Abstract
The mammalian retina comprises six major neuronal cell types and one glial type that are further classified into multiple subtypes based on their anatomical and functional differences. Nevertheless, how these subtypes arise remains largely unknown at the molecular level. Here, we demonstrate that the expression of Bhlhb5, a bHLH transcription factor of the Olig family, is tightly associated with the generation of selective GABAergic amacrine and Type 2 OFF-cone bipolar subtypes throughout retinogenesis. Targeted deletion of Bhlhb5 results in a significant reduction in the generation of these selective bipolar and amacrine subtypes. Furthermore, although a Bhlhb5-null mutation has no effect on the expression of bHLH-class retinogenic genes, Bhlhb5 expression overlaps with that of the pan-amacrine factor NeuroD and the expression of Bhlhb5 and NeuroD is negatively regulated by ganglion cell-competence factor Math5. Our results reveal that a bHLH transcription factor cascade is involved in regulating retinal cell differentiation and imply that Bhlhb5 functions downstream of retinogenic factors to specify bipolar and amacrine subtypes.
Keywords: Bhlhb5 (Beta3), bHLH, Math5 (Atoh7), NeuroD (Neurod1), Math3 (Neurod4), Amacrine cell, Bipolar cell, Retina, Neurogenesis, Transcription factors, Mouse
INTRODUCTION
In the vertebrate retina, the six major neuronal cell types and one type of Müller glia are arranged in a well-defined laminar structure. Whereas photoreceptors (rod and cones) are located in the outer nuclear layer (ONL), the three types of interneurons (bipolar, horizontal and amacrine cells) and the Müller glial cells comprise the inner nuclear layer (INL). The ganglion cell layer (GCL) is made of retinal ganglion cells (RGCs) and displaced amacrine cells. Based on their anatomical and functional disparities, retinal neurons are further divided into a number of subtypes (Masland, 2001a; Masland, 2001b). For example, there is one rod bipolar (RB) subtype and nine cone bipolar (CB) subtypes in mouse retina based on their synaptic inputs from rod or cone photoreceptors and their sub-laminar localization (Ghosh et al., 2004). Bipolar cells can be further classified as ON and OFF subtypes according to their responses to visual stimuli. RB cells are all of the ON subtype and relay visual information to RGCs indirectly through the AII subtype of amacrine cells. CB cells synapse directly with RGCs and convey both rod- and cone-generated visual information to RGCs. Certain OFF-CB cells also contact rod photoreceptors directly (DeVries and Baylor, 1995; Hack et al., 1999). Amacrine cells are mainly localized to the INL and the GCL, and make synapses with bipolar cells and RGCs. They constitute the most diverse cell type within the retina where they function to modulate synaptic activity between bipolar and ganglion cells (Masland, 2001a; Masland, 2001b). Similarly, amacrine cells are classified into subtypes based on criteria such as sub-laminar localization [the inner plexiform layer (IPL), the GCL, and the innermost layer of the INL known as the amacrine cell layer (ACL)], morphology (e.g. starburst, parasol and midget) and neurotransmitter type (e.g. GABAergic, dopaminergic and cholinergic).
Transcription factors play important roles in retinal cell fate determination and genetic manipulation of these factors often leads to a loss or an alteration of one or more cell fates in the retina (Cepko, 1999; Marquardt, 2003; Marquardt and Gruss, 2002). Bipolar cell fate specification is dependent on the combined action of the homeodomain protein Chx10 and the bHLH factors Mash1 and Math3 (Ascl1 and Neurod4, respectively – both Mouse Genome Informatics) (Burmeister et al., 1996; Hatakeyama et al., 2001). Chx10-null or Mash1-Math3 double-null mice exhibit a complete loss of bipolar cells (Burmeister et al., 1996; Hatakeyama et al., 2001). Studies of Mash1-Math3 double-null retinas also show that Mash1 and Math3 function redundantly to regulate bipolar cell genesis and that loss of the two genes leads to an absence of bipolar cells (Hatakeyama et al., 2001). Gain-of-function studies demonstrate that misexpression of Chx10, Mash1 or Math3 alone does not promote bipolar cell genesis. By contrast, ectopic expression of Mash1 or Math3 together with Chx10 increases the population of mature bipolar cells. Thus, it is hypothesized that whereas Chx10 provides the INL-specific identity, Mash1 and Math3 determine bipolar cell fate (Hatakeyama et al., 2001). Recently, targeted deletion studies have revealed that Vsx1, a paired-like homeodomain factor, and Bhlhb4, an Olig family bHLH factor, are required for the development of the CB and RB subtypes, respectively. Mice lacking Vsx1 are able to generate CB cells. However, the terminal differentiation of CB cells in Vsx1-null retina is incomplete, indicating that Vsx1 is required solely for the late differentiation of OFF-CB cells (Chow et al., 2004; Ohtoshi et al., 2004). Similarly, Bhlhb4 plays no role in the initial generation of bipolar cells, and loss of Bhlhb4 prevents nascent RB cells from maturating and eventually results in their apoptosis (Bramblett et al., 2004).
The expression of Math3, NeuroD (Neurod1 – Mouse Genome Informatics) and Pax6 is associated with differentiating amacrine cells as well as other retinal cells (Jones et al., 1998; Morrow et al., 1999; Nishina et al., 1999). Mutation of NeuroD, Pax6 or Math3 alone does not impair the differentiation of amacrine cells. Conditional deletion of Pax6 in retina results in the formation of only amacrine cells (Marquardt et al., 2001). Although the development of amacrine cells is unaffected in mice lacking either NeuroD or Math3, mice deficient for both Math3 and NeuroD are devoid of amacrine cells (Inoue et al., 2002). Cell lineage analysis studies have demonstrated that retinal progenitors of the Math3 and NeuroD double-null mice fail to differentiate into amacrine cells. Instead of undergoing programmed cell death, these progenitors trans-differentiate into ganglion and Müller glial cells (Inoue et al., 2002). Misexpression of Math3 or NeuroD alone in developing retina does not promote the differentiation of amacrine cells, but results in the formation of rod cells (Inoue et al., 2002). Recently, it has been shown that the winged helix/forkhead transcription factor Foxn4 plays an essential role in the formation of amacrine and horizontal cells (Li et al., 2004). Loss of Foxn4 function leads to the complete absence of horizontal cells and to a reduction in amacrine cells. Interestingly, the expression of NeuroD and Math3 are reduced in Foxn4-null retina, suggesting that Foxn4 acts upstream of NeuroD and Math3 in the amacrine differentiation pathway. The above genetic studies reveal the roles of these transcription factors as pan-amacrine-determining factors. However, the transcription factors responsible for retinal subtype specification remain elusive.
