Abstract
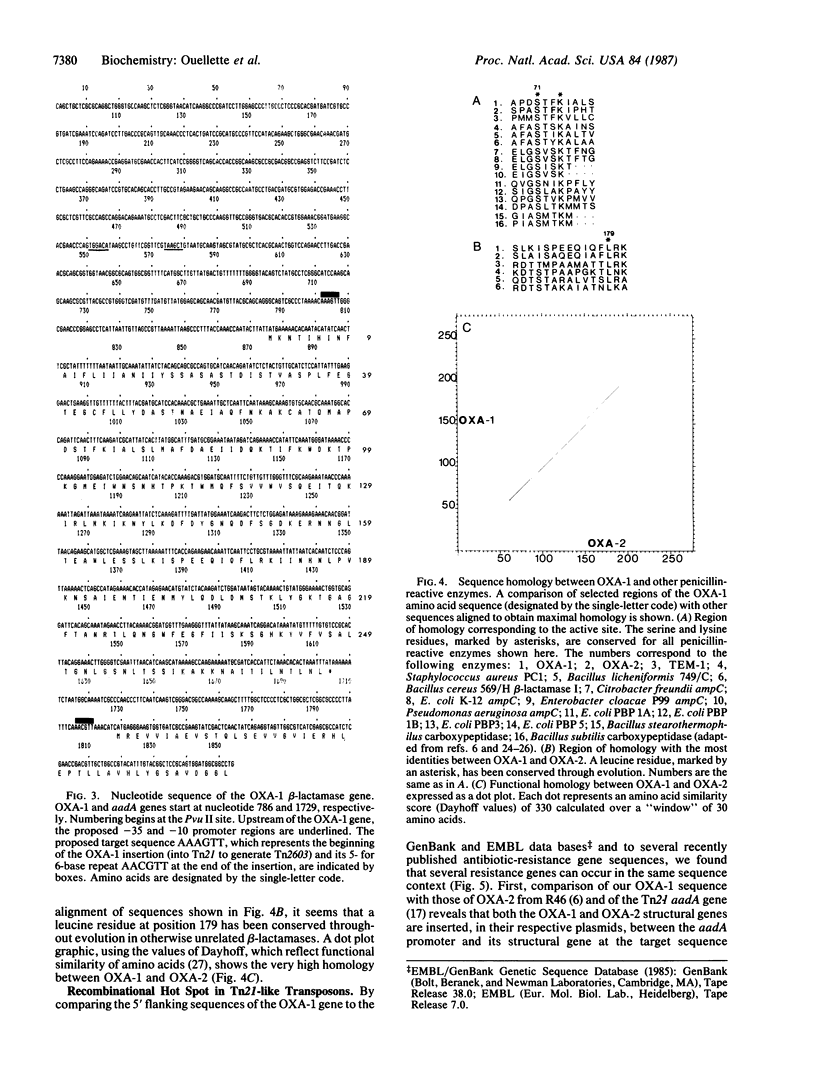
Several plasmid-encoded beta-lactamases are on multiresistance transposable elements. The OXA-1 beta-lactamase gene is part of Tn2603, which is borne on the R plasmid RGN238. We report here the complete nucleotide sequence of the OXA-1 beta-lactamase gene and flanking sequences. The OXA-1 gene shows a greater than 50% sequence divergence from the OXA-2 gene, yet there is significant functional similarity at the peptide level. Analysis of 5' and 3' flanking sequences shows that Tn2603 differs from its probable precursor, Tn21, by a precise 1004-base-pair insertion, containing the OXA-1 structural gene, at the target sequence AAAGTT, which is located between the Tn21 streptomycin/spectinomycin (aadA) promoter and its structural gene. A 5- for 6-base repeat of the target sequence is found at the end of the insertion. The same precise insertion and repeat of the target sequence are found for the OXA-2 gene from R46. The 5' flanking regions of two other genes, the trimethoprim-resistance gene from R388 and the gentamicin resistance (aadB) gene from pDGO100, are greater than 98% homologous to the 5' flanking sequences of the OXA-1, OXA-2, and aadA genes until they diverge at the target sequence. From the available sequence data a recombinational hot spot is defined at the nucleotide level 5' of the aadA gene of Tn21, and a second potential hot spot is proposed 3' of this gene.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Groot Obbink D. J., Ackerman V. P., Hall R. M. Nucleotide sequence of the AAD(2'') aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986 Nov 11;14(21):8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinault A. C., Blakesley V. A., Roessler E., Willis D. G., Smith C. A., Cook R. G., Fenwick R. G., Jr Characterization of transferable plasmids from Shigella flexneri 2a that confer resistance to trimethoprim, streptomycin, and sulfonamides. Plasmid. 1986 Mar;15(2):119–131. doi: 10.1016/0147-619x(86)90048-x. [DOI] [PubMed] [Google Scholar]
- Dale J. W., Godwin D., Mossakowska D., Stephenson P., Wall S. Sequence of the OXA2 beta-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 1985 Oct 21;191(1):39–44. doi: 10.1016/0014-5793(85)80989-3. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fling M. E., Kopf J., Richards C. Nucleotide sequence of the transposon Tn7 gene encoding an aminoglycoside-modifying enzyme, 3"(9)-O-nucleotidyltransferase. Nucleic Acids Res. 1985 Oct 11;13(19):7095–7106. doi: 10.1093/nar/13.19.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1974 Jan;117(1):56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S., Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985 Jan;13(1):17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- Hyde D. R., Tu C. P. tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell. 1985 Sep;42(2):629–638. doi: 10.1016/0092-8674(85)90120-5. [DOI] [PubMed] [Google Scholar]
