Abstract
Epidemiologic observations that have been made in the context of the current pandemic influenza virus include a stable virulence phenotype and a lack of propensity to reassort with seasonal strains. In an attempt to determine whether either of these observations could change in the future, we coinfected differentiated human airway cells with seasonal oseltamivir-resistant A/New Jersey/15/07 and pandemic A/Tennessee/1-560/09 (H1N1) viruses in three ratios (10:90, 50:50, and 90:10) and examined the resulting progeny viruses after 10 sequential passages. When the pandemic virus was initially present at multiplicities of infection equal to or greater than those for the seasonal virus, only pandemic virus genotypes were detected. These adapted pandemic strains did, however, contain two nonsynonymous mutations (hemagglutinin K154Q and polymerase acidic protein L295P) that conferred a more virulent phenotype, both in cell cultures and in ferrets, than their parental strains. The polymerase acidic protein mutation increased polymerase activity at 37°C, and the hemagglutinin change affected binding of the virus to α2,6-sialyl receptors. When the seasonal A/H1N1 virus was initially present in excess, the dominant progeny virus was a reassortant containing the hemagglutinin gene from the seasonal strain and the remaining genes from the pandemic virus. Our study demonstrates that the emergence of an A/H1N1 pandemic strain of higher virulence is possible and that, despite their lack of detection thus far in humans, viable seasonal/pandemic virus reassortants can be generated.
IMPORTANCE
This report supplies a key piece of information for investigating future evolution scenarios of pandemic A/H1N1 influenza in the human population. We report that the emergence of an A/H1N1 pandemic strain of higher virulence is possible and that, despite their lack of detection thus far in humans, viable seasonal/pandemic virus reassortants can be generated.
INTRODUCTION
Influenza A viruses of the H1N1 subtype have had a significant epidemiologic impact in humans by causing seasonal epidemics of various degrees of severity and two pandemics in 1918 and 2009 (1, 2). Despite the recent availability of complete genome sequence data, many aspects of the evolutionary and epidemiologic dynamics of the A/H1N1 virus remain unknown. How different influenza subtypes interact with each other and why one subtype replaces its counterpart over a season or over decades are currently unclear. Since 2009, two main lineages of A/H1N1 have been circulating in humans: the new swine-origin pandemic lineage and a seasonal lineage. Seasonal A/H1N1 viruses became spontaneously resistant to the neuraminidase (NA) inhibitor oseltamivir, the primary treatment for influenza virus-infected patients, after the 2007-2008 season and then spread rapidly from Europe around the globe (3). A recent study identified 53 areas of cocirculation of oseltamivir-resistant seasonal and pandemic A/H1N1 (4). Despite limited testing, there are a few reports of mixed infections in China and in at least 11 patients in New Zealand (4, 5). This lends support for the possibility of reassortment between pandemic and seasonal influenza viruses, with the likelihood of emergence of an NA inhibitor-resistant pandemic-like virus if the N1 NA segment from the oseltamivir-resistant seasonal ancestor were to reassort with the pandemic strain.
Epidemiologic observations that have been made in the context of the current pandemic influenza virus include a stable virulence phenotype and a lack of propensity to reassort with seasonal strains. Indeed, a recent study of A/H1N1 coinfection in ferrets found no reassortment and predicted the dominance of the pandemic virus (6). Additionally, seasonal A/H1N1 strains are more and more rarely reported worldwide, whereas pandemic A/H1N1 seems to have almost replaced its seasonal counterpart (7). Thus, in this study, we made an attempt to determine (i) the genomic-scale interaction between the pandemic and seasonal viruses, (ii) the pattern of further adaptation of pandemic A/H1N1 influenza to humans, and (iii) whether such adaptation could lead to substantially increased virulence.
RESULTS
Coinfection of seasonal and pandemic A/H1N1 influenza virus strains in differentiated human cells.