Bhlhb5 is a member of the Olig subfamily of bHLH transcription factors. It has previously been shown to be expressed in a restricted manner in the developing central nervous system (CNS), sensory organs, kidney and hair follicles (Bramblett et al., 2002; Brunelli et al., 2003; Kim et al., 2002; McLellan et al., 2002; Peyton et al., 1996). Due to its inability to bind DNA alone or in combination with other bHLH proteins, Bhlhb5 is thought to function as a negative regulator of other bHLH proteins (Peyton et al., 1996). In vitro evidence has shown that Bhlhb5 represses Pax6 promoter activity through a non-DNA-binding mechanism (Xu et al., 2002). However, the role of Bhlhb5 in embryonic development remains unknown. In this report, we demonstrate that Bhlhb5 is predominantly expressed in post-mitotic cells in the developing mouse retina. Co-localization of Bhlhb5 and retinal cell type-specific markers reveals that Bhlhb5 expression is restricted to selective GABAergic amacrine and Type 2 OFF-CB cells. Targeted deletion of Bhlhb5 leads to a loss of Type 2 OFF-CB and selective GABAergic amacrine cells, implying that Bhlhb5 is indispensable for the formation of these cells. Expression studies of early embryonic retinas have demonstrated that Bhlhb5 is mostly expressed in cells of the NeuroD+ and Math5+ lineage, and that the increased formation of displaced amacrine cells in Math5-null retina is associated with a substantial increase in cells expressing Bhlhb5 and NeuroD. Furthermore, whereas loss of Bhlhb5 has no effect on the expression of NeuroD, and Math5, Mash1 and Ngn1 (Atoh7, Ascl1 and Neurog2, respectively – all Mouse Genome Informatics), Math3 or Chx10, it does lead to the reduced generation of GABAergic amacrine and OFF-CB subtypes. These studies strongly argue for the crucial role of Bhlhb5 as a factor downstream of the bHLH-class of retinogenic factors in the specification of amacrine and bipolar subtypes.
MATERIALS AND METHODS
Animals
The Math5lacZ and Math5GFP knock-in mice were generated previously (Wang et al., 2001). To generate the Bhlhb5lacZ allele, genomic sequences were isolated from a mouse 129S6 (formally 129SvEvTac) BAC library (CHORI) using Bhlhb5 coding sequences as a probe. The Bhlhb5lacZ targeting construct was generated by inserting 4.0 kb of 5′-flanking sequences that ends immediately upstream of the translational initiation codon ATG and 3.6 kb of 3′-flanking sequences into the HindIII and the XbaI sites of the pKI-lacZ vector, respectively (see Fig. S2 in the supplementary material). The knock-in construct removed the entire Bhlhb5 coding sequences and placed the lacZ reporter gene under the control of Bhlhb5 regulatory sequences. To generate Bhlhb5lacZ knock-in mice, an AscI-linearized Bhlhb5lacZ targeting construct was electroporated into AB1 embryonic stem (ES) cells (a gift from Dr A. Bradley, Baylor College of Medicine, Houston, USA). Three targeted ES clones were obtained from a total of 192 G418- and FIAU-resistant ES clones. Genotypes of targeted clones were confirmed by Southern blotting. Two targeted ES clones were injected into C57BL/6J blastocysts to generate mouse chimeras and heterozygous Bhlhb5lacZ/+ mice were generated in a 129S6 and C57BL/6J mixed background as previously described (Gan et al., 1999; Gan et al., 1996). PCR methods were used to genotype mice from subsequent breeding of Bhlhb5lacZ/+ heterozygotes. The PCR primers used to identify the wild-type Bhlhb5 mice were Bhlhb5-2F 5′-GACAGCGACGGCCGCT-3′ and Bhlhb5-2R 5′-GTGCACTGTTTGCAG-3′, and the lacZ knock-in allele 5′-AGGGCCGCAAGAAAACTATCC-3′ and 5′-ACTTCGGCACCTTACGCTTCTTCT-3′. Embryos were designated as E0.5 at noon on the day at which vaginal plugs were observed. All animal procedures in this study were approved by the University Committee of Animal Resources (UCAR) at the University of Rochester.
Histochemistry, immunohistochemistry, in situ hybridization, BrdU labeling and X-Gal staining
Staged mouse embryos were dissected and fixed in 4% paraformaldehyde/PBS (PFA) at 4°C for 1–2 hours. The samples were then embedded and frozen in OCT medium (Tissue-Tek) and sectioned at 14 μm. BrdU pulse-labeling and Hematoxylin and Eosin staining were performed as described (Pan et al., 2005). β-Galactosidase activity was determined by X-Gal staining as previously described (Gan et al., 1999). For in situ hybridization experiments, 20 μm cryosections were used as previously described (Li and Joyner, 2001). The working dilutions and sources of antibodies used in this study were: goat anti-Bhlhb5 (1:1000, Santa Cruz), mouse anti-bromodeoxyuridine (BrdU) (1:50, Developmental Studies Hybridoma Bank), mouse anti-Brn3a (1:100, Santa Cruz), goat anti-Brn3b (1:2000, Santa Cruz), mouse anti-calbindin (1:5000, Sigma), rabbit anti-calretinin (1:2000, Oncogene), rabbit anti-activated caspase-3 (1:200, BD Pharmingen), anti-ChAT (choline acetyltransferase, 1:200, Chemicon), sheep anti-Chx10 (1:200, Exalpha), rabbit anti-cyclin D3 (1:200, Santa Cruz), mouse anti-GAD65 (glutamate decarboxylase 65 kD, 1:200, BD Biosciences), mouse anti-β-galactosidase (LacZ) (1:500, DSHB), rabbit anti-LacZ (1:500, Chemicon), mouse anti-Isl1/2 (1:400, DSHB), goat anti-NeuroD (1:500, E-17, Santa Cruz), rabbit anti-NK3R (1:200) (Chow et al., 2001), rabbit anti-parvalbulmin (1:200, Sigma), rabbit anti-phosphorylated histone H3 (1:400, Santa Cruz), and mouse anti-Pax6 (1:200, DSHB), rabbit anti-PKCα (1:5000, Sigma), rabbit anti-Prox1 (1:1000, Covance), rabbit anti-recoverin (1:200) (Chow et al., 2001), mouse anti-syntaxin (1:5000, Sigma), rabbit anti-TH (tyrosine hydroxylase, 1:200, Chemicon), rabbit anti-Vsx1 (1:100) (Chow et al., 2001). Alexa-conjugated secondary antibodies (Molecular Probes) were used at a dilution of 1:1000. Images were captured with a Zeiss Axioplan microscope. Whole-Mount immunostaining was performed as described (Wang et al., 2001), and was scored using a confocal microscope. To quantify the number of different cell type-specific markers on sections of retina, three or more age-matched retinas were analyzed for each cell type. All data are presented as the mean±s.d. Statistical analysis was performed using two-sample Student’s t test for unequal variances.