- Jaurin B., Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck W., Glauner B., Schwarz U., Broome-Smith J. K., Spratt B. G. Sequences of the active-site peptides of three of the high-Mr penicillin-binding proteins of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1999–2003. doi: 10.1073/pnas.82.7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz J., Schmidt F., Wiedemann B. Characterization of Tn2411 and Tn2410, two transposons derived from R-plasmid R1767 and related to Tn2603 and Tn21. J Bacteriol. 1983 Sep;155(3):1333–1342. doi: 10.1128/jb.155.3.1333-1342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Normark S. Sequence of the Citrobacter freundii OS60 chromosomal ampC beta-lactamase gene. Eur J Biochem. 1986 May 2;156(3):441–445. doi: 10.1111/j.1432-1033.1986.tb09601.x. [DOI] [PubMed] [Google Scholar]
- Matthew M. Plasmid-mediated beta-lactamases of Gram-negative bacteria: properties and distribution. J Antimicrob Chemother. 1979 Jul;5(4):349–358. doi: 10.1093/jac/5.4.349. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A. Beta-lactamases. Br Med Bull. 1984 Jan;40(1):18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A., Cohenford M., Jacoby G. A. Five novel plasmid-determined beta-lactamases. Antimicrob Agents Chemother. 1985 May;27(5):715–719. doi: 10.1128/aac.27.5.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. F., Nies B. A., Wiedemann B. Amikacin resistance mediated by multiresistance transposon Tn2424. J Bacteriol. 1983 Aug;155(2):755–760. doi: 10.1128/jb.155.2.755-760.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Roy P. H. Analysis by using DNA probes of the OXA-1 beta-lactamase gene and its transposon. Antimicrob Agents Chemother. 1986 Jul;30(1):46–51. doi: 10.1128/aac.30.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders J. R. Genetics and evolution of antibiotic resistance. Br Med Bull. 1984 Jan;40(1):54–60. doi: 10.1093/oxfordjournals.bmb.a071948. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Klopfer-Kaul I. Evolutionary relationship between Tn21-like elements and pBP201, a plasmid from Klebsiella pneumoniae mediating resistance to gentamicin and eight other drugs. Mol Gen Genet. 1984;197(1):109–119. doi: 10.1007/BF00327930. [DOI] [PubMed] [Google Scholar]
- Schmidt F. The role of insertions, deletions, and substitutions in the evolution of R6 related plasmids encoding aminoglycoside transferase ANT-(2"). Mol Gen Genet. 1984;194(1-2):248–259. doi: 10.1007/BF00383524. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G., Hedge P. J., te Heesen S., Edelman A., Broome-Smith J. K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41(2-3):337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift G., McCarthy B. J., Heffron F. DNA sequence of a plasmid-encoded dihydrofolate reductase. Mol Gen Genet. 1981;181(4):441–447. doi: 10.1007/BF00428733. [DOI] [PubMed] [Google Scholar]
- Tait R. C., Rempel H., Rodriguez R. L., Kado C. I. The aminoglycoside-resistance operon of the plasmid pSa: nucleotide sequence of the streptomycin-spectinomycin resistance gene. Gene. 1985;36(1-2):97–104. doi: 10.1016/0378-1119(85)90073-3. [DOI] [PubMed] [Google Scholar]
- Wiedemann B., Meyer J. F., Zühlsdorf M. T. Insertions of resistance genes into Tn21-like transposons. J Antimicrob Chemother. 1986 Oct;18 (Suppl 100):85–92. doi: 10.1093/jac/18.supplement_c.85. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Baba R., Yamagishi S. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol Gen Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737. [DOI] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]