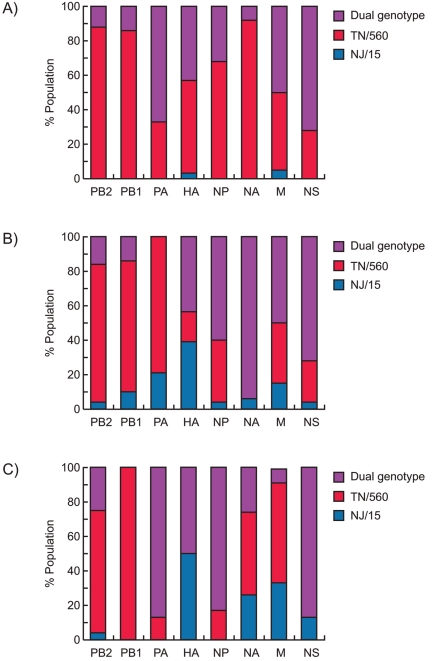
We used differentiated normal human bronchial epithelial (NHBE) cells as a model to mimic A/H1N1 virus evolution in humans. NHBE cells were coinfected with oseltamivir-resistant seasonal A/New Jersey/15/07 (NJ/15) and pandemic A/Tennessee/1-560/09 (TN/560) (H1N1) viruses in different ratios (10:90, 50:50, and 90:10). The virus mixtures were serially passaged 10 times to provide an opportunity for selection of efficiently adapted genotypes. After three sequential passages, viruses isolated from all coinfection groups were plaque purified in Madin-Darby canine kidney (MDCK) cells, and 25 clones per group were genotyped to establish the origin of each of the eight genes that constitute the influenza virus (Fig. 1) (8, 9). The method was not quantitative and was used as a rough estimate of virus genes present. Among all the samples analyzed, TN/560 dominated over NJ/15, although dual genotypes (plaques containing the same genes of both parental viruses) were still present for all genes and in all three groups. Notably, viruses carrying the NJ/15 hemagglutinin (HA) gene were isolated at higher frequencies (39 to 50%) from NHBE cultures that were initially coinfected with a proportion of seasonal A/H1N1 equal to or higher than that of the pandemic virus (Fig. 1B and C).
FIG 1 .
Genotypes of virus populations recovered after three sequential passages in cultures of differentiated normal human bronchial epithelial (NHBE) cells coinfected with 10% NJ/15-90% TN/560 (A), 50% NJ/15-50% TN/560 (B), and 90% NJ/15-10% TN/560 (C) mixtures of seasonal and pandemic H1N1 influenza viruses. Results are shown for genotypes of NJ/15 or TN/560 or dual genotypes of plaque-purified viruses (n = 25).
We assayed whether pandemic/seasonal reassortants could be favored by analyzing viral genomes (n = 30) after 10 passages in NHBE cells (Table 1). A unique reassortant carrying NJ/15 HA in a TN/560 backbone was systemically selected (92%) in the group with an initial predominance of NJ/15. In the two other coinfection groups, all recovered viruses possessed all eight genes from the pandemic TN/560 virus (Table 1). The proportions of HA and NA RNAs of each virus after each passage in coinfected NHBE cells were quantified by multiplex real-time reverse transcriptase PCR (RT-PCR) (see Fig. S1 in the supplemental material). Both HA and NA RNAs of TN/560 dominated early in the groups, with initial proportions of the pandemic virus that were equal to or higher than those of the seasonal virus (Fig. S1A and B). Evidence of reassortment between seasonal and pandemic strains was confirmed for the 90:10 NJ/15-TN/560 coinfection group starting from passage 1 (Fig. S1C). Taken together, our findings showed that pandemic A/H1N1 dominated over its seasonal counterpart in the NHBE cell model used. However, the emergence of a reassortant between two A/H1N1 viruses was also observed, but only in the group with an initial dominance of the seasonal strain. The fact that such reassortant viruses acquired the HA gene from the seasonal A/H1N1 virus suggests that the pandemic HA gene segment may not yet be optimal for viral replication in humans.
TABLE 1 .
H1N1 reassortant genotypes recovered after 10 sequential passages from differentiated NHBE cultures coinfected with seasonal NJ/15 and pandemic TN/560 (H1N1) influenza viruses
| Coinfection group | Genotypea |
No. of isolates with the described genotype/total no. of viruses examined | Abbreviation | Molecular change(s) during coinfectionb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | ||||
| 10% NJ/15-90% TN/560 | TN | TN | TN | TN | TN | TN | TN | TN | 30/30 | G1 | PA L295P; HA K154Q |
| 50% NJ/15-50% TN/560 | TN | TN | TN | TN | TN | TN | TN | TN | 31/31 | PA L295P | |
| 90% NJ/15-10% TN/560 | TN | TN | TN | NJ | TN | TN | TN | TN | 24/26c | G2 | PB2 D87N |
NJ, gene derived from the NJ/15 parental virus; TN, gene derived from the TN/560 parental virus.
Molecular changes are shown based on the sequences of the ~20 to 25% of plaque-purified viruses isolated from NHBE culture supernatants from three coinfection groups (H1 HA numbering) (20).
