helicase loading in vivo, and potentially the regulation of their interactions, allows for both temporal and spatial control and provides a mechanism by which oriC, the membrane, and the replicative helicase are coordinately brought together for replication initiation.
Experimental Procedures
Media and growth conditions
Cells were grown in LB, or defined minimal medium with 0.1% glutamate, supplemented with required amino acids (typically trp and phe), 1% glucose as a carbon source, and 1mM IPTG as inducer as necessary. For strains carrying a xylose-inducible construct (Pxyl), glucose was replaced with arabinose. Xylose was added to 1% to induce expression from Pxyl, as indicated. Single crossover constructs were maintained under antibiotic selection throughout the experiments.
Strains
B. subtilis strains used are listed in Table 1. All are isogenic and contain the trpC2 and pheA1 alleles. dnaA1, dnaB134, dnaD23 and dnaI2 (Karamata & Gross, 1970; Moriya et al., 1990; Bruand et al., 2001; Bruand et al., 2005) are temperature sensitive alleles that prevent replication initiation at the non-permissive temperature. The transposon insertions Tn917ΩHU163, Tn917ΩHU151, and zhb83∷Tn917 are linked to dnaA, dnaD, and the dnaB-dnaI operon, respectively.
Table 1.
B. subtilis strains used.
| Strain | Relevant genotype (reference) |
|---|---|
| KG844 | amyE∷{Pspank-sspB, spc} (Griffith & Grossman, 2008) |
| KG1096 | lacA∷{Pxyl-sspB, tet} (Griffith & Grossman, 2008) |
| KG1098 | amyE∷{Pspank(-7TA)-sspB, spc} (Griffith & Grossman, 2008) |
| KPL2 | dnaA1(ts)-Tn917ΩHU163 (mls) (Burkholder et al., 2001) |
| KPL69 | dnaB134(ts)-zhb83∷Tn917 (mls) (Rokop et al., 2004; Wang et al., 2007) |
| KPL73 | dnaD23(ts)-Tn917ΩHU151 (mls) (Lemon et al., 2000; Rokop et al., 2004; Goranov et al., 2005) |
| KPL147 | dnaI2(ts)-zhb83∷Tn917 (mls) |
| KPL205 | dnaI∷pKL94 (spc) {dnaI-myc} |
| MER454 | dnaD∷pDnaDmyc (spc) {dnaD-myc} (Rokop et al., 2004) |
| WKS8 | dnaI∷pGCS-dnaI (cat) {dnaI-ssrA*} |
| WKS29 | dnaC∷pGCS-dnaC (cat) {dnaC-ssrA*} |
| WKS40 | dnaD∷pGCS-dnaD (cat) {dnaD-ssrA*} |
| WKS66 | amyE∷{Pspank-sspB, spc}, dnaC∷pGCS-dnaC (cat) {dnaC-ssrA*} |
| WKS265 | amyE∷{Pspank(-7TA)-sspB, spc}, dnaD∷pGCS-dnaD (cat) {dnaD-ssrA*} |
| WKS404 | dnaB134-zhb83∷Tn917 (mls), dnaD∷pDnaDmyc (spc) {dnaD-myc} |
| WKS406 | dnaB134-zhb83∷Tn917 (mls), dnaI∷pKL94 (spc) {dnaI-myc} |
| WKS567 | sacA∷{PrpsF-ssb-myc, kan} |
| WKS581 | dnaB∷p1292-dnaB (mls) {dnaB-ssrA* Pspac-dnaI} |
| WKS588 | dnaA1(ts)-Tn917ΩHU163 (mls), sacA∷{PrpsF-ssb-myc, kan} |
| WKS649 | lacA∷{Pxyl-sspB, tet}, dnaB∷p1292-dnaB (mls) {dnaB-ssrA* Pspac-dnaI} |
| WKS738 | amyE∷{Pspank-sspB, spc}, dnaI∷pGCS-dnaI (cat) {dnaI-ssrA*} |
An Ssb-myc strain was constructed by cloning a PCR product carrying the operon promoter (PrpsF), the first gene in the operon, rpsF, and ssb from an ssb-gfp plasmid (Berkmen & Grossman, 2006) and fused in-frame to a linker and a 3xmyc tag and cloned into pSac-Kan (Middleton & Hofmeister, 2004). This construct was integrated into the B. subtilis chromosome at sacA by a double crossover.
