SUMMARY
N-alkylated polyamine analogues have potential as anticancer and antiparasitic drugs. However, their metabolism in the host has remained incompletely defined thus potentially limiting their utility. Here, we have studied the degradation of three different spermine analogues N,N′-bis-(3-ethylaminopropyl)butane-1,4-diamine (DESPM), N-(3-benzyl-aminopropyl)-N'-(3-ethylaminopropyl)butane-1,4-diamine (BnEtSPM) and N,N′-bis-(3-benzylaminopropyl)butane-1,4-diamine (DBSPM) and related mono-alkylated derivatives as substrates of recombinant human polyamine oxidase (APAO) and spermine oxidase (SMO). APAO and SMO metabolized DESPM to EtSPD (Km(APAO)=10μM, kcat(APAO)=1.1s−1 and Km(SMO)=28μM, kcat(SMO)=0.8s−1, respectively), metabolized BnEtSPM to EtSPD (Km(APAO)=0.9 μM, kcat(APAO)=1.1s−1 and Km(SMO)=51μM, kcat(SMO)=0.4s−1, respectively), and metabolized DBSPM to BnSPD (Km(APAO)=5.4μM, kcat(APAO)= 2.0s−1 and Km(SMO)=33μM, kcat(SMO)=0.3s−1, respectively). Interestingly, mono-alkylated spermine derivatives were metabolized by APAO and SMO to SPD (EtSPM Km(APAO)=16μM, kcat(APAO)=1.5s−1; Km(SMO)=25μM, kcat(SMO) =8.2s−1; BnSPM Km(APAO)=6.0μM, kcat(APAO)=2.8s−1; Km(SMO)=19μM, kcat(SMO)=0.8s−1, respectively). Surprisingly, E t S P D ( Km(APAO)=37μM, kcat(APAO)=0.1s−1; Km(SMO)=48μM, kcat(SMO)=0.05s−1) and BnSPD (Km(APAO)=2.5μM, kcat(APAO)=3.5s−1; Km(SMO)=60μM, kcat(SMO)=0.54s−1) were metabolized to SPD by both the oxidases. Furthermore, we studied the degradation of DESPM, BnEtSPM or DBSPM in the DU145 prostate carcinoma cell line. The same major metabolites EtSPD and/or BnSPD were detected both in the culture medium and intracellularly after 48 hours of culture. Moreover, EtSPM and BnSPM were detected from cell samples. Present data shows that inducible SMO parallel with APAO could play an important role in polyamine based drug action, i.e. degradation of parent drug and its metabolites, having significant impact on efficiency of these drugs, and hence for the development of novel N-alkylated polyamine analogues.
Keywords: Polyamines, N-alkylated polyamine analogues, Flavin-dependent amino-oxidoreductases, Spermine oxidase, Polyamine oxidase
INTRODUCTION
The polyamines (PA) spermidine (SPD), spermine (SPM) and their diamine precursor putrescine (PUT) are small organic bases, which are ubiquitous in mammalian cells (Tabor and Tabor 1984). Their intracellular levels are strictly regulated by several enzymes and the cell membrane transport system (Seiler 2004). Dysregulated polyamine metabolism has been associated with neoplastic transformation and cancer cell growth (Pegg 1988; Pegg and Feith 2007). Parasitic diseases are an enormous health problem, especially in many developing countries, requiring cost effective drugs. Fortunately, the polyamine metabolic pathway has been found to contain several potential drug targets in parasites (Heby et al. 2007; Kaiser et al. 2006; Muller et al. 2008; Reguera et al. 2005). Thus, targeting differences in polyamine metabolism between host and parasite, or healthy and malignant tissue may offer a selective advantage and avoid significant host toxicity. Therefore, polyamine metabolism or their cellular targets offers a rational basis for drug design against both parasitic diseases and diseases caused by uncontrolled proliferation (Casero and Marton 2007; Heby et al. 2007).
Various polyamine analogues and polyamine synthesis inhibitors have been investigated as chemopreventive, chemotherapeutic or antiparasitic agents (Pegg et al. 1995; Seiler 2003; Williams 1997). It is known that relatively modest structural modifications in the PA backbone or incorporation of terminal N-alkyl substituent(s) may evoke significant differences in their chemical and biological behaviour (Bergeron et al. 1997; Byers et al. 1990). Some of the N-alkylated polyamine analogues have already shown promising results in various model systems of chemotherapy (Casero and Marton 2007; Seiler 2003). N-ethyl substituted polyamines, like diethylnorspermine (DENSPM) have displayed cytotoxic activity and are potential therapeutics for cancer, while N-benzyl substituted polyamines have shown to be effective in the treatment for malaria in a mouse model (Bitonti et al. 1989; Casero and Marton 2007; Edwards et al. 1991). Close structural PA analogues mimic their natural counterparts in regulatory functions, deplete natural polyamines in cells and most importantly block the compensatory uptake of polyamines (Bergeron et al. 1997). Unfortunately, most of the clinical trials have failed due to apparent toxicity and/or low efficacy of the studied compounds. One reason for low cell response to antitumor polyamine analogues may be the catabolism of the active drug by different oxidases (Lawson et al. 2002). Interestingly, at the same time, the biological activity of specific polyamine analogues has been shown to originate from the active catabolism of cellular polyamines by SMO to cytotoxic hydrogen peroxide (Pledgie et al. 2005).
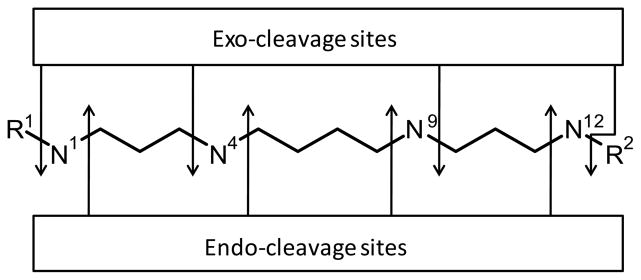
APAO and SMO are two of the key enzymes of polyamine catabolism. APAO was characterized in 1977 when Hölttä purified rat liver APAO enzyme (Hölttä 1977). Since then, APAO has been purified from very few other sources and the human enzyme was cloned in 2003 (Libby and Porter 1987; Tsukada et al. 1988; Wu et al. 2003). SMO (PAOh1) was initially cloned and characterized in 2001 (Wang et al. 2001). Cloning of both the enzymes has made them readily available for recombinant protein production that has facilitated further testing of their substrate properties (Järvinen et al. 2006a; Wang et al. 2003; Wang et al. 2005a). During polyamine catabolism, SPM and SPD are first acetylated by spermidine/spermine N1-acetyltransferase (SSAT) and subsequently oxidized by APAO to produce SPD and PUT, respectively, and to yield stoichiometric amounts of hydrogen peroxide and 3-acetamidopropanal. APAO prefers acetylated polyamines over non-acetylated polyamines, and its natural substrates are N1,N12-DiAcSPM, N1-AcSPM and N1-AcSPD (Seiler 1995; Wang et al. 2005a). In addition to the natural polyamines, some terminally N-alkylated polyamines are degraded by APAO, and some of the otherwise metabolically stable polyamine analogues are catabolized when certain aldehydes are present in the reaction mixture (Järvinen et al. 2005; Järvinen et al. 2006a; Vujcic et al. 2003; Wu et al. 2003). By contrast, SMO possesses very different substrate specificities for the natural polyamines as compared with APAO. It prefers non-acetylated polyamines, efficiently oxidizing SPM, less efficiently N1-AcSPM, and does not use SPD as a substrate (Wang et al. 2003). In addition to APAO and SMO, also other oxidases (such as mono and diamine oxidases) can metabolize polyamines by selectively removing primary N-terminal amine, which will lead to subsequent spontaneous cleavage at the exo side of the N4-amine of SPM (Fig. 1.) (Lee and Sayre 1998; Seiler 2004; Agostinelli et al 2004).