2 H1N1 clones had a mixed NJ/TN genotype for the HA gene.
To monitor the emergence of adaptive amino acid changes, the complete genomes of six clones isolated from each group were sequenced. HA K154Q (H1 numbering) and/or polymerase acid (PA) protein L295P mutations were observed in two groups with an initial 50% or 90% proportion of TN/560 (Table 1). A polymerase basic 2 (PB2) protein D87N mutation was observed in all the clones recovered from the group with an initial predominance of NJ/15. The two A/H1N1 strains that represented two major genotypes isolated from coinfected NHBE cells were designated G1 (TN/560-like virus with PA L295P and HA K154Q) and G2 (reassortant NJ/15 HA in a TN/560 backbone with PB2 D87N) and were further analyzed (Table 1).
Replication and pathogenicity of G1 and G2 viruses in cell culture and in ferrets.
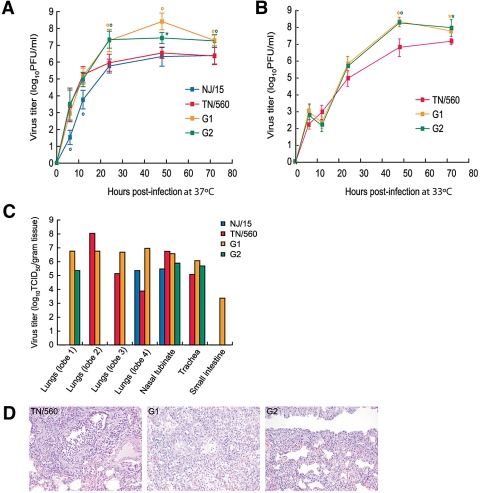
We first studied the growth of G1 and G2 viruses both in eggs and in MDCK cells (see Table S1 in the supplemental material). The yields of both viruses in cell culture were significantly higher (~1.5 logs) than those of their parental TN/560 and NJ/15 strains (P < 0.01). G1 and G2 formed larger plaques (diameter, 1.5 to 2.2 mm) than the parental counterparts (diameter, 0.6 to 1.1 mm) (Table S1). To assess the relative replication efficiencies of the G1 and G2 viruses, the multiple-cycle replication kinetics of these viruses in comparison with the levels for TN/560 and NJ/15 in NHBE cells were examined. The replication levels of G1 and G2 were significantly higher than those of both parents 24, 48, and 72 h after infection at 37°C (>0.9 to 1.8 logs; P < 0.05) (Fig. 2A). Higher titers of G1 and G2 than of TN/560 were also observed 48 and 72 hours after infection at 33°C (>0.6 to 1.3 logs; P < 0.05) (Fig. 2B).
FIG 2 .
Replication of NJ/15, TN/560, and two recovered H1N1 genotypes, G1 and G2, in NHBE cells and ferrets. (A and B) NHBE cultures were infected via the apical side with each virus at an MOI of 0.1 at either 37°C (A) or 33°C (B). The progeny viruses released from the apical surfaces of infected cultures were collected at the indicated time points and titrated in MDCK cells by performing a plaque assay. Representative results expressed as log10 numbers of PFU/ml from three independent experiments are shown. *, P < 0.05; °, P <0.01 compared with the value for TN/560 virus (one-way ANOVA). (C) Four ferrets were intranasally inoculated with 106 PFU of each virus, and 3 days later, tissues were collected and virus titers were determined. Virus titers are expressed as log10 TCID50/gram tissue for one ferret. (D) The images shown are hematoxylin-and-eosin-stained sections of lung tissue from donor ferrets inoculated with TN/560, G1, or G2 (H1N1) influenza viruses obtained on day 3 postinoculation. The TN/560 lung infection was primary restricted to the bronchioles and consisted of epithelial necrosis and regeneration associated with intraluminal sloughed epithelial cells and mononuclear inflammatory cells. The G1 infection involved the bronchioles and alveoli and consisted of bronchiolar epithelial necrosis and regeneration associated with alveolar pneumocytes hyperplasia and mononuclear inflammatory cell infiltrates. The pathologic changes in the G2-infected lungs were restricted to the alveolar interstitium only. The alveolar spaces were free of inflammatory cells, whereas the alveolar interstitial walls were hypercellular with mononuclear cells. Magnification, ×20.