ssrA* is a modified Escherichia coli ssrA-tag that allows for user-controlled degradation of the tagged protein by ClpXP when a heterologous adapter protein (SspB) is expressed (Griffith & Grossman, 2008). PCR products carrying a C-terminal fragment of dnaC, dnaD and dnaI were cloned into pKG1268 (Griffith & Grossman, 2008) to give plasmids pGCS-dnaC, pGCS-dnaD and pGCS-dnaI. For dnaB, a similar product was cloned into p1292, a derivative of pMutin2 (Vagner et al., 1998) deleted for lacZ and carrying an ssrA* tag, resulting in p1292-dnaB. The plasmids were integrated by single crossover after natural transformation into wild type B. subtilis (AG174). Chromosomal DNA from these strains was used to introduce the constructs into strains carrying loci for the controlled expression of SspB, and transformants were screened for a growth defect on LB plates with 1mM IPTG (DnaC, DnaD, DnaI) or 1% xylose (DnaB). Although growth rates of tagged and untagged strains were within 10–15% of each other, we found that cultures of dnaD-ssrA* and to lesser extent dnaI-ssrA* mutants accumulated suppressors that were resistant to IPTG. For that reason, experiments were carried out using fresh transformants. Strains containing ts alleles were routinely checked for temperature sensitivity. Expression of SspB in cells without tags caused no detectable phenotypes (Griffith & Grossman, 2008).
Antibodies and chromatin immunoprecipitation (ChIP)
The presence of various proteins in cell lysates was assayed by Western blotting as described (Griffith & Grossman, 2008). Chromatin immunoprecipitation of DNA bound to the various proteins was done essentially as described (Goranov et al., 2005), except that DNA was precipitated in the presence of glycogen (20 µg) as a carrier. For DnaB, DnaC, and DnaD, polyclonal antibodies from rabbit were used (Covance). For Ssb-myc monoclonal anti-cMyc antibodies (Zymed) were used.
We chose to use polyclonal antibodies as we found that epitope-tagged replication proteins, while functional, were not completely wild type and sometimes had synthetic phenotypes with other replication mutations. In addition, the use of polyclonal antibodies allowed us to immunoprecipitate multiple proteins from the same cell extracts, helping to minimize experimental variation. We verified that the antibodies recognized the protein of interest on Western blots comparing the mobilities of untagged and tagged protein and comparing signals before and after degradation of ssrA*-tagged proteins. We also verified that following formaldehyde-mediated crosslinking, each antiserum was able to deplete the protein from a cell extract under ChIP conditions. Furthermore, and most importantly, for all of the replication initiation proteins, association of the protein of interest with oriC was severely decreased or eliminated when the protein was inactivated or degraded (see above).
Quantative real time PCR (qRT-PCR)
qRT-PCR was performed on a Roche LightCycler 480 II. 2 µl samples of immunoprecipitated DNA were analyzed in triplicate in a 20 µl reaction volume that contained Sybr green, using primers designed for oriC (5’-GGAGGACGTGATCATACGA-3’ and 5’-TAGGGCCTGTGGATTTGTG-3’) or the yabM locus (5’-TAGGCGTTAAACGGCATTGG-3’ and 5’-GACAGCATGACCGCAATACC-3’) for which no binding was expected (Breier & Grossman, 2009). Signals were analyzed using the LightCycler 480 SW 1.5 software (Roche), according to the manufacturer (Advanced RelQuant; 2nd derivative of Max, using Median Cp for calculation). Signals were normalized against standard curves of a dilution series of total chromosomal DNA obtained from dnaB134ts (KPL69) arrested cells.
Measurement of DNA replication
Replication rates were determined by pulse-labeling exponentially growing cells with 3H-thymidine (70–90Ci/mmol; 1.0 mCi/ml; Perkin-Elmer; 7 µl with 200 µl of culture) for 1 min at 37°C essentially as described (Wang et al., 2007). Trichloroacetic acid-precipitable counts were determined and background was subtracted.