Fig 1.
Endo- and exo-cleavage sites of N-alkylated PA analogue are shown with arrows. R1, R2 = Et and/or Bn.
Although APAO and SMO may significantly affect the efficacy of polyamine-based drugs, metabolic studies with purified enzymes and N-alkylated PA analogues have been very limited (Lawson et al. 2002; Wang et al. 2003; Wang et al. 2005a; Wu et al. 2003). A key for detailed metabolic studies has been the development of a novel accurate LC-MS/MS method for determining polyamines, N-alkylated analogues and their metabolites from biological samples. Using LC-MS/MS method we were able to verify the complicity of APAO-mediated degradation of DESPM (Häkkinen et al. 2007; Häkkinen et al. 2008).
In this study we have thoroughly investigated the substrate properties of N,N′-bis-(3-ethylaminopropyl)butane-1,4-diamine (DESPM), N-(3-benzyl-aminopropyl)-N'-(3-ethylaminopropyl)butane-1,4-diamine (BnEtSPM) and N,N′-bis-(3-benzylaminopropyl)butane-1,4-diamine (DBSPM) and their predicted secondary metabolites with APAO and SMO. In addition, experiments were performed to evaluate the cellular accumulation, catabolism, and excretion of the metabolites of N,N’-bis-alkylated spermine analogues in DU145 prostate cancer cells. Results of this study provide important information of the substrate properties of these two catabolic enzymes that will benefit future drug design.
MATERIALS AND METHODS
Reagents
Ultra-gradient HPLC-grade acetonitrile (ACN) was from J.T. Baker, heptafluorobutyric acid (HFBA, >99%) from Fluka, benzonitrile from Lancaster, and succinonitrile-d4 and benzylamine from Aldrich. Formic acid, glycine and NaOH were from Sigma. Ultrapure water was prepared using a Milli-Q Gradient system (Millipore, Milford, MA, USA). Human recombinant APAO and SMO were produced as described earlier (Järvinen et al. 2005). APAO and SMO were stored frozen in 20% glycerol at –80°C, until use. Some loss of activity was detected during storage, thus 250 μM SPM or N1-AcSPD activity controls in triplicates were always included for all the analyses. Propane-1,3-diamine dihydrochloride (DAP, 98%) was from Aldrich. Butane-1,4-diamine dihydrochloride (PUT, 98%), N1-(3-aminopropyl)butane-1,4-diamine trihydrochloride (SPD, 98%), N,N’-bis-(3-aminopropyl)butane-1,4-diamine tetrahydro-chloride (SPM, >95%) were from Sigma. All the deuterated reference compounds except those described below were prepared as previously described (Häkkinen et al. 2009). Benzylamine hydrochloride (BnNH2) was prepared from benzylamine and recrystallized from ethanol-ethyl acetate.
α,α-2H2-Benzylamine hydrochloride (BnNH2-2D) was prepared from benzonitrile (1 g, 9.7 mmol) and LiAlD4 (815 mg, 19.4 mmol) using the previously described methods (Häkkinen et al. 2009) to give BnNH2-2D as colourless crystals. (974 mg, 69 %). 1H NMR (D2O): δ 7.57-7.43 (5H, m); 13C NMR (D2O): δ 135.3, 132.1 (4C), 131.7, 45.4 (quintet, J = 22 Hz).
1,1,2,2,3,3,4,4-2H8-Butane-1,4-diamine dihydrochloride (PUT-8D) was prepared from succinonitrile-d4 (200 mg, 2.38 mmol) by catalytic deuteration essentially as described in (Smith and Daves 1978) to give PUT-8D as colourless crystals. (158 mg, 39 %). 13C NMR (D2O): δ 41.0 (2C, quintet, J = 22 Hz), 25.6 (2C, quintet, J = 19 Hz)
N1-(3-Benzylamino-1,1,2,2-2H4-propyl)butane-1,4-diamine trihydrochloride (BnSPD-4D) was prepared from N1-benzyl-bis-N1,N3-(2-nitrobenzenesulfonyl)-2,2,3,3-2H4-propane-1,3-diamine as described in (Häkkinen et al. 2009) (808 mg, 1.5 mmol) and N-(4-iodobutyl)phthalimide (Järvinen et al. 2006b) (545 mg, 1.65 mmol) using method described in (Häkkinen et al. 2009) to give BnSPD-4D (193 mg, 37 %) as a colourless solid. 1H NMR (D2O): δ 7.55-7.48 (5H, m), 4.29 (2H, s), 3.20 (2H, s), 3.16-3.10 (2H, m), 3.10-3.03 (2H, m), 1.85-1.73 (4H, m); 13C NMR (D2O): δ 133.3, 132.7 (2C), 132.6, 132.2 (2C), 54.1, 49.8, 46.6 (2C, s + m), 41.7, 26.7, 25.6, 24.7 (quintet, J = 20 Hz).
N,N′-bis-(3-Benzylaminopropyl)butane-1,4-diamine tetrahydrochloride (DBSPM) was prepared from N1-benzyl-bis-N1,N3-(2-nitro-benzenesulfonyl)propane-1,3-diamine (Häkkinen et al. 2009) (1 g, 1.87 mmol) and 1,4-diiodobutane (276 mg, 0.89 mmol) using methods described in (Häkkinen et al. 2009) to give DBSPM (211 mg, 45 %) as a colourless solid. 1H NMR chemical shifts as described earlier (Bergeron et al. 2001); 13C NMR (D2O): δ 133.4 (2C), 132.8 (4C), 132.7 (2C), 132.2 (4C), 54.1 (2C), 49.9 (2C), 47.4 (2C), 46.8 (2C), 25.6 (2C), 25.5 (2C).
N,N′-bis-(3-Benzylamino-1,1,2,2-2H4-propyl)butane-1,4-diamine tetrahydrochloride (DBSPM-8D) was prepared from N1-benzyl-bis-N1,N3-(2-nitrobenzenesulfonyl)-2,2,3,3-2H4-propane-1,3-diamine (Häkkinen et al. 2009) (1.13 g, 2.1 mmol) and 1,4-diiodobutane (310 mg, 1 mmol) using the methods described in (Häkkinen et al. 2009) to give DBSPD-8D (318 mg, 59 %) as a colourless solid. 1H NMR (D2O): δ 7.56-7.47 (10H, m), 4.29 (4H, s), 3.20 (4H, s), 3.17-3.08 (4H, m), 1.85-1.73 (4H, m); 13C NMR (D2O): δ 133.3 (2C), 132.7 (4C), 132.6 (2C), 132.2 (4C), 54.1 (2C), 49.8 (2C), 46.6 (4C, s + m), 25.6 (2C), 24.7 (2C, quintet J = 19 Hz).
Instrumentation
LC separations, MS/MS detection and analysis of the compounds were achieved with Agilent 6410 Triple Quad LC/MS equipped with Agilent 1200 Series Binary Pump SL pumping system and Agilent 1200 Autosampler. Data acquisition and analysis were performed using an Agilent MassHunter Workstation software (Agilent Corporation, MA, USA). 1H and 13C NMR spectra were measured on a Bruker Avance (Bruker, Rheinstetter, Germany) 500 DRX spectrometer as described earlier (Häkkinen et al. 2009).