RESULTS
Expression of Bhlhb5 during retinal development
The onset of Bhlhb5 expression was detected at E11.5 within the neuroblast layer (NBL) of the central retina (Fig. 1A). As retinogenesis progressed from central to peripheral retina from E12 to E15.5, Bhlhb5 expression expanded to the entire retina with the majority of Bhlhb5+ cells being detected in the proliferating NBL (Fig. 1B–D,J–M). From E17.5 to P0, Bhlhb5 expression became restricted to the GCL and to the inner boundary of the NBL, presumably the newly formed ACL (Fig. 1E,F,N,O). In situ hybridization and immunostaining experiments revealed identical expression profiles for Bhlhb5 mRNA and protein, indicating that Bhlhb5 expression is mostly regulated at the transcriptional level. At P7, Bhlhb5 expression became localized to three distinctive rows: two in the INL and one in the GCL, and this expression pattern was maintained in the adult retina (Fig. 1G–I, arrowheads). In the INL, Bhlhb5+ cells were divided into two groups based on their expression levels and sub-laminar locations: a higher expression level in cells at the inner boundary (presumptive amacrine cells) and a lower level in cells at the outer boundary (presumptive bipolar and/or horizontal cells).
Fig. 1. Expression profile of Bhlhb5 in retinogenesis.

Retinal sections from the indicated developmental stages were immunolabeled with anti-Bhlhb5 (green) and the nuclei counter-stained with Propidium Iodide (PI, red) (A–I) or probed with a Bhlhb5 in situ probe (J–O). The onset of Bhlhb5 expression starts at E11.5 in the central retina (A). At E12.5 to E15.5, Bhlhb5 expression expands toward the peripheral retina and is mostly detected in cells in the NBL (B–D,J–M). At E17.5 to P0, Bhlhb5 expression becomes localized in the inner boundary of the NBL and the GCL (E,F,N,O). Inserts in E and F show the enlarged view of the corresponding boxed regions. At P7 to P28, two rows of Bhlhb5 expression are seen in the INL and one in the GCL (arrowheads, G–I). (P–U) Bhlhb5 expression is mostly observed in post-mitotic cells of the developing retina. Anti-Bhlhb5 (green) labeling of E12.5 retina shows Bhlhb5 in nuclei of cells in the NBL (P) and anti-BrdU (red) labels the nuclei of proliferating cells at S-phase (Q). (R) Overlay image of P and Q. (S-U) Anti-Bhlhb5 (S, red) and anti-phosphorylated histone H3 (PH3) (T, green) show that Bhlhb5+ cells are mostly negative for PH3 labeling. Abbreviations for this and other figures: L, lens; NBL, neuroblast layer; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer. Scale bars: 100 μm.
To test whether Bhlhb5 is expressed in progenitors or in nascent neurons, co-localization of Bhlhb5 with bromodeoxyuridine (BrdU), an S-phase marker, and with phosphorylated histone 3 (Ser-10), an M-phase marker, was performed in E12.5 and E13.5 retinas. Bhlhb5 expression was absent in the vast majority of proliferating cells in Sand M-phase (Fig. 1P–U). The expression of Bhlhb5 in post-mitotic cells in the NBL, and its expression in selective groups of cells of the INL and the GCL at later stages, suggest that Bhlhb5 could play a role in the differentiation of specific retinal subtypes.
Restricted expression of Bhlhb5 in GABAergic amacrine and OFF-CB subtypes
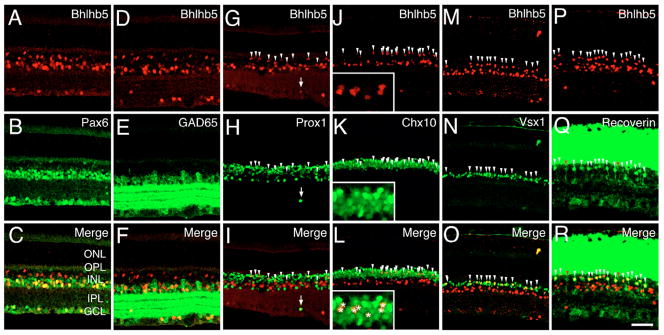
We further determined the identities of Bhlhb5+ cells by co-immunolabeling adult mouse retinas with anti-Bhlhb5 and cell type-specific markers. Strongly anti-Bhlhb5-labeled cells were mostly detected in the ACL and the GCL, and somewhat weakly labeled cells within the outer boundary of the INL (Fig. 2A). Co-labeling of Bhlhb5 with the pan-amacrine cell marker Pax6 demonstrated that all Bhlhb5+ cells in the ACL and in the GCL were Pax6+ (Fig. 2A–C), implying their amacrine identity. The absence of Brn3a (Pou4f1 – Mouse Genome Informatics) and cyclin D3 expression in Bhllhb5+ cells further excluded their identity as ganglion and Müller cells (see Fig. S1A-A″,I-I″ in the supplementary material). To further define the subtypes of Bhlhb5+ amacrine cells, we double-labeled retinas with anti-Bhlhb5 and amacrine subtype-specific markers. Whereas none of the Bhlhb5+ amacrine cells expressed markers for cholinergic amacrine (Isl1 and ChAT) (see Fig. S1B-B″,C-C″ in the supplementary material), dopaminergic amacrine (TH) (see Fig. S1D-D″ in the supplementary material), calretinin (Fig. S1E-E″ in the supplementary material), or AII amacrine (parvalbumin) (see Fig. S1F-F″ in the supplementary material) subtypes, a majority of Bhlhb5+ cells expressed the GABAergic marker GAD65 (Gad2 – Mouse Genome Informatics), indicating that the Bhlhb5+ amacrine cells were mostly of the GABAergic subtype (Fig. 2D–F). In addition, the Prox1+ displaced amacrine cells in the GCL were Bhlhb5+ (Fig. 2G–I, arrows). The co-localization of Prox1+ and Bhlhb5+ cells in the outer boundary of the INL implied their identities as bipolar or horizontal cells (Fig. 2G–I, arrowheads). Nevertheless, Bhlhb5 was not co-expressed with the horizontal cell marker calbindin-28K (see Fig. S1H-H″ in the supplementary material). Co-immunolabeling with anti-Bhlhb5 and an antibody to Chx10, a pan-bipolar cell marker, demonstrated that Bhlhb5 was expressed in bipolar cells (Fig. 2J–L, arrowheads). Careful examination of the double-labeled bipolar cells revealed that Bhlhb5 was expressed in those bipolar cells with a lower level of Chx10 but not those with a higher level of Chx10 (Fig. 2J–L inserts, asterisks). Additional labeling experiments showed that Bhlhb5+ cells did not express the RB cell-specific marker, PKCα (see Fig. S1G-G″ in the supplementary material). Rather, all of these Bhlhb5+ bipolar cells were labeled with the CB-specific marker Vsx1 and represented approximately one third (37%) of the Vsx1+ CB cell population (Fig. 2M–O, arrowheads). As Vsx1 is expressed in 60–70% of all CB cells (Chow et al., 2004), Bhlhb5 is expressed in approximately 21–25% of CB cells. Furthermore, whereas not all recoverin+ Type 2 OFF-CB cells (red arrowheads) expressed Bhlhb5 (white arrowheads), all Bhlhb5+ CB cells were recoverin+ (Fig. 2P–R), confirming these Bhlhb5+ bipolar cells as Type 2 OFF-CB cells. In conclusion, Bhlhb5 is expressed selectively in GABAergic amacrine and Type 2 OFF-CB cells in the adult retina.