Acknowledgements
We thank K. Griffith for strains, G. Wright for HPUra, S.P. Bell, C. Lee, P. Soultanas, C. Bonilla, and H. Merrikh for comments on the manuscript, and members of the Grossman lab for useful discussions. This work was supported, in part, by a Rubicon fellowship from the Netherlands Organization for Scientific Research to WKS and Public Health Service grant GM41934 from the NIH to ADG.
References
- Atlung T, Clausen ES, Hansen FG. Autoregulation of the dnaA gene of Escherichia coli K12. Mol Gen Genet. 1985;200:442–450. doi: 10.1007/BF00425729. [DOI] [PubMed] [Google Scholar]
- Baker TA, Funnell BE, Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987;262:6877–6885. [PubMed] [Google Scholar]
- Baker TA, Sekimizu K, Funnell BE, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45:53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- Berkmen MB, Grossman AD. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol. 2006;62:57–71. doi: 10.1111/j.1365-2958.2006.05356.x. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Braun RE, O'Day K, Wright A. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell. 1985;40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- Breier AM, Grossman AD. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J Bacteriol. 2009;191:486–493. doi: 10.1128/JB.01294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NC. 6-(p-hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc Natl Acad Sci U S A. 1970;67:1454–1461. doi: 10.1073/pnas.67.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD, Janniere L. Primosome assembly site in Bacillus subtilis. Embo J. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol Microbiol. 2001;42:245–255. doi: 10.1046/j.1365-2958.2001.02631.x. [DOI] [PubMed] [Google Scholar]
- Bruand C, Velten M, McGovern S, Marsin S, Serena C, Ehrlich SD, Polard P. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol Microbiol. 2005;55:1138–1150. doi: 10.1111/j.1365-2958.2004.04451.x. [DOI] [PubMed] [Google Scholar]
- Bruck I, O'Donnell M. The DNA replication machine of a gram-positive organism. J Biol Chem. 2000;275:28971–28983. doi: 10.1074/jbc.M003565200. [DOI] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, O'Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- Garner J, Durrer P, Kitchen J, Brunner J, Crooke E. Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J Biol Chem. 1998;273:5167–5173. doi: 10.1074/jbc.273.9.5167. [DOI] [PubMed] [Google Scholar]
- Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci U S A. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, Grossman AD. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol Microbiol. 2008;70:1012–1025. doi: 10.1111/j.1365-2958.2008.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, McKenzie T, Schmidt S, Tanaka T, Sueoka N. Nucleotide sequence of Bacillus subtilis dnaB: a gene essential for DNA replication initiation and membrane attachment. Proc Natl Acad Sci U S A. 1987;84:653–657. doi: 10.1073/pnas.84.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Ogasawara N, Ishigo-Oka D, Kadoya R, Daito T, Moriya S. Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol Microbiol. 2000;36:1037–1048. doi: 10.1046/j.1365-2958.2000.01928.x. [DOI] [PubMed] [Google Scholar]
- Ioannou C, Schaeffer PM, Dixon NE, Soultanas P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006;34:5247–5258. doi: 10.1093/nar/gkl690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Akimitsu N, Kashioka T, Hatano M, Kubota T, Ogata Y, Sekimizu K, Katayama T. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J Biol Chem. 2004;279:45546–45555. doi: 10.1074/jbc.M402762200. [DOI] [PubMed] [Google Scholar]
- Ishigo-Oka D, Ogasawara N, Moriya S. DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J Bacteriol. 2001;183:2148–2150. doi: 10.1128/JB.183.6.2148-2150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- Karamata D, Gross JD. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol Gen Genet. 1970;108:277–287. doi: 10.1007/BF00283358. [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. Embo J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su'etsugu M, Fujimitsu K, Kagawa W, Yokoyama S, Kurumizaka H, Katayama T. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. University Science Books; 1992. p. 931. [Google Scholar]