To determine whether the G1 and G2 variants acquired increased virulence in vivo, we further characterized the pathogenicity and transmissibility potentials of A/H1N1 viruses in ferrets. Four groups of two ferrets (Mustela putorius furo) each were inoculated with 106 PFU of TN/560, NJ/15, G1, or G2 (Table 2; see also Fig. S2 in the supplemental material). After 24 h, two naïve ferrets (contacts) were housed in the same cage with two inoculated ferrets (donors) to assess direct-contact transmission. All A/H1N1 viruses caused only mild clinical signs (relative inactivity index [RII], ≈0.1), without marked weight changes (~4 to 10%) (Table 2). The body temperature elevation was slightly higher in animals inoculated with G1 and G2 than in those inoculated with parental viruses (mean peak increases of 1.6°C and 0.4°C for donors and contacts, respectively). The donor ferrets inoculated with G1 had significantly higher nasal wash titers on day 1 postinoculation (p.i.) than the TN/560-infected animals had (P < 0.01). The duration of infectious-virus shedding was 2 days longer for animals inoculated with G1 or G2 viruses than for animals inoculated with either TN/560 or NJ/15 (Fig. S2A). Although all four A/H1N1 strains transmitted efficiently, as shown by virus shedding as well as by seroconversion (Table 2), G1 and G2 contact ferrets exhibited significantly higher virus titers in the upper respiratory tract than the TN/560 group on all days studied (>1.5 to 3.8 logs; P < 0.01) (Fig. S2A). Comparison of the protein concentrations and total cell counts in the nasal washes showed a statistically significant difference between G2 and all other viruses on day 2 p.i. (P < 0.01), suggesting an increased inflammation in the upper respiratory tract (Fig. S2D and E).
TABLE 2 .
Replication and transmission of NJ/15, TN/560, G1, and G2 (H1N1) influenza viruses in ferrets
| Virus | Animal group | Clinical signs | Virus detection | Seroconversion values (serum HI titer/neutralizing titer)d |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Wt loss (%)a | Increase in temp (°C) (day of onset) | Peak virus titer (log10 TCID50/ml) (day of onset)b | Last day of sheddingc | NJ/15 | TN/560 | G1 | G2 | ||
| NJ/15 | Donors | 10.5 | 1.2 (2) | 5.6e (2) | 6 | 160/40 | </< | </< | 160/40 |
| Contacts | 4.5 | 0.9 (1) | 5.4 (3) | 9 | 160/20 | </< | </< | 240/40 | |
| TN/560 | Donors | 8.9 | 1.0 (1) | 6.7 (2) | 6 | </< | 320/120 | 320/1,280 | </< |
| Contacts | 2.0 | 0.6 (3) | 5.2 (3) | 7 | </< | 320/240 | 320/640 | </< | |
| G1 | Donors | 9.1 | 2.5 (2) | 7.2 (2) | 8 | </< | 160/20 | 640/2,560 | </< |
| Contacts | 2.2 | 1.3 (3) | 7.4° (3) | 9 | </< | 160/60 | 940/1,280 | </< | |
| G2 | Donors | 4.1 | 2.8 (2) | 6.1 (2) | 8 | 320/80 | </< | </< | 320/40 |
| Contacts | 2.5 | 1.0 (4) | 6.7 (3) | 9 | 640/200 | </< | </< | 640/180 | |
The maximum percent weight loss during the 21 days p.i.
Virus titer in nasal washes.
The first day of observation on which virus was not detected.
Hemagglutination inhibition (HI) antibody titers in ferret sera were determined at 21 days p.i.; values are expressed as reciprocals of serum that inhibited 4 hemagglutination units of H1N1 influenza virus. Neutralizing antibodies to H1N1 viruses (100 TCID50) were titrated in MDCK cells. Data (arithmetic mean titers) are shown for 1 ferret (for donors) or groups of 2 ferrets (for contacts). Homologous HI/neutralizing titers are shown in bold type. <, titer of <1:10.
P < 0.01 compared with the nasal wash virus titer for TN/560 virus (donors compared with donors and contacts with contacts; one-way ANOVA).