Fig. 2. Expression of Bhlhb5 in GABAergic amacrine and OFF-cone bipolar subtypes.
Sections from P28 mouse retinas were double-immunolabeled with anti-Bhlhb5 (red) and subtype-specific markers (green) as indicated. (A–C) The Bhlhb5+ cells in the GCL and the ACL are Pax6+ amacrine cells. (D–F) Bhlhb5 is expressed in GAD65+ GABAergic amacrine cells. (G–I) Bhlhb5 is co-expressed with Prox1 in displaced amacrine cells (arrows) in the GCL and bipolar cells (arrowheads) in the INL. (J–L) Bhlhb5 is co-expressed with Chx10 in bipolar cells (arrowheads). Inserts show the double-labeling of bipolar cells (asterisks) at high magnification. (M–O) All Bhlhb5+ bipolar cells are Vsx1+ cone bipolar cells (white arrowheads). (P–R) All Bhlhb5+ bipolar cells (white arrowheads) are recoverin+ Type 2 OFF-bipolar cells; red arrowheads indicate the OFF-bipolar cells immunoreactive to recoverin only. Scale bar: 50 μm.
Retinal defects in Bhlhb5-null mice
To investigate the role of Bhlhb5 in retinogenesis in vivo, we generated a targeted deletion allele of Bhlhb5 by removing the entire Bhlhb5 ORF (see Fig. S2 in the supplementary material). The resulting heterozygous Bhlhb5lacZ/+ mice were normal and showed no discernible defects. The homozygous Bhlhb5lacZ/lacZ mutants were born indistinguishable from the wild-type or heterozygous littermates at birth. Examination of the offsprings from heterozygous intercrosses revealed that the null mutants were fertile and were born in a normal Mendelian ratio with 46 wild type (19.1%), 130 Bhlhb5lacZ/+ (53.9%) and 65 Bhlhb5lacZ/lacZ (27.0%). However, Bhlhb5-null mice displayed signs of slower weight gain than the wild-type or heterozygous littermates at approximately 3 weeks of age, and developed skin lesions between 1–2 months of age (data not shown).
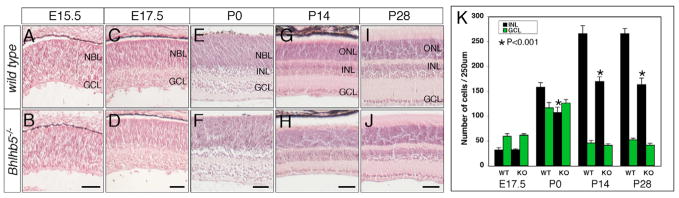
To determine the role of Bhlhb5 in retinogenesis, we first examined the retinas by Hematoxylin and Eosin staining. Whereas no noticeable change in the thickness and laminar organization was found in Bhlhb5-null retinas during embryogenesis (Fig. 3A–F,K), there was a significant reduction in the thickness of the INL postnatally (Fig. 3G–K). Given the restriction of Bhlhb5 expression to selective amacrine and CB subtypes, the decrease in the INL of Bhlhb5-null mice indicated a loss of these interneuron subtypes. Therefore, we analyzed the changes in specific retinal subtypes using subtype-specific markers at P21, a time when all retinal cells are generated and mature in mice. Anti-Pax6 labeling revealed that the total number of amacrine cells was reduced in the INL and the GCL (Fig. 4A,G). Immunolabeling studies demonstrated a loss of 44.2±6% GABAergic amacrine cells, a significant reduction of TH+ amacrine cells, and the absence of Prox1+ displaced amacrine cells, respectively (Fig. 4B–D,H–J and see Fig. S3 in the supplementary material). Although the anti-ChAT immunolabeling was somewhat weak in Bhlhb5-null retina (Fig. 4E,K), the number of ChAT+ cells was unchanged (Fig. 4M,S and see Fig. S3 in the supplementary material). Similarly, no overt changes in the number of AII (Prox1+) and calretinin+ amacrine subtypes were observed (Fig. 4D,F,J,L, and see Fig. S3 in the supplementary material). Among the bipolar subtypes, the Vsx1+ CB subtype was reduced by 36.3±7% (Fig. 4N,T and see Fig. S3 in the supplementary material) whereas the PKCα+ and Goα+ bipolar cells (Fig. 4O,P,U,V) and the Brn3b+ and Brn3a+ RGCs were unaffected (Fig. 4Q,R,W,X). Altogether, our results demonstrated that targeted deletion of Bhlhb5 resulted in the reduction of specific retinal subtypes, particularly the CB, GABAergic and displaced amacrine subtypes that normally express Bhlhb5.
Fig. 3. Developmental abnormality of Bhlhb5-null retinas.
(A–J) Retinal sections from Bhlhb5−/− and wild-type control retinas at indicated developmental stages were stained with Haemotoxylin and Eosin. Compared with the control retina (A,C,E), no overt change in retinal thickness and laminar organization is seen in the mutant (B,D,F) from E15.5-P0. At P14 to P28, the INL of Bhlhb5-null retinas is thinner and the number of cells in the INL is reduced by approximately 40% (G–J). (K) Quantitation of cells in the INL and the GCL per 250 μm length of retinal section at E17.5 to P28. Each bar represents the mean±s.d. for three or more retinas. Scale bars: 50 μm.
Fig. 4. Selective loss of retinal cell subtypes in Bhlhb5-null retinas.

(A–X) Sections from P21 mouse retinas were immunolabeled with subtype-specific markers (green) and nuclear-counterstained with PI (red). Loss of Bhlhb5 leads to a severe loss of amacrine cells immunoreactive for Pax6 (A,G) and GAD65 (B,H) and to an absence of TH+ (C,I) and Prox1+ (D,J) amacrine cells. There is no overt change in the number of amacrine cells immunoreactive to ChAT (E,K), calretinin (F,L) and Isl1 (M,S). A significant loss of Vsx1+ CB cells was observed in Bhlhb5-null retina (N,T). However, no discernible change is seen in the number of PKCα+ RB (O,U) and Goα+ ON-bipolar (P,V) cells, and Brn3b+ (Q,W) and Brn3a+ (R,X) ganglion cells. Scale bar: 100 μm.