Analytical conditions
LC-MS/MS experiments were based on selected reaction monitoring (SRM) analysis and were performed essentially as described earlier (Häkkinen et al. 2008). The chromatographic separations were carried out using a Phenomenex Gemini reversed phase C18 column (3 μm, 50 mm × 2 mm, 110 Å) protected with a Phenomenex C18 guard column (4 mm × 2 mm). A gradient solvent system consisting of 0.1% (v/v) HFBA in water (solvent A) and 0.1 % (v/v) HFBA in ACN (solvent B) was used as before (Häkkinen et al. 2008), but the gradient was increased from 2–50 % B over 12 min at a flow rate of 0.2 mL/min. Two time segments were used, turning point being 7.5 min. In the first time segment (0–7.5 min) dwell times were set to 100 (10 ion transitions), and after 7.5 min dwell time were 20 (30 ion transitions). The precursor and the product spectra of each polyamine were recorded similarly as described earlier (Häkkinen et al. 2007). Fragmentor voltage value was set to 60 V for DAP, DAP-2D, PUT, PUT-8D, BnNH2 and BnNH2-2D, and to 90 V for rest of the analytes. Precursor ions, selected product ions for quantification and qualification, and collision energy values for all analytes used in the quantitative SRM analysis are given in Table 1. DAP and PUT have only one product ion.
Table 1.
LC-MS/MS properties of polyamines used in this study
| Amine standard | Precursor [M+H]+ | QT | CID (eV) | QL | CID (eV) | Deuterated internal standard | Precursor [M+H]+ | QT | CID (eV) | Retention time |
|---|---|---|---|---|---|---|---|---|---|---|
| DAP | 75 | 58 | 5 | - | - | DAP-2D | 77 | 60 | 5 | 4.4 |
| PUT | 89 | 72 | 5 | - | - | PUT-8D | 97 | 80 | 5 | 4.6 |
| EtDAP | 103 | 86 | 5 | 58 | 15 | EtDAP-2D | 105 | 88 | 5 | 5.7 |
| BnNH2 | 108 | 91 | 10 | 65 | 20 | BnNH2-2D | 110 | 93 | 10 | 6.7 |
| SPD | 146 | 72 | 15 | 112 | 10 | SPD-2D | 148 | 114 | 10 | 7.8 |
| EtSPD | 174 | 72 | 15 | 86 | 15 | EtSPD-2D | 176 | 88 | 15 | 8.2 |
| BnDAP | 165 | 148 | 5 | 91 | 20 | BnDAP-4D | 169 | 152 | 5 | 8.3 |
| SPM | 203 | 129 | 5 | 112 | 15 | SPM-4D | 207 | 133 | 5 | 8.9 |
| EtSPM | 231 | 157 | 10 | 129 | 10 | EtSPM-2D | 233 | 159 | 10 | 9.1 |
| DESPM | 259 | 157 | 10 | 112 | 20 | DESPM-4D | 263 | 159 | 10 | 9.3 |
| BnSPD | 236 | 148 | 15 | 112 | 15 | BnSPD-4D | 240 | 152 | 15 | 9.4 |
| BnSPM | 293 | 219 | 10 | 112 | 20 | BnSPM-4D | 297 | 223 | 10 | 9.9 |
| BnEtSPM | 321 | 219 | 10 | 112 | 20 | BnEtSPM-8D | 329 | 223 | 10 | 10.1 |
| DBSPM | 383 | 219 | 15 | 112 | 20 | DBSPM-8D | 391 | 223 | 15 | 10.8 |
Chemical structures with abbreviated names are shown in Table 2, and the chemical names are given in “Materials and Methods”. QT, quantifier ion; QL, qualifier ion; CID, collision energy
Preparation of standards and quality controls
Standard working solutions (concentrations of 0.03, 0.1, 0.3, 1, 3, 10, 30 and 60 μM) and working solutions for quality control (QC) samples (concentrations of 0.05, 0.2, 2 and 20 μM) in 90 mM glycine-NaOH-formic acid buffer, and internal standard (IS) working solution containing 1 μM of each 14 deuterated amine in water, were prepared essentially as described earlier (Häkkinen et al. 2008).
Two types of calibration standards and QC samples were prepared using these working solutions by adding 50 μl of each STD working solution or QC working solution, 50 μl of 1 μM IS working solution and 25 μl 0.5 % HFBA (calibration curve 1 for enzyme kinetic analyses) or 10 % HFBA (calibration curve 2 for cell culture experiments). Additional HFBA was used to maintain the chromatographic performance of samples containing sulphosalisylic acid. Samples were transferred into Agilent polypropylene vial inserts for the LC-MS/MS analysis. The calibration curves included also a blank sample and a “zero” sample as described earlier (Häkkinen et al. 2008).
Calibration curves and assay validation
Calibration curves were constructed from accurate concentrations of working solutions of each analyte in x-axis, and peak-area ratio sample vs. internal standard in y-axis using 1/x weighted linear or quadratic (for SPD and EtSPM) least-squares regression model. Assay validation was performed according to the FDA guideline for bioanalytical method validation (FDA 2001) essentially as described earlier (Häkkinen et al. 2008).
Enzyme Kinetic Analyses with Human Recombinant APAO and SMO
The kinetic studies were carried out twice in triplicates at 4 to 7 different (SMO 25–250 μM and 100–1000 μM; APAO 10–1000 μM) substrate concentrations. The enzyme stock solution was diluted with 50 mM sodium phosphatebuffer (pH 8.0) (containing 0.1% Triton X-100) before kineticstudies to yield 0.1–2.0 μg/10 μl of APAO or SMO for each reaction mixture. Reactions were carried out in a total volume of 180 μl in the 100 mM glycine-NaOH at pH 9.5 buffer and were allowed to proceed for 2 to 60 min at 37 °C before addition of 20 μl of50 % (v/v) formic acid. All kinetic determinations included a T1/2 reference (incubated half of the reaction time of actual samples to monitor linearity of reaction) and the reaction mixture without enzyme supplements both with the highest studied drug concentration treated identically as analysed sample to exclude non-enzymatic degradation of studied compounds. Moreover, with some selected reaction mixtures preincubation with 100 μM MDL 72,527 was used to inactivate SMO in order to rule out any nonenzymatic degradation of tested drug. LC-MS/MS wasused to determine the concentrations of the polyamines and their analogues as described above. Prior to LC-MS/MS analysis, concentrated samples ( > 50 μM) were diluted with 90 mM glycine-NaOH-formic acid buffer and all samples were passed through a 0.22 μm filter. LC-MS/MS samples were prepared similarly to calibration standards and QC samples (Häkkinen et al. 2008).
SMO activity at different pH in Bis-Tris propane buffer
Fixed 500 μM SPM in 170 mM Bis-Tris propane at pH 7.5, 8.0, 8.5, 9.0, 9.5 or 10.0 was used with 0.2 μg of SMO in total volume of 180 μl, incubated for 5 or 2.5 minutes at 37°C water bath. Reactions carried out twice in triplicates were stopped by adding 20 μl of 50 % sulphosalicylic acid containing 100 μM 1,7-diaminoheptane and the SPD content was determined by using HPLC method described earlier (Hyvönen et al. 1992).
Cell culture
The prostate carcinoma cell line DU145 was obtained from American Type Culture Collection, USA. The cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10 % fetal bovine serum and 50 μg/ml gentamycin under conditions of +37°C and 10 % CO2. The cells were harvested by trypsinization, counted electronically (Coulter Counter model Z1) and divided to six well plates. Cells were allowed to attach and after 24h, medium supplemented with or without 50 μM DESPM, BnEtSPM or BnSPM was added. Samples were collected at 24 and 48 hours post-treatment and analyzed for polyamines and N-alkylated analogues or their metabolites. LC-MS/MS samples were prepared similarly as calibration standards for calibration curve 2, and QC samples. Assays were carried out twice in triplicates.
Statistical Analyses
The data are expressed as the mean ± S.D. A software package, GraphPad Prism 4.03 (GraphPad Software, Inc., San Diego, CA) was used for the analyses of enzyme kinetic data using Michaelis-Menten equation with nonlinear fitting (Mr of 55,382 for APAO and Mr of 68,000 for SMO were used in calculations).