- Lemon KP, Kurtser I, Wu J, Grossman AD. Control of initiation of sporulation by replication initiation genes in Bacillus subtilis. J Bacteriol. 2000;182:2989–2991. doi: 10.1128/jb.182.10.2989-2991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kurokawa K, Matsuo M, Fukuhara N, Murakami K, Sekimizu K. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol Genet Genomics. 2004;271:447–457. doi: 10.1007/s00438-004-0996-6. [DOI] [PubMed] [Google Scholar]
- Li Y, Kurokawa K, Reutimann L, Mizumura H, Matsuo M, Sekimizu K. DnaB and DnaI temperature-sensitive mutants of Staphylococcus aureus: evidence for involvement of DnaB and DnaI in synchrony regulation of chromosome replication. Microbiology. 2007;153:3370–3379. doi: 10.1099/mic.0.2007/009001-0. [DOI] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. Early steps of Bacillus subtilis primosome assembly. J Biol Chem. 2001;276:45818–45825. doi: 10.1074/jbc.M101996200. [DOI] [PubMed] [Google Scholar]
- Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- Messer W, Blaesing F, Jakimowicz D, Krause M, Majka J, Nardmann J, Schaper S, Seitz H, Speck C, Weigel C, Wegrzyn G, Welzeck M, Zakrzewska-Czerwinska J. Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading. Biochimie. 2001;83:5–12. doi: 10.1016/s0300-9084(00)01216-5. [DOI] [PubMed] [Google Scholar]
- Middleton R, Hofmeister A. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid. 2004;51:238–245. doi: 10.1016/j.plasmid.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Moriya S, Kato K, Yoshikawa H, Ogasawara N. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells' initiation potential. Embo J. 1990;9:2905–2910. doi: 10.1002/j.1460-2075.1990.tb07481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P. An expanded view of bacterial DNA replication. Proc Natl Acad Sci U S A. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot-Gros MF, Velten M, Yoshimura M, McGovern S, Morimoto T, Ehrlich SD, Ogasawara N, Polard P, Noirot P. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc Natl Acad Sci U S A. 2006;103:2368–2373. doi: 10.1073/pnas.0506914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Imai Y, Ogasawara N, Moriya S. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P, Marsin S, McGovern S, Velten M, Wigley DB, Ehrlich SD, Bruand C. Restart of DNA replication in Gram-positive bacteria: functional characterisation of the Bacillus subtilis PriA initiator. Nucleic Acids Res. 2002;30:1593–1605. doi: 10.1093/nar/30.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Nakai S, Moriya S, Yoshikawa H, Ogasawara N. The Bacillus subtilis dnaC gene encodes a protein homologous to the DnaB helicase of Escherichia coli. Microbiology. 1995;141(Pt 3):641–644. doi: 10.1099/13500872-141-3-641. [DOI] [PubMed] [Google Scholar]
- Sivaprasad U, Dutta A, Bell SP. Assembly of pre-replication complexes. In: Depamphilis M, editor. DNA replication and human disease. Cold Spring Harbor, NY: CSHL Press; 2006. pp. 63–88. [Google Scholar]
- Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. Embo J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Cell membrane and chromosome replication in Bacillus subtilis. Prog Nucleic Acid Res Mol Biol. 1998;59:35–53. doi: 10.1016/s0079-6603(08)61028-4. [DOI] [PubMed] [Google Scholar]
- Turner IJ, Scott DJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD protein: a putative link between DNA remodeling and initiation of DNA replication. FEBS Lett. 2004;577:460–464. doi: 10.1016/j.febslet.2004.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144(Pt 11):3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional Control of Elongation of DNA Replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe K, Forough R. Effects of temperature-sensitive variants of the Bacillus subtilis dnaB gene on the replication of a low-copy-number plasmid. J Bacteriol. 1987;169:4141–4146. doi: 10.1128/jb.169.9.4141-4146.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston S, Sueoka N. DNA-membrane association is necessary for initiation of chromosomal and plasmid replication in Bacillus subtilis. Proc Natl Acad Sci U S A. 1980;77:2834–2838. doi: 10.1073/pnas.77.5.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis primosomal protein DnaD untwists supercoiled DNA. J Bacteriol. 2006;188:5487–5493. doi: 10.1128/JB.00339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Carneiro MJ, Turner IJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J Mol Biol. 2005;351:66–75. doi: 10.1016/j.jmb.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]