Impaired genesis of GABAergic amacrine and CB cells in the absence of Bhlhb5
The close association of Bhlhb5 expression with the genesis of amacrine and bipolar cells suggested a role for Bhlhb5 in regulating retinal cell differentiation. We then tested whether the formation of amacrine and bipolar subtypes was impaired in the absence of Bhlhb5. In mice, most bipolar cells are generated postnatally; Vsx1 expression is first detected in presumptive CB cells from P5 to P6 (Chow et al., 2001), thus serving as a suitable early marker for CB cells. Whereas no overt change in Chx10 expression was observed in bipolar cells and progenitors in Bhlhb5-null retinas (Fig. 5D,H), loss of Bhlhb5 resulted in a reduction of approximately 35% in Vsx1+ CB cells (Fig. 5A,E), a value comparable to the reduction in P21 retinas (Fig. 4N,T). A comparable loss of cells labeled with antibody to recoverin, a Type 2 OFF-bipolar cell marker, and antibody to NK3R (Tacr3 – Mouse Genome Informatics), a Type 1 and 2 OFF-bipolar cell marker (Chow et al., 2001), was also seen in Bhlhb5-null retinas (Fig. 5B,C,F,G). Similarly, immunolabeling of retinal sections at P0 and P6 revealed the agenesis of Prox1+ displaced amacrine cells in the GCL of Bhlhb5-null mice, whereas the generation of Prox1+ horizontal, bipolar and amacrine cells in the INL was not affected (Fig. 5I,J,M,N). Additionally, the number of amacrine cells immunoreactive for Pax6 and GAD65 were greatly reduced in P6 and P7 retinas (Fig. 5K,L,O,P), implying a reduced generation of selective GABAergic subtypes. We also tested the possibility that GABAergic amacrine and OFF-CB subtypes were initially generated but later died of apoptosis in Bhlhb5-null retinas. Anti-activated-caspase-3 immunolabeling revealed no increase in apoptotic cells in Bhlhb5-null retinas from E15.5 to P10 (see Fig. S4A–D in the supplementary material and data not shown). Taken together, targeted deletion of Bhlhb5 specifically diminished the generation of selective OFF-CB and GABAergic amacrine subtypes.
Fig. 5. Decreased genesis of Type 2 cone bipolar, GABAergic and displaced amacrine cells.

(A–H) Immunolabeling of retinal sections at P6 with bipolar subtype-specific markers reveals a dramatic drop in the genesis of CB cells immunoreactive for Vsx1 (A,E), recoverin (B,F, white arrowheads) and NK3R (C,G, white arrowheads) in Bhlhb5-null mice, whereas the total number of bipolar cells labeled by Chx10 is unchanged (D,H). (I,J,M,N) Anti-Prox1 labeling of retinal sections at P0 (I,M) and P6 (J,N) demonstrates the absence of displaced amacrine genesis in the GCL (white arrowheads) in Bhlhb5-null mice, whereas the Prox1+ horizontal cells (red arrowheads) in Bhlhb5-null mice are formed normally. (K,L,O,P) Anti-Pax6 (K,O) and anti-GAD65 (L,P) labeling also demonstrate a significant decrease in the genesis of amacrine cells in the INL. Scale bar: 100 μm.
Upregulation of Bhlhb5 and NeuroD in math5-null retinas
Previous studies have indicated that NeuroD and Math3 play redundant roles in the differentiation of amacrine cells (Inoue et al., 2002). To determine the genetic relationship of Bhlhb5 and NeuroD in the amacrine differentiation pathway, we investigated whether Bhlhb5 co-expressed with NeuroD in developing retinas. Immunolabeling experiments demonstrated that although both NeuroD- and Bhlhb5-expressing cells were similarly distributed throughout the NBL of E13.5 retina, NeuroD was detected in a greater number of cells than Bhlhb5 (Fig. 6A–D), and virtually all Bhlhb5+ cells expressed NeuroD (Fig. 6E,F). We then tested whether the absence of Bhlhb5 could affect the expression of NeuroD and Math3. As shown by anti-NeuroD labeling and in situ hybridization for Math3, the expression of both NeuroD and Math3 was detected mostly in the NBL of developing retinas and was unaltered in Bhlhb5-null retinas (Fig. 6G,H,L,M). Similarly, the expression of other retinogenic factors such as Math5, Ngn2 and Mash1 was unaffected by the targeted deletion of Bhlhb5 (Fig. 6I,K,N-P). Therefore, it is unlikely that Bhlhb5 functions upstream of these retinogenic factors during retinal neurogenesis. Rather, it could act downstream of these factors to control the differentiation of retinal subtypes.
Fig. 6. Normal expression of retinogenic bHLH factors in Bhlhb5-null retinas.
(A–F) Immunolabeling shows a largely overlapping expression of Bhlhb5 (green) and NeuroD (red) in E13 wild-type retina. B, D and F show the enlarged view of the corresponding boxed regions in A, C and E, respectively. (G,L) Anti-NeuroD labeling reveals no change in NeuroD expression in Bhlhb5-null retinas at E13. (H–K,M–P) Similarly, the expression of Math3 (H,M), Ngn2 (I,N), Math5 (J,O) and Mash1 (K,P) is unaffected in Bhlhb5-null retina at E14.5 as assessed by in situ hybridization. Scale bars: 100 μm.
Previously published studies have shown that the loss of RGC-determining factor Math5 in mice, or of lakritz in zebrafish, results in an increase in displaced amacrine cells, suggesting that the expression of Math5 or lakritz suppresses the amacrine cell fate during normal retinogenesis and that null mutations in these genes cause a cell fate conversion from RGC to displaced amacrine cell fates (Kay et al., 2001; Wang et al., 2001; Yang et al., 2003). Bhlhb5 expression in displaced amacrine cell lineages provided us with a tool to examine the molecular basis of this cell fate change. Whole-mount immunolabeling of normal and Math5-null retinas at P21 showed that although the Bhlhb5+ CB cells in the INL were dramatically reduced in Math5-null retinas, the number of Bhlhb5+ displaced amacrine cells was increased sevenfold (Fig. 7A–H). This increased number of Bhlhb5+ cells in Math5-null retina was detected throughout embryogenesis (see Fig. S5 in the supplementary material). We further examined whether Math5 could suppress the generation of amacrine cells by negatively regulating the expression of Bhlhb5 and NeuroD within the Math5+ cell lineage. The expression of Math5 mRNA is transiently detected in progenitors of the NBL and is not suitable to trace Math5+ lineage (Brown et al., 1998; Yang et al., 2003). We have previously shown that the nuclear Math5-LacZ knock-in reporter protein is relatively stable, serving as a suitable marker to trace Math5+ cells in the NBL and in nascent RGCs (Wang et al., 2001). Co-localization of Bhlhb5 and LacZ revealed that loss of Math5 resulted in a significant increase in the number of cells expressing Bhlhb5 in E13 retina and that the majority of these Bhlhb5+ cells expressed LacZ (Fig. 7I–P). A similar increase in retinal cells expressing LacZ and NeuroD was observed in E13.5 Math5-null retina (Fig. 7Q–X). Therefore, the increased generation of displaced amacrine cells in Math5-null retinas corresponded with the premature expression of NeuroD and Bhlhb5 in the Math5+ cell lineage.
Fig. 7. Upregulation of Bhlhb5 and NeuroD expression in the Math5-LacZ cell lineage in Math5-null retina.