RESULTS
LC-MS/MS method used for polyamine and polyamine analogue quantification
Polyamine quantification was based on the previously described LC-MS/MS method (Häkkinen et al. 2007; Häkkinen et al. 2008). Structures and abbreviations for the studied polyamines are shown in Table 2 together with their deuterium-labelled derivatives used in the LC-MS/MS measurements as the internal standards (Häkkinen et al. 2009). All of the 14 amines tested were quantifiable in a single run (Fig. 2). The dynamic range for each analyte was 0.03–60 μM, except for DAP which was 0.03–10 μM. The correlation coefficients (R2) were always > 0.995 for all analytes. Inter-day accuracy of the assay for all analytes ranged between 88.8 and 109.8 % and the inter-day precision of the method was always < 13.4 %.
Table 2.
Structures of the amine compounds in the study and their deuterated analogues as internal standards in LC-MS/MS quantification.
| Structure and abbreviation | Internal standard (IS) | ||
|---|---|---|---|
| DAP | DAP-2D | ||
| PUT | PUT-8D | 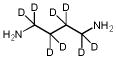 |
|
| SPD | SPD-2D | ||
| SPM | SPM-4D | ||
| EtDAP | EtDAP-2D | 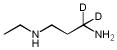 |
|
| EtSPD | EtSPD-2D | ||
| EtSPM | EtSPM-2D | ||
| DESPM | DESPM-4D | ||
| BnNH2 | BnNH2-2D | 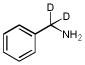 |
|
| BnDAP | BnDAP-4D |  |
|
| BnSPD | BnSPD-4D | ||
| BnSPM | BnSPM-4D | ||
| BnEtSPM | BnEtSPM-8D | ||
| DBSPM | DBSPM-8D |
Fig. 2.
Overlaid RP-LC-MS/MS SRM chromatograms of all the tested 14 amines, which were able to quantify in a single run. Interday accuracy for all was between 88.8–109.8 % and precision < 13.4 %.
SMO activity at various pH
SMO activity was the highest between pH 8.5 and 9.5 (Fig. 3). Therefore, all the kinetic determinations were carried out at pH 9.5 being near optimal for both APAO and SMO (Hölttä 1977; Royo and Fitzpatrick 2005).
Fig. 3.
Reaction velocities (μmol/min) for SMO were determined by using 500 μM SPM as a substrate at different pH in 170 mM Bis-Tris propane buffer.
Terminally N,N’-bis-alkylated polyamine analogues as substrates for recombinant humanAPAO and SMO
As shown in Table 3, DESPM was catabolized to EtSPD by SMO and APAO. Minor N4-endo cleavage pathways resulting in formation of EtDAP was detected with both oxidases. Moreover, very minor de-ethylation takes place with APAO. BnEtSPM was likewise catabolized to EtSPD. However, in the case of SMO some competing debenzylation occurred and evolved EtSPM was readily being cleaved to SPD. Moreover, some other cleavage site products were detected that were formed from secondary metabolites by both the oxidases. DBSPM was mainly metabolized to BnSPD by APAO. However, DBSPM was metabolized to BnSPM and BnSPD by SMO (Table 3).
Table 3.
Kinetic values of N-alkylated polyamine analogues and their predicted secondary metabolites for recombinant polyamine and spermine oxidases
| Substrate | Metabolites | APAO | SMO | ||||
|---|---|---|---|---|---|---|---|
| Vmax | Km | kcat | Vmax | Km | kcat | ||
| DAP | 36.9 | 84 | 0.62 | NA | NA | NA | |
| PUT | 18.5 | 0.4 | 0.31 | 0.33 | 54 | 0.01 | |
| SPD | 209 | 2.5 | 3.49 | 32.2 | 60 | 0.54 | |
| BnDAP | NA | NA | NA | 0.30 | 75 | 0.01 | |
| BnNH2 | 26.1 | 5.0 | 0.44 | 13.2 | 9.8 | 0.22 | |
| SPD | 166 | 6.0 | 2.77 | 44.9 | 19 | 0.75 | |
| SPM | 3.79 | 12 | 0.06 | NA | NA | NA | |
| BnSPD | 7.51 | 20 | 0.12 | NA | NA | NA | |
| BnNH2 | 5.59 | 1.4 | 0.09 | 9.79 | 8.3 | 0.16 | |
| BnDAP | 3.17 | 6.3 | 0.05 | 4.53 | 34 | 0.08 | |
| BnSPD | 121 | 5.4 | 2.02 | 17.8 | 33 | 0.30 | |
| BnSPM | 5.45 | 17 | 0.09 | 16.7 | 117 | 0.28 | |
| PUT | 0.38 | 36 | 0.01 | 0.28 | 47 | 0.00 | |
| SPD | 5.90 | 37 | 0.01 | 2.83 | 48 | 0.05 | |
| EtDAP | 0.23 | 48 | 0.00 | 0.09 | 79 | 0.00 | |
| SPD | 87.6 | 16 | 1.46 | 490 | 25 | 8.17 | |
| SPM | 1.73 | 11 | 0.03 | NA | NA | NA | |
| EtSPD | 2.51 | 25 | 0.04 | NA | NA | NA | |
| EtDAP | 1.48 | 4.7 | 0.02 | NA | NA | NA | |
| EtSPD | 64.2 | 10 | 1.07 | 45.6 | 28 | 0.76 | |
| EtDAP | 4.05 | 6.6 | 0.07 | 1.56 | 33 | 0.03 | |
| EtSPM | 4.66 | 17 | 0.08 | NA | NA | NA | |
| EtSPM | 4.17 | 1.9 | 0.07 | 8.28 | 651 | 0.14 | |
| EtSPD | 63.5 | 0.9 | 1.06 | 21.1 | 51 | 0.35 | |
| EtDAP | 5.00 | 0.2 | 0.08 | 1.73 | 27 | 0.03 | |
| BnNH2 | 5.96 | 1.0 | 0.09 | 8.89 | 49 | 0.15 | |
| SPD | NA | NA | NA | 8.44 | 5.7 | 0.14 | |
| BnSPD | NA | NA | NA | 4.95 | 34 | 0.08 | |
| BnDAP | NA | NA | NA | 1.51 | 28 | 0.02 | |
Kinetic data was obtained as described in “Materials and Methods” by using human recombinant PAO or SMO. The abbreviations and structures are shown in Table 2. Apparent Km(μM), Vmax (μmol/min/μmol protein), kcat (1/s). NA = not applicable. Reference activity for SMO by using 250 μM SPM as a substrate was 460 ± 62 μmol/min and for APAO 270 ± 15 μmol/min by using 250 μM N1-AcSPD as a substrate.
Metabolism of predicted secondary metabolites by APAO and SMO
As shown in Table 3, EtSPM was readily degraded to SPD by both oxidases. APAO metabolized EtSPM variably, other competing pathways being less efficient. BnSPM was readily metabolized to SPD by APAO and SMO. APAO showed again more versatile substrate usage in comparison with SMO. Benzylamine is formed either from direct N1-endo or N4-exo cleavage and subsequent chemical decomposition of benzylaminopropanal under alkaline conditions (Fig. 1). BnSPD was debenzylated to yield SPD by both oxidases. EtSPD was catabolized to SPD inefficiently by both the oxidases. Interestingly, EtDAP was not metabolized by either of the oxidases, but APAO was able to debenzylate BnDAP to DAP with reasonable catalytic efficiency (Table 3).
Metabolism of N,N’-bis-alkylated polyamine analogues in DU145 cells
The effects of 50 μM analogues on intracellular polyamine levels in DU145 cells are shown in Table 4. DESPM accumulated readily inside the cells and most efficiently depleted the natural polyamine pools. EtSPD was detected as the major metabolite in conjunction with trace of EtSPM. BnEtSPM accumulated as efficiently as DESPM inside the cells, but did not deplete polyamine pools as efficiently as the treatment with DESPM did. EtSPD and EtSPM were detected as the major metabolites and some BnSPD accumulated during 48 hours in the BnEtSPM-treated cells. DBSPM was taken up least efficiently among the studied analogues. Moreover, it distorted intracellular polyamine pools less efficiently in comparison with DESPM and BnEtSPM. BnSPM and BnSPD were detected as the major metabolites from DBSPM. In culture medium from cell samples treated with DESPM or BnEtSPM, only EtSPD was detected together with intact drugs (data not shown). BnSPD and a trace amount of BnDAP (48 hour time point) were detected as the metabolites of DBSPM treatment. Total excretion of each metabolite from the cells was less than 0.2 μM (equals to 400 pmol/metabolite in two ml of medium) in culture medium within 48 hours with all the drugs studied. Some PUT and SPD, at less than 0.8 μM concentrations, were detected in control and drug supplemented culture media in 24 hour and 48 hour samples (2 ml medium/well, data not shown). Interestingly, no SPM was detected in any of the culture media samples (data not shown). Moreover, SPD was present at 2.5–5 times higher concentrations in drug-treated medium samples in comparison with control medium samples (data not shown). This could imply that SPM is metabolized to SPD prior to being excreted from the cells.
Table 4.
Intracellular polyamine and analogue levels in DU145 cells.
| cell number×106 | PUT | SPD | SPM | Analogue | EtSPM | BnSPM | EtSPD | BnSPD | |
|---|---|---|---|---|---|---|---|---|---|
| (pmol/106 cells) | |||||||||
| Control 0h | 0.63 ± 0.03 | 396 ± 65 | 2409 ± 320 | 1916 ± 138 | NA | NA | NA | NA | NA |
| Control 24h | 1.06 ± 0.08 | 246 ± 16 | 2194 ± 68 | 2226 ± 72 | NA | NA | NA | NA | NA |
| Control 48h | 2.29 ± 0.11 | 117 ± 18 | 1626 ± 224 | 1759 ± 140 | NA | NA | NA | NA | NA |
| DESPM 24h | 1.01 ± 0.02 | ND | 179 ± 12 | 488 ± 30 | 3799 ± 270 | ND | NA | 31 ± 3 | NA |
| DESPM 48h | 1.50 ± 0.06 | ND | 35 ± 4 | 111 ± 14 | 3706 ± 351 | 3.7 ± 0.3 | NA | 41 ± 6 | NA |
| BnEtSPM 24h | 0.97 ± 0.08 | ND | 430 ± 41 | 1543 ± 74 | 3794 ± 202 | 18 ± 1 | ND | 34 ± 3 | ND |
| BnEtSpm 48h | 1.72 ± 0.08 | ND | 282 ± 19 | 777 ± 72 | 3929 ± 144 | 16 ± 1 | ND | 29 ± 2 | 5.1 ± 0.3 |
| DBSPM 24h | 1.04 ± 0.05 | 98 ± 9 | 1359 ± 119 | 2419 ± 149 | 962 ± 39 | NA | 50 ± 3 | NA | 14 ± 1.6 |
| DBSPM 48h | 0.86 ± 0.07 | 34 ± 5 | 822 ± 50 | 1824 ± 52 | 1047 ± 72 | NA | 36 ± 2 | NA | 12 ± 0.3 |
NA = not applicable, ND = not detectable
DISCUSSION
N-Alkylated PA-analogues have been shown to be metabolized by cellular oxidase(s) which are sensitive to MDL 72,527 (Bergeron et al. 1995; Bergeron et al. 1997; Bitonti et al. 1990; Bolkenius and Seiler 1989). The oxidase responsible was thought to be APAO but recent cloning and characterization of SMO has complicated the picture of how these drugs are being metabolized in vivo and in vitro. It has been clearly shown that human and murine recombinant APAO is capable of metabolizing DENSPM (Häkkinen et al. 2008; Vujcic et al. 2003; Wu et al. 2003). Some unsymmetrically N-substituted norspermine analogues are substrates of APAO (Wang et al. 2005a). In the case of SMO, studies have shown the affinity of many of the studied analogues to the enzyme and many of them have been thought to be inhibitors, not substrates (Vujcic et al. 2002; Wang et al. 2003). The assay method used has been based on fluorometric detection of hydrogen peroxide in a dual enzyme assay system (Wang et al. 2003). The fluorometric detection method is sensitive, but lacks the ability to distinguish which drug in a combination of analogues/polyamine is used as a substrate and how it is being metabolized in the reaction mixtures. However, other methods are valid for testing substrate properties of tested compounds and can be used in inhibition studies. A recent application of using 13C NMR for the detection of oxidase-mediated catabolism of labelled tracer drug in vitro offers means to characterize different reaction products (Bacchi et al. 2009). NMR is a feasible method but lacks sensitivity, and mixtures of polyamine analogues and reaction products are sometimes hard to interpret accordingly. We have recently developed a novel LC-MS/MS method that was exploited to reveal versatile cleavage of DESPM by APAO and shed some light on earlier controversy related to degradation pathways of diethylated SPM analogues (Häkkinen et al. 2007; Häkkinen et al. 2008). Now, we have further extended these studies to analyze the metabolism of three different SPM analogues in vitro and in situ.
The LC-MS/MS method proved to be functional for analyzes of N-alkylated polyamine analogues and their metabolites in in vitro enzyme assay mixtures. Moreover, the method was sufficiently sensitive for quantifying polyamines and their N-alkylated analogues in cell and medium samples i.e. different sample matrix. The validity of the assay is based on the use of deuterated reference compounds with LC-MS/MS analysis of each compound. Advantages over existing HPLC methods are the increased sensitivity, and exact quantification and identification of each analyte (Häkkinen et al. 2008).
APAO- and SMO-mediated catabolism of N-alkylated polyamine analogues was mediated through the same major catabolic pathways. Both enzymes showed ability for versatile cleavages of the studied substrates in the pilot studies with DESPM, BnEtSPM or DBSPM. Thus, a systemic study with all predicted metabolites of DESPM, BnEtSPM or DBSPM was required to dissociate direct metabolism from the metabolism of secondary metabolites. SMO and APAO metabolized DESPM to EtSPD, thus mimicking the degradation of N1,N12-DiAcSPM by APAO. The aromatic benzyl end of BnEtSPM was strongly preferred by SMO and APAO, resulting in formation of EtSPD. However, with SMO there was an active debenzylation pathway present, and evolved EtSPM was effectively further metabolized to SPD. In the case of SMO, N1-exo and N4-exo cleavage were equally active when DBSPM was used as a substrate. Furthermore, some BnDAP was detected indicating the presence of N4-endo cleavage. APAO strongly preferred N4-exo cleavage yielding BnSPD with DBSPM as substrate and other competing pathways were less than 5 % active when compared to the catalytic-centre activity of the major pathway.
EtSPM was metabolized very efficiently to SPD by SMO. It has been shown that SMO is able to degrade N1-AcSPM and it seems that ethyl substituent that retains the amine as positively charged strongly accelerates reaction velocity (Wang et al. 2003). Similarly, SPD was a major metabolite with APAO, but some other competing pathways were also detected. Interestingly, with charge retaining EtSPM and DESPM the reaction velocity is slower when compared with N1-AcSPM and N1,N12-DiAcSPM, respectively (Järvinen et al. 2006a). BnSPM was metabolized to SPD by both oxidases, APAO being more efficient than SMO. Some other competing pathways were also present. EtSPD was a very weak substrate for both oxidases and was de-ethylated to SPD. However, BnSPD was very efficiently debenzylated by APAO to yield SPD, and with a 15 % reaction velocity by SMO in comparison to APAO. Moreover, APAO was capable in debenzylating BnDAP to DAP with a reasonable catalytic efficiency. SPD has been shown to be an inhibitor for SMO, but it seems that the aromatic N-terminal substituent evokes catalytic activity with SMO.