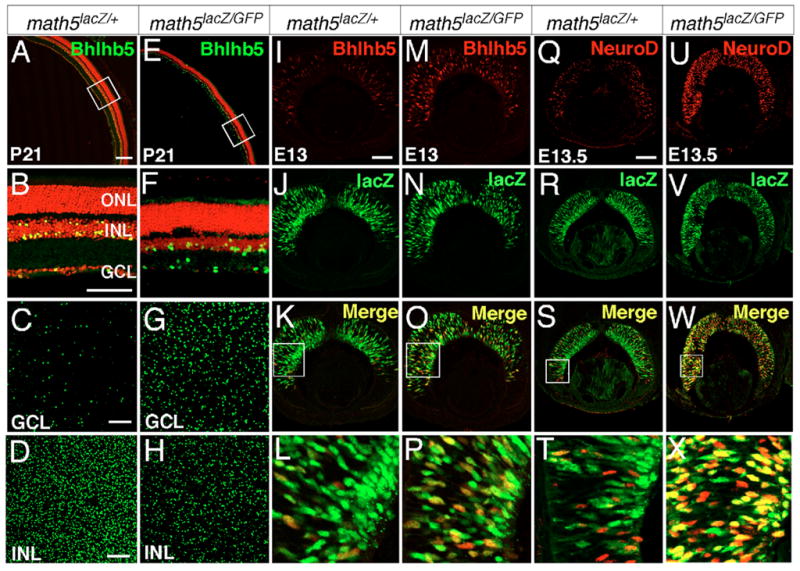
(A–H) Retinal sections immunolabeled with anti-Bhlhb5 (green) and nuclei counterstained with PI (red) show that loss of Math5 results in a large increase in Bhlhb5+ displaced amacrine cells and a significant decrease in Bhlhb5+ bipolar cells. (A,E) Low magnification of retinas at P21. (B,F) Enlarged view of the corresponding boxed regions in A and E, respectively. (C,G) Confocal sections of Bhlhb5+ displaced amacrine cells in the GCL. (D,H) Confocal sections of Bhlhb5+ cells in the INL. (I–P) Co-immunolabeling of Bhlhb5 (red) and LacZ (green) indicates that loss of Math5 results in a significant increase in the number of cells expressing Bhlhb5 in E13 retina and that a majority of these Bhlhb5+ cells express Math5-LacZ. The boxed areas in K and O are shown at high magnification in L and P. (Q–X) Similarly, an increase in retinal cells expressing Math5-LacZ and NeuroD is observed in E13.5 Math5-null mice. The boxed areas of S and W are shown at high magnification in T and X. The Math5-GFP fluorescence is undetectable under the fixation and detection conditions used and all green signals are derived from either LacZ or anti-Bhlhb5 staining. Scale bars: in A, 200 μm for A and E; in all other panels, 100 μm.
DISCUSSION
The expression of specific retinogenic bHLH factors plays crucial roles in the cell fate selection of retinal progenitors. In this report, we have identified that a crucial aspect of retinogenesis, namely the formation of specific amacrine and bipolar subtypes, depends on the activity of Bhlhb5. Targeted disruption of Bhlhb5 causes the selective loss of GABAergic amacrine and Type 2 OFF-cone bipolar cells. Although loss of Bhlhb5 has no impact on the expression of retinogenic bHLH factors, Bhlhb5 expression co-localizes with that of NeuroD. Moreover, Math5 negatively regulates the expression of Bhlhb5 and NeuroD. Therefore, our findings establish a key step in the molecular mechanism of retinogenesis and define a bHLH cascade that connects the regulation of pan-neuronal type determination by retinogenic factors with the subsequent formation of retinal neuronal subtypes.
Bhlhb5 expression as an early, specific marker for GABAergic amacrine and OFF-CB subtypes
Retinal neurons are born in the NBL and migrate to their defined laminar layers within the retina soon after their birth. The early onset and the spatiotemporal profile of Bhlhb5 expression during embryogenesis and early postnatal development coincide with the genesis of amacrine and bipolar cells, respectively (Fig. 1). Our expression studies demonstrate that the expression of Bhlhb5 is unambiguously restricted to selective GABAergic amacrine and Type 2 OFF-CB subtypes (Fig. 2). To the best of our knowledge, Bhlhb5 is the earliest transcription factor to be specifically expressed in these amacrine and bipolar subtypes during early retinogenesis. Thus, Bhlhb5 expression serves as the earliest subtype-specific cellular marker for these cells. Additionally, because not all recoverin+ Type 2 OFF-CB cells express Bhlhb5 or are affected in Bhlhb5-null retinas, Type 2 bipolar cells can be further divided into two subtypes based on Bhlhb5 expression.
Requirement for Bhlhb5 in the generation of Type 2 OFF-CB and selective GABAergic amacrine subtypes
Previous studies have shown that Math3 and Mash1, along with Chx10, play an essential role in regulating the generation of all bipolar cells, but not in defining bipolar subtypes (Burmeister et al., 1996; Hatakeyama et al., 2001). Targeted mutagenesis studies have also revealed that though not playing a role in the initial bipolar genesis, Vsx1 and Bhlhb4 are required for the terminal differentiation and maturation of CB and RB subtypes, respectively (Bramblett et al., 2002; Cheng et al., 2005; Chow et al., 2004; Ohtoshi et al., 2004). A recent study shows that the null mutation of the Iroquois homeobox gene Irx5 in mice causes a partial loss of Type 2 and Type 3 OFF CB cells. However, it is unclear whether these defects result from a failure in their specification or differentiation (Cheng et al., 2005). In this study, we demonstrate that Bhlhb5 expression in the INL is closely associated with the period of bipolar generation in the first postnatal week and that its expression is tightly restricted in Type 2 OFF-CB cells. In Bhlhb5-null retinas, there is a significant reduction in Vsx1+ and recoverin+ Type 2 CB cells (Figs 4, 5), indicating that Bhlhb5 is indeed required for the genesis of a majority of Type 2 OFF-CB cells and that it acts upstream of Vsx1 during CB development. Although the genetic relationship between Bhlhb5 and Mash1 or Math3 needs to be further examined in mice lacking Mash1 and Math3, the unaltered expression of Mash1 and Math3 in Bhlhb5-null retinas suggests that Bhlhb5 is unlikely to function upstream of Mash1 and Math3 (Fig. 6). Instead, given the requirement for Mash1 and Math3 in pan-bipolar development, Bhlhb5 can conceivably play a role downstream of Mash1 and Math3 to regulate the formation of bipolar subtypes.