The metabolite formation was linear with all tested metabolites except for BnNH2, which may also be formed via further chemical degradation of produced benzylaminopropanal liberating acrolein in the incubation solutions of high pH. This was noted as an increased reaction velocity of the formation of BnNH2 after a longer (2x) incubation time in comparison to velocities obtained with T1/2 reference samples. Thus, rapid acidification of reaction mixture stabilizes benzylaminopropanal and prevents its chemical decomposition to BnNH2 and acrolein. The previous data rules out N1-endo cleavage as a source of BnNH2.
APAO is constitutively expressed in various tissues but SMO activity has been shown to be inducible by some of the N-alkylated polyamine analogues in certain cell lines (Wang et al. 2001; Devereux et al. 2003; Wang et al. 2005b). Significant APAO activity has been determined from post mortem human specimens (Suzuki et al. 1984) and furthermore SMO and APAO expression could be estimated by using virtual Northern analysis of EST databases as described in (Vujcic et al. 2003). Evaluation of the previous data strongly supports our view that the role of N-alkylated analogue degradation in determining the drug effect should be systematically studied. The basal activity of 431 ± 92 pmol/mg/h for SMO in DU145 cell has been determined earlier (Hyvönen et al. 2007). We did not determine the induction of SMO or APAO activity by N-alkylated analogues in our current study. Cell culture study was included to test the applicability of LC-MS/MS method for quantification of PA, N-alkylated PA analogues and their metabolites from different sample matrix than in vitro enzyme assay mixture.
Recently, SMO was shown to be responsible for drug-induced apoptosis acting as the primary source of hydrogen peroxide, and thus, playing an important role in polyamine catabolism-mediated cytotoxic response (Pledgie et al. 2005). Our present data clearly shows that inducible SMO is capable of metabolizing N-alkylated polyamine analogues. This is not in agreement with earlier publications showing that none of the studied N,N’-bis-alkylated PA analogues were metabolized by SMO (Vujcic et al. 2002; Wang et al. 2003). Discrepancy could be explained by structural difference (3-3-3) or (4-4-4), instead of (3-4-3) carbon backbone in conjunction with different conditions in in vitro assay system. We did use the same cDNA described in (Vujcic et al. 2002) for production of recombinant SMO. However, Vucjic et al. did not purify enzyme instead they used cell lysates that could have caused DESPM behave like an inhibitor of SMO, not substrate like EtSPM. Thus, some apparent toxicity or low efficacies of the polyamine-based drugs in some cell lines may be explained by the ability of these two enzymes metabolize these drugs and their metabolites. It should be noted that some of the metabolites generated from N-alkylated PA analogues by SMO or APAO could be substrates for other cellular oxidases e.g. mono- or diamine oxidase. Although cellular APAO and SMO activities might have a significant impact on drug efficacy, kinetic studies of DESPM, BnEtSPM or DBSPM and their predicted secondary metabolites using purified polyamine oxidases have not been done until now. It is clear that present data should be taken into a consideration when novel N-alkylated polyamine analogues are being developed and used for the treatment of proliferative disorders or parasitic diseases. Further studies are clearly needed in order to assess the role of APAO and SMO in polyamine analogue-mediated drug response. Large versatility in their substrate properties and flexible cleavage of their substrates could implicate that SMO and APAO may have other natural substrates in addition to natural polyamines (Bacchi et al. 2009; Lentini et al. 2007). It should be noted that the studied analogues resembled SPM (3-4-3) carbon backbone. Many analogues based on norspermine (3-3-3) or homospermine (4-4-4) carbon backbone and several analogues having a variable length carbon chain between charged amines have been synthesized (Bergeron et al. 1997; Casero and Marton 2007). Alterations in carbon backbone could have a profound effect on their substrate properties with APAO and SMO and should be systematically studied in future. This work may require synthesis of appropriate reference compounds for exact quantification of reaction products by using LC-MS/MS. However, for screening of reaction mixtures just to detect any of the metabolites derived from the tested drugs could be carried out without internal reference compounds.
In summary, here we show for the first time that both APAO and SMO are capable of metabolizing several N-alkylated polyamine derivatives. This suggests that they might have a role in drug-mediated cytotoxic response, especially after the natural polyamines have been depleted. Although, exact mechanism of drug action of N-alkylated PA-analogues remains obscure, polyamine metabolism is still an attractive target for drug design. It is evident that further studies including substrate properties on analogues with APAO and SMO are required.
Acknowledgments
We thank Ms. Helena Vepsäläinen and Ms. Maritta Salminkoski, Department of Biosciences, Laboratory of Chemistry, University of Kuopio, for their help with LC-MS/MS sample preparation and in the synthesis work. Ms. Anne Karppinen and Ms. Tuula Reponen at A. I. Virtanen Institute, University of Kuopio, are acknowledged for their help with enzyme and cell experiments. This work was supported by Academy of Finland (projects 1241851 and 1287022), NIH (USA) CA984544, and the Russian Foundation for Basic Research (project 08-04-917775).
The abbreviations and trivial names
- ACN
acetonitrile
- APAO
(acetyl)polyamine oxidase (exo-N4-amino) [EC 1.5.3.11]
- BnDAP
N1-Benzyl-propane-1,3-diamine
- BnDAP-4D
N1-Benzyl-2,2,3,3-2H4-propane-1,3-diamine
- BnEtSPM
N-(3-benzylaminopropyl)-N'-(3-ethylaminopropyl)butane-1,4-diamine
- BnEtSPM-8D
N-(3-Benzylamino-1,1,2,2-2H4-propyl)-N'-(3-ethylamino-1,1,2,2-2H4-propyl)butane-1,4-diamine
- BnNH2
Benzylamine
- BnNH2-2D
α,α-2H2-Benzylamine
- BnSPD
N1-(3-Benzylaminopropyl)butane-1,4-diamine
- BnSPD-4D
N1-(3-Benzylamino-1,1,2,2-2H4-propyl)butane-1,4-diamine
- BnSPM
N-(3-Aminopropyl)-N'-(3-Benzylaminopropyl)butane-1,4-diamine
- BnSPM-4D
N-(3-Aminopropyl)-N'-(3-Benzylamino-1,1,2,2-2H4-propyl)butane-1,4-diamine
- CID
collision energy
- DAP
Propane-1,3-diamine
- DAP-2D
1,1-2H2-Propane-1,3-diamine
- DBSPM
N,N′-bis-(3-Benzylaminopropyl)butane-1,4-diamine
- DBSPM-8D
N,N′-bis-(3-Benzylamino-1,1,2,2-2H4-propyl)butane-1,4-diamine
- DENSPM
diethylnorspermine, N,N'-bis-(3-Ethylaminopropyl)-propane-1,3-diamine
- DESPM
N,N′-bis-(3-ethylaminopropyl)butane-1,4-diamine
- DESPM-4D
N,N′-bis-(3-Ethylamino-1,1-2H2-propyl)butane-1,4-diamine
- EtDAP
N1-Ethylpropane-1,3-diamine
- EtDAP-2D
N1-Ethyl-3,3-2H2-propane-1,3-diamine
- EtSPD
N1-(3-Ethylaminopropyl)butane-1,4-diamine trihydrochloride
- EtSPD-2D
N1-(3-Ethylamino-1,1-2H2-propyl)butane-1,4-diamine
- EtSPM
N-(3-Aminopropyl)-N'-(3-ethylaminopropyl)butane-1,4-diamine
- EtSPM-2D
N-(3-Aminopropyl)-N'-(3-ethylamino-1,1-2H2-propyl)butane-1,4-diamine
- FDA
U.S. Food and Drug Administration, U.S.A
- HFBA
heptafluorobutyric acid
- HPLC
high pressure liquid chromatography
- IS
internal standard
- LC-MS/MS
Liquid chromatography-electrospray ionization-tandem mass spectrometry
- MDL 72
527, N,N'-bis-(2,3-butadienyl)-1,4-butanediamine
- NMR
nuclear magnetic resonance
- N1-AcSPD
N1,N12-Diacetylspermine
- N1-AcSPM
N1,N12-Diacetylspermidine
- N1
N12-DiAcSPM, N1,N12-Diacetylspermine
- PA
polyamine
- PUT
putrescine, Butane-1,4-diamine
- PUT-8D
1,1,2,2,3,3,4,4-2H8-Butane-1,4-diamine
- SMO
(PAOh1), spermine oxidase [EC 1.5.3.-]
- SPD
spermidine, N1-(3-aminopropyl)butane-1,4-diamine
- SPD-2D
N1-(3-Amino-1,1-2H2-propyl)butane-1,4-diamine
- SPM
spermine, N,N’-bis-(3-aminopropyl)butane-1,4-diamine
- SPM-4D
N-(3-Amino-1,1,2,2-2H4-propyl)-N'-(3-aminopropylbutane-1,4-diamine
- SRM
selected reaction monitoring
- SSAT
spermidine/spermine N1-acetyltransferase [EC 2.3.1.57]
- STD
standard
- QC
quality control
- QL
qualifier ion
- QT
quantifier ion
References
- Agostinelli E, Arancia G, Dalla Vedova I, Belli F, Marra M, Salvi M, Toninello A. The biological functions of polyamine oxidation products by amine oxidases: Perspectives of clinical applications. Amino Acids. 2004;3–4:347–358. doi: 10.1007/s00726-004-0114-4. [DOI] [PubMed] [Google Scholar]
- Bacchi CJ, Yarlett N, Faciane E, Bi X, Rattendi D, Weiss LM, Woster PM. Metabolism of an Alkyl Polyamine Analog by a Polyamine Oxidase from the Microsporidian Encephalitozoon cuniculi. Antimicrob Agents Chemother. 2009;53:2599–2604. doi: 10.1128/AAC.00267-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron RJ, Weimar WR, Luchetta G, Streiff RR, Wiegand SJ, Perrin J, Schreier KM, Porter C, Yao GW, Dimova H. Metabolism and pharmacokinetics of N1, N11-diethylnorspermine. Drug Metab Dispos. 1995;23:1117–1125. [PubMed] [Google Scholar]
- Bergeron RJ, Feng Y, Weimar WR, McManis JS, Dimova H, Porter C, Raisler B, Phanstiel O. A comparison of structure-activity relationships between spermidine and spermine analogue antineoplastics. J Med Chem. 1997;40:1475–1494. doi: 10.1021/jm960849j. [DOI] [PubMed] [Google Scholar]
- Bergeron RJ, Wiegand J, McManis JS, Weimar WR, Smith RE, Algee SE, Fannin TL, Slusher MA, Snyder PS. Polyamine Analogue Antidiarrheals: A Structure-Activity Study. J Med Chem. 2001;44:232–244. doi: 10.1021/jm000277+. [DOI] [PubMed] [Google Scholar]
- Bitonti AJ, Dumont JA, Bush TL, Edwards ML, Stemerick DM, McCann PP, Sjoerdsma A. Bis(benzyl)polyamine analogs inhibit the growth of chloroquine-resistant human malaria parasites (Plasmodium falciparum) in vitro and in combination with a-difluoromethylornithine cure murine malaria. Proc Natl Acad Sci USA. 1989;86:651–655. doi: 10.1073/pnas.86.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitonti AJ, Dumont JA, Bush TL, Stemerick DM, Edwards ML, McCann PP. Bis(benzyl)polyamine analogs as novel substrates for polyamine oxidase. J Biol Chem. 1990;265:382–388. [PubMed] [Google Scholar]
- Bolkenius FN, Seiler N. New substrates of polyamine oxidase. Dealkylation of N-alkyl-a, w-diamines. Biol Chem Hoppe-Seyler. 1989;370:525–531. doi: 10.1515/bchm3.1989.370.1.525. [DOI] [PubMed] [Google Scholar]
- Byers TL, Bitonti AJ, McCann PP. Bis(benzyl)polyamine analogues are substrates for a mammalian cell-transport system which is distinct from the polyamine-transport system. Biochem J. 1990;269:35–40. doi: 10.1042/bj2690035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Devereux W, Wang Y, Stewart TM, Hacker A, Smith R, Frydman B, Valasinas AL, Reddy VK, Marton LJ, Ward TD, Woster PM, Casero RA. Induction of the PAOh1/SMO polyamine oxidase by polyamine analogues in human lung carcinoma cells. Cancer Chemother Pharmacol. 2003;52:383–390. doi: 10.1007/s00280-003-0662-4. [DOI] [PubMed] [Google Scholar]
- Edwards ML, Stemerick DM, Bitonti AJ, Dumont JA, McCann PP, Bey P, Sjoerdsma A. Antimalarial polyamine analogues. J Med Chem. 1991;34:569–574. doi: 10.1021/jm00106a015. [DOI] [PubMed] [Google Scholar]
- FDA Guidance for Industry, Bioanalytical Method Validation, US Department of Health and Human Services Food and Drug Administration, Center for Drug Evaluation and Research (CDER) 2001 Available: http://www.fda.gov/cder/guidance/4252fnl.pdf.
- Heby O, Persson L, Rentala M. Targeting the polyamine biosynthetic enzymes: a promising approach to therapy of African sleeping sickness, Chagas’ disease, and leishmaniasis. Amino Acids. 2007;33:359–366. doi: 10.1007/s00726-007-0537-9. [DOI] [PubMed] [Google Scholar]
- Hyvönen T, Keinänen TA, Khomutov AR, Khomutov RM, Eloranta TO. Monitoring of the uptake and metabolism of aminooxy analogues of polyamines in cultured cells by high-performance liquid chromatography. J Chromatogr. 1992;574:17–21. doi: 10.1016/0378-4347(92)80093-6. [DOI] [PubMed] [Google Scholar]
- Hyvönen MT, Keinänen TA, Cerrada-Gimenez M, Sinervirta R, Grigorenko N, Khomutov AR, Vepsäläinen J, Alhonen L, Jänne J. Role of hypusinated eukaryotic translation initiation factor 5A in polyamine depletion-induced cytostasis. J Biol Chem. 2007;282:34700–34706. doi: 10.1074/jbc.M704282200. [DOI] [PubMed] [Google Scholar]
- Häkkinen MR, Keinänen TA, Vepsäläinen J, Khomutov AR, Alhonen L, Jänne J, Auriola S. Analysis of underivatized polyamines by reversed phase liquid chromatography with electrospray tandem mass spectrometry. J Pharm Biomed Anal. 2007;45:625–634. doi: 10.1016/j.jpba.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Häkkinen MR, Keinänen TA, Vepsäläinen J, Khomutov AR, Alhonen L, Jänne J, Auriola S. Quantitative determination of underivatized polyamines by using isotope dilution RP-LC-ESI-MS/MS. J Pharm Biomed Anal. 2008;48:414–421. doi: 10.1016/j.jpba.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Häkkinen MR, Keinänen TA, Khomutov AR, Auriola S, Weisell J, Alhonen L, Jänne J, Vepsäläinen J. Synthesis of novel deuterium labelled derivatives of N-alkylated polyamines. Tetrahedron. 2009;65:547–562. [Google Scholar]
- Hölttä E. Oxidation of spermidine and spermine in rat liver: purification and properties of polyamine oxidase. Biochemistry. 1977;16:91–100. doi: 10.1021/bi00620a015. [DOI] [PubMed] [Google Scholar]