Similarly, whereas NeuroD and Math3 are essential for the generation of amacrine cells, our studies have shown that Bhlhb5 is expressed only in selective GABAergic and displaced amacrine subtypes, and that targeted deletion of Bhlhb5 leads to a specific reduction in these amacrine subtypes (Fig. 4 and see Fig. S3 in the supplementary material). During early retinogenesis, Bhlhb5 expression in the NBL is mostly confined to cells that express NeuroD and loss of Bhlhb5 does not alter the retinal expression of NeuroD and Math3 (Fig. 6), suggesting that Bhlhb5 is unlikely to function upstream of NeuroD and Math3 in retinogenesis. Although further studies are needed to test whether Bhlhb5 is downstream of NeuroD and Math3 and, in particular, whether its expression in retina is reduced in mice deficient for NeuroD and Math3, it is plausible that Bhlhb5 is expressed in a subset of NeuroD+ cells after they acquire a pan-amacrine identity to render these cells a GABAergic amacrine subtype identity.
During retinal development, all retinal cell types are generated from the same pool of multipotent progenitors. The bHLH-class retinogenic factors have been shown to play crucial roles in regulating the cell fate choices of progenitors and loss of these factors frequently results in cell fate phenotypes. Math3 and NeuroD double-null retinas lack amacrine cells but gain more RGCs and Müller cells (Inoue et al., 2002). Loss of both Mash1 and Math3 leads to a cell fate switch from bipolar to Müller cells (Tomita et al., 2000). In this study, the loss of Type 2 OFF-CB and selective GABAergic amacrine cells in Bhlhb5-null retinas was not accompanied by an overt increase in other retinal cell types (Fig. 4). One explanation for the lack of a readily identifiable cell fate switch is that the amacrine and CB subtypes affected in Bhlhb5-null retinas only represent a small population of retinal cells and that any consequent cell fate change would be less obvious. Additionally, analysis of cell proliferation during retinogenesis revealed a slight decrease in the number of proliferating cells in postnatal Bhlhb5-null retinas (see Fig. S4E–J in the supplementary material). It is possible that the reduced cell proliferation could also contribute to the loss of late-born cells in the INL of Bhlhb5-null retinas. To detect a possible cell fate change, Cre recombinase could be used to replace the Bhlhb5 allele. Cell lineage analysis using Bhlhb5Cre knock-in and lineage-reporter mice could be used to trace the fates of Bhlhb5-expressing cells. Comparison of the retinal cell fates of Bhlhb5+ lineage in the presence and absence of Bhlhb5 would accurately reveal whether Bhlhb5 is exclusively expressed in selective OFF-CB and GABAergic amacrine lineages, and whether Bhlhb5-expressing cells switch fates in the absence of Bhlhb5. Our results have also shown that the Bhlhb5-null mutation leads to a reduction in TH+ dopaminergic amacrine cells (Fig. 4). It is possible that such a reduction results, as a non-cell-autonomous mechanism of Bhlhb5, from the loss of other amacrine and bipolar subtypes. Alternatively, the loss could be through a cell-autonomous mechanism as Bhlhb5 could be transiently expressed in the dopaminergic amacrine cell lineage and be essential for their development. Due to the lack of embryonic and early postnatal markers for dopaminergic amacrine cells, we are unable to distinguish these two possibilities in this study. Future cell lineage analysis with Bhlhb5Cre knock-in mice could effectively show whether Bhlhb5 is expressed in dopaminergic amacrine lineage.
Genetic cascade of bHLH transcription factors in the determination of retinal cell types
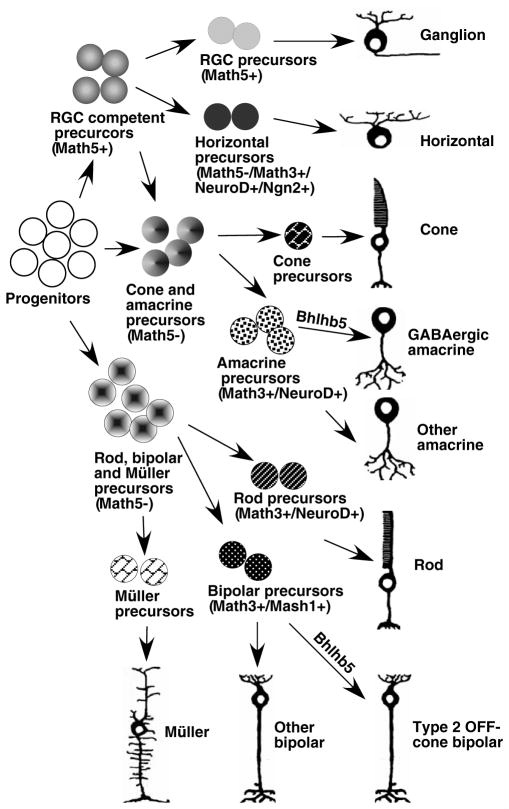
We have previously shown that during normal retinal development, Math5+ cell lineage contributes to retinal cell types including ganglion, cone, horizontal and amacrine cells and that targeted deletion of Math5 leads to a cell fate conversion from RGC to amacrine cells (Wang et al., 2001; Yang et al., 2003). We have hypothesized that in addition to promoting RGC differentiation, Math5 negatively regulates amacrine differentiation pathways by suppressing the expression of key transcription factors, particularly the bHLH-class factors. The upregulation of amacrine factors Bhlhb5 and NeuroD in the absence of Math5 (Fig. 7) provides experimental evidence to support our hypothesis. Moreover, we have demonstrated that removal of Math5 leads to alleviation of inhibition of NeuroD and Bhlhb5 expression in the cells of Math5+ lineage (Fig. 7). Therefore, the suppression of the amacrine differentiation pathway by Math5 could be mediated through its negative regulation of NeuroD and Bhlhb5 expression. Mu et al. have also recently reported that NeuroD expression is upregulated in Math5-null retina (Mu et al., 2005). Furthermore, our expression studies show that a group of cells exists that express NeuroD and Bhlhb5 but not Math5-LacZ in normal and Math5-null retinas. It is not clear whether these cells expressing NeuroD or Bhlhb5 alone arise from Math5+ lineage as Math5-LacZ expression only transiently labels the cells of Math5+ lineage (Wang et al., 2001; Yang et al., 2003). By Math5-Cre-mediated lineage tracing, we have demonstrated that although Math5+ cell lineage produces nearly all RGCs and horizontal cells and a limited number of photoreceptor and amacrine cells, none of the bipolar cells are derived from Math5+ cell lineage. Thus, these Math5− and NeuroD+ or Bhlhb5+ cells are probably derived from a separate, Math5-independent progenitor pool. Taken together, our expression and targeted deletion analyses suggest the following model of retinogenesis (Fig. 8). As a selective pool of retinal progenitors exit the cell cycle, the transient expression of Math5 in these post-mitotic precursors endows them with a short period of RGC competence. During this competence period, Math5 activates a network of transcription factors including Brn3b and Isl1 to initiate the RGC differentiation program (Yang et al., 2003). Additionally, Math5 suppresses the non-RGC differentiation pathways by negatively regulating the non-RGC-specifying factors such as NeuroD and Bhlhb5. As Math5 expression diminishes in the precursors uncommitted to RGC fate, these precursors lose RGC competence, start to express non-RGC-specifying factors and become competent to adopt amacrine, horizontal and cone cell fates. Whereas Math5+ lineage is mostly limited to selective amacrine, horizontal and cone cells, and no bipolar, rod or Müller cells are derived from Math5+ lineage (Yang et al., 2003) (data not shown), there must exist a separate pool of Math5− precursors that never express Math5 but express other retinal cell fate-specifying factors. The highly restricted expression of Bhlhb5 in amacrine and CB subtypes suggests that Bhlhb5 could play a role, downstream of retinogenic bHLH factors, in allowing amacrine- and bipolar-competent precursors to adopt subtype identities by activating the expression of subtype-specific genes such as those encoding GAD65 in GABAergic amacrine cells, and Vsx1, NK3R and recoverin in Type 2 OFF-CB cells. How Bhlhb5 functions at the transcriptional level remains unknown. It is conceivable that Bhlhb5 could dimerize with retinogenic or other bHLH factors to fine-tune their roles in cell differentiation and to allow for subtype distinction. It remains unknown what factors make selective Math5+ cells adopt RGC or non-RGC fates and whether retinal cells generated from Math5+ lineages belong to specific retinal subtypes.