- Järvinen A, Grigorenko N, Khomutov AR, Hyvönen MT, Uimari A, Vepsäläinen J, Sinervirta R, Keinänen TA, Vujcic S, Alhonen L, Porter CW, Jänne J. Metabolic stability of alpha -methylated polyamine derivatives and their use as substitutes for the natural polyamines. J Biol Chem. 2005;280:6595–6601. doi: 10.1074/jbc.M412788200. [DOI] [PubMed] [Google Scholar]
- Järvinen A, Keinänen TA, Grigorenko NA, Khomutov AR, Uimari A, Vepsäläinen J, Alhonen L, Jänne J. Guide molecule-driven stereospecific degradation of alpha-methylpolyamines by polyamine oxidase. J Biol Chem. 2006a;281:4589–4595. doi: 10.1074/jbc.M509959200. [DOI] [PubMed] [Google Scholar]
- Järvinen AJ, Cerrada-Gimenez M, Grigorenko NA, Khomutov AR, Vepsäläinen JJ, Sinervirta RM, Keinänen TA, Alhonen LI, Jänne JE. Alpha-Methyl polyamines: Efficient synthesis and tolerance studies in vivo and in vitro. First evidence for dormant stereospecificity of polyamine oxidase. J Med Chem. 2006b;49:399–406. doi: 10.1021/jm050872h. [DOI] [PubMed] [Google Scholar]
- Kaiser A, Ulmer D, Goebel T, Holzgrabe U, Saeftel M, Hoerauf A. Inhibition of hypusine biosynthesis in plasmodium: a possible, new strategy in prevention and therapy of malaria. Mini Rev Med Chem. 2006;6:1231–1241. doi: 10.2174/138955706778742795. [DOI] [PubMed] [Google Scholar]
- Lawson KR, Marek S, Linehan JA, Woster PM, Casero RA, Jr, Payne CM, Gerner EW. Detoxification of the polyamine analogue N1-ethyl-N11-[(cycloheptyl)methy]-4,8-diazaundecane (CHENSpm) by polyamine oxidase. Clin Cancer Res. 2002;8:1241–1247. [PubMed] [Google Scholar]
- Lee Y, Sayre LM. Reaffirmation that metabolism of polyamines by bovine plasma amine oxidase occurs strictly at the primary amino termini. J Biol Chem. 1998;273:19490–19494. doi: 10.1074/jbc.273.31.19490. [DOI] [PubMed] [Google Scholar]
- Lentini A, Mattioli P, Provenzano B, Abbruzzese A, Caraglia M, Beninati S. Role of the FAD-dependent polyamine oxidase in the selective formation of N(1), N(8)-bis(gamma-glutamyl)spermidine protein cross-links. Biochem Soc Trans. 2007;35:396–400. doi: 10.1042/BST0350396. [DOI] [PubMed] [Google Scholar]
- Libby PR, Porter CW. Separation of two isozymes of polyamine oxidase from murine L1210 leukemia cells. Biochem Biophys Res Commun. 1987;144:528–535. doi: 10.1016/s0006-291x(87)80541-7. [DOI] [PubMed] [Google Scholar]
- Muller IB, Das Gupta R, Luersen K, Wrenger C, Walter RD. Assessing the polyamine metabolism of Plasmodium falciparum as chemotherapeutic target. Mol Biochem Parasitol. 2008;160:1–7. doi: 10.1016/j.molbiopara.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy. Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- Pegg AE, Poulin R, Coward JK. Use of aminopropyltransferase inhibitors and of non-metabolizable analogs to study polyamine regulation and function. Int J Biochem Cell Biol. 1995;27:425–442. doi: 10.1016/1357-2725(95)00007-c. [DOI] [PubMed] [Google Scholar]
- Pegg AE, Feith DJ. Polyamines and neoplastic growth. Biochem Soc Trans. 2007;35:295–299. doi: 10.1042/BST0350295. [DOI] [PubMed] [Google Scholar]
- Pledgie A, Huang Y, Hacker A, Zhang Z, Woster PM, Davidson NE, Casero RA., Jr Spermine oxidase SMO(PAOh1), Not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J Biol Chem. 2005;280:39843–39851. doi: 10.1074/jbc.M508177200. [DOI] [PubMed] [Google Scholar]
- Reguera RM, Tekwani BL, Balana-Fouce R. Polyamine transport in parasites: a potential target for new antiparasitic drug development. Comp Biochem Physiol C Toxicol Pharmacol. 2005;140:151–164. doi: 10.1016/j.cca.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Royo M, Fitzpatrick PF. Mechanistic studies of mouse polyamine oxidase with N1, N12-bisethylspermine as a substrate. Biochemistry. 2005;44:7079–7084. doi: 10.1021/bi050347k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler N. Polyamine oxidase, properties and functions. Prog Brain Res. 1995;106:333–344. doi: 10.1016/s0079-6123(08)61229-7. [DOI] [PubMed] [Google Scholar]
- Seiler N. Thirty years of polyamine-related approaches to cancer therapy. Retrospect and prospect. Part 2. Structural analogues and derivatives. Curr Drug Targets. 2003;4:565–585. doi: 10.2174/1389450033490876. [DOI] [PubMed] [Google Scholar]
- Seiler N. Catabolism of polyamines. Amino Acids. 2004;26:217–233. doi: 10.1007/s00726-004-0070-z. [DOI] [PubMed] [Google Scholar]
- Smith RG, Daves DG., Jr New Mass Spectrometric Rearrangements Involving Silicon. A Study of Trimethylsilylated Di- and Polyamines and Their Isotopically Labeled Analogues. J Org Chem. 1978;43:2178–2183. [Google Scholar]
- Suzuki O, Matsumoto T, Katsumata Y. Determination of polyamine oxidase activities in human tissues. Experientia. 1984;40:838–839. doi: 10.1007/BF01951981. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines. Ann Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Tsukada T, Furusako S, Maekawa S, Hibasami H, Nakashima K. Purification by affinity chromatography and characterization of porcine liver cytoplasmic polyamine oxidase. Int J Biochem. 1988;20:695–702. doi: 10.1016/0020-711x(88)90164-4. [DOI] [PubMed] [Google Scholar]
- Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem J. 2002;367:665–675. doi: 10.1042/BJ20020720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. Genomic identification and biochemical characterization of the mammalian polyamine oxidase involved in polyamine back-conversion. Biochem J. 2003;370:19–28. doi: 10.1042/BJ20021779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA., Jr Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61:5370–5373. [PubMed] [Google Scholar]
- Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM, Casero RA., Jr Properties of purified recombinant human polyamine oxidase, PAOh1/SMO. Biochem Biophys Res Commun. 2003;304:605–611. doi: 10.1016/s0006-291x(03)00636-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hacker A, Murray-Stewart T, Frydman B, Valasinas A, Fraser AV, Woster PM, Casero RA., Jr Properties of recombinant human N1-acetylpolyamine oxidase (hPAO): potential role in determining drug sensitivity. Cancer Chemother Pharmacol. 2005a;56:83–90. doi: 10.1007/s00280-004-0936-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hacker A, Murray-Stewart T, Fleischer JG, Woster PM, Casero RA., Jr Induction of human spermine oxidase SMO(PAOh1) is regulated at the levels of new mRNA synthesis, mRNA stabilization and newly synthesized protein. Biochem J. 2005b;386:543–547. doi: 10.1042/BJ20041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. Interactions of polyamines with ion channels. Biochem J. 1997;325:289–297. doi: 10.1042/bj3250289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Yankovskaya V, McIntire WS. Cloning, sequencing, and heterologous expression of the murine peroxisomal flavoprotein, N1-acetylated polyamine oxidase. J Biol Chem. 2003;278:20514–20525. doi: 10.1074/jbc.M302149200. [DOI] [PubMed] [Google Scholar]