Fig. 8. A model for the role of Bhlhb5 in the generation of GABAergic amacrine and Type 2 CB cells.
The retinal progenitors exit the cell cycle and are divided into the Math5+ and Math5− precursor pools based on the expression of Math5. Precursors with the transient activation of Math5 are RGC-competent. Some of these precursors choose the RGC differentiation pathway and generate nearly all RGCs. The remaining precursors lose RGC-competence when Math5 expression ceases, express other retinogenic bHLH factors and generate horizontal cells or, along with precursors from the Math5− pool, produce amacrine and cone cells. Together with NeuroD and Math3, Bhlhb5 determines the genesis GABAergic amacrine cells. The rod, bipolar and Müller cells are derived from the Math5− pool of precursors and Bhlhb5 expression provides the precursors with competence to differentiate into Type 2 bipolar cells.
Supplementary Material
Acknowledgments
We thank the members of the L.G.’s laboratory for helpful discussions and technical assistance. This work was supported by NIH grants EY013426 and EY015551 to L.G., a Rochester Eye Bank research grant to L.F., the Research to Prevent Blindness unrestricted grant to the Department of Ophthalmology at the University of Rochester, and funding from the Canada Research Chairs Program and the New Investigator Award from the Foundation Fighting Blindness, Canada to R.L.C.
Footnotes
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/133/24/4815/DC1
References
- Bramblett DE, Copeland NG, Jenkins NA, Tsai MJ. BHLHB4 is a bHLH transcriptional regulator in pancreas and brain that marks the dimesencephalic boundary. Genomics. 2002;79:402–412. doi: 10.1006/geno.2002.6708. [DOI] [PubMed] [Google Scholar]
- Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43:779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brunelli S, Innocenzi A, Cossu G. Bhlhb5 is expressed in the CNS and sensory organs during mouse embryonic development. Gene Expr Patterns. 2003;3:755–759. doi: 10.1016/s1567-133x(03)00135-2. [DOI] [PubMed] [Google Scholar]
- Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Chow RL, Lebel M, Sakuma R, Cheung HO, Thanabalasingham V, Zhang X, Bruneau BG, Birch DG, Hui CC, et al. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev Biol. 2005;287:48–60. doi: 10.1016/j.ydbio.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Chow RL, Snow B, Novak J, Looser J, Freund C, Vidgen D, Ploder L, McInnes RR. Vsx1, a rapidly evolving paired-like homeobox gene expressed in cone bipolar cells. Mech Dev. 2001;109:315–322. doi: 10.1016/s0925-4773(01)00585-8. [DOI] [PubMed] [Google Scholar]
- Chow RL, Volgyi B, Szilard RK, Ng D, McKerlie C, Bloomfield SA, Birch DG, McInnes RR. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci USA. 2004;101:1754–1759. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries SH, Baylor DA. An alternative pathway for signal flow from rod photoreceptors to ganglion cells in mammalian retina. Proc Natl Acad Sci USA. 1995;92:10658–10662. doi: 10.1073/pnas.92.23.10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Wang SW, Huang Z, Klein WH. POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol. 1999;210:469–480. doi: 10.1006/dbio.1999.9280. [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wassle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- Hack I, Peichl L, Brandstatter JH. An alternative pathway for rod signals in the rodent retina: rod photoreceptors, cone bipolar cells, and the localization of glutamate receptors. Proc Natl Acad Sci USA. 1999;96:14130–14135. doi: 10.1073/pnas.96.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Jones SE, Jomary C, Grist J, Thomas MR, Neal MJ. Expression of Pax-6 mRNA in the retinal degeneration (rd) mouse. Biochem Biophys Res Commun. 1998;252:236–240. doi: 10.1006/bbrc.1998.9631. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a zebrafish atonal Homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kim MH, Gunnersen J, Augustine C, Tan SS. Region-specific expression of the helix-loop-helix gene BETA3 in developing and adult brains. Mech Dev. 2002;114:125–128. doi: 10.1016/s0925-4773(02)00036-9. [DOI] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Marquardt T. Transcriptional control of neuronal diversification in the retina. Prog Retin Eye Res. 2003;22:567–577. doi: 10.1016/s1350-9462(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001a;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001b;11:431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- McLellan AS, Langlands K, Kealey T. Exhaustive identification of human class II basic helix-loop-helix proteins by virtual library screening. Gene Expr Patterns. 2002;2:329–335. doi: 10.1016/s0925-4773(02)00390-8. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Furukawa T, Lee JE, Cepko CL. NeuroD regulates multiple functions in the developing neural retina in rodent. Development. 1999;126:23–36. doi: 10.1242/dev.126.1.23. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Nishina S, Kohsaka S, Yamaguchi Y, Handa H, Kawakami A, Fujisawa H, Azuma N. PAX6 expression in the developing human eye. Br J Ophthalmol. 1999;83:723–727. doi: 10.1136/bjo.83.6.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtoshi A, Wang SW, Maeda H, Saszik SM, Frishman LJ, Klein WH, Behringer RR. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–536. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Pan L, Yang Z, Feng L, Gan L. Functional equivalence of Brn3 POU-domain transcription factors in mouse retinal neurogenesis. Development. 2005;132:703–712. doi: 10.1242/dev.01646. [DOI] [PubMed] [Google Scholar]
- Peyton M, Stellrecht CM, Naya FJ, Huang HP, Samora PJ, Tsai MJ. BETA3, a novel helix-loop-helix protein, can act as a negative regulator of BETA2 and MyoD-responsive genes. Mol Cell Biol. 1996;16:626–633. doi: 10.1128/mcb.16.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZP, Dutra A, Stellrecht CM, Wu C, Piatigorsky J, Saunders GF. Functional and structural characterization of the human gene BHLHB5, encoding a basic helix-loop-helix transcription factor. Genomics. 2002;80:311–318. doi: 10.1006/geno.2002.6833. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.