To evaluate virus replication and tissue tropism of A/H1N1 viruses in ferrets, we determined virus titers in the lungs, nasal turbinate, trachea, spleen, liver, and small intestine of one inoculated ferret per group on day 3 p.i. (Fig. 2C). While no evidence of infectious virus was found in the liver or spleen tissues of any animals tested, all four viruses were detected in the lungs and nasal turbinates. Consistent with the more-pronounced clinical signs of disease, G1 exhibited the widest tissue tropism and was detected at higher titers in three out of four lobes of the lungs (~6.8 log10 50% tissue culture infective doses [TCID50]/gram tissue) and small intestine (~3.4 log10 TCID50/gram tissue) (Fig. 2C). Histopathologic analysis revealed that the lungs of donor ferrets inoculated with G1 had significantly more necrotizing bronchiolitis and alveolitis than the lungs from animals infected with TN/560, NJ/15, or G2, which exhibited infection restricted either to the bronchioles or to the alveolar interstitium (Fig. 2D). Taken together, our findings showed that two variants selected by coinfection of human cells acquired increased replicative fitness and virulence both in vitro and in vivo.
Effect of observed mutations on viral polymerase activity, receptor specificity, and antigenicity.
A luciferase minigenome assay was used to study the role of ribonucleoprotein (RNP) complex mutations in the viral replication, comparing G1 and G2 to TN/560. The G1 RNP complex, carrying the PA L295P mutation, showed ~20% more polymerase activity than TN/560 at 37°C (and comparable activities at 33°C and 39°C) (Fig. 3A). The G2 RNP complex also had a significantly higher polymerase activity at 33°C (+30%) but a slightly lower one at 39°C (−10%) than TN/560 RNP (P < 0.05) (Fig. 3A). Interestingly, growth curves at 33°C showed that both G1 and G2 grew faster than TN/560 at later time points (Fig. 2B), suggesting an advantage for the adapted viruses for replication in the upper respiratory tract. At least in the case of G2, it appears that the PB2 D87N mutation may play a role in viral replication and that this mutation functions by increasing polymerase activity. The discrepancy between comparative growth curve and polymerase assay data at 33°C for G1, however, clearly shows that polymerase activity is not the sole reason for increased viral production from human airway cells, at least at this temperature.
FIG 3 .
Polymerase activity and receptor specificity of H1N1 influenza viruses. (A) Polymerase activity of ribonucleoprotein complexes of TN/560, G1, and G2 (H1N1) viruses as determined by a dual-luciferase reporter assay in three independent experiments. 293T cells were transfected in triplicate with luciferase and Renilla reporter plasmids, together with plasmids expressing PB2, PB1, PA, and NP from either TN/560, G1, or G2 viruses. Cells were incubated at 33°C (orange bars), 37°C (red bars), or 39°C (purple bars) for 24 h, and cell lysates were analyzed to measure firefly luciferase and Renilla activities. The latter was used to normalize transfection efficiency. Values shown represent the activities of each ribonucleoprotein (RNP) complex relative to that of TN/560. *, P < 0.05; °, P <0.01 compared with the value for TN/560 virus (one-way ANOVA). (B) Receptor specificity of NJ/15, TN/560, G1, and G2 (H1N1) influenza viruses. Association constants (Kass) of virus complexes with “human-type” sialylglycopolymers conjugated to 6′-SL and 6′-SLN are shown. Higher Kass values indicate stronger binding. Values are the means from four independent experiments. *, P < 0.05; °, P <0.01 compared with the value for TN/560 virus (one-way ANOVA).
The receptor specificities of all four A/H1N1 viruses were also compared (Fig. 3B) (9). The patterns of binding to fetuin were identical among all isolates (association constant [Kass], ≈6.9 ± 1.4; 1/µM sialic acid) and negligible to the “avian-type” 3′-sialylglycopolymer (3′-SL). NJ/15 and G2 viruses showed significantly less binding to both “human-type” 6′-sialylglycopolymer (6′-SL) and 6′-sialyllactosamine (6′-SLN) receptors than did the pandemic TN/560 strain (~3-fold difference; P < 0.05) (Fig. 3B). In contrast, the G1 variant (with the HA K154Q substitution) exhibited an affinity for the 6′-SLN sialic polymer similar to that observed for TN/560 but was unable to bind to 6′-SL, indicating that G1 discriminated both α2,6-linked receptors (Fig. 3B). Given the preferential binding of human H1N1 viruses to a 6′-SLN receptor (10), our findings suggest that the HA K154Q mutation is a mammal-associated change that leads to better refinement of receptor-binding properties of currently circulating pandemic A/H1N1 viruses.
To test whether the acquired HA K154Q mutation changed the antigenic specificity of the pandemic TN/560 virus, we determined the antigenic properties of all A/H1N1 viruses in a hemagglutination inhibition (HI) test using postinfection ferret antisera and monoclonal antibodies (MAbs) raised against a selection of currently circulating seasonal, pandemic, and swine H1 isolates (see Table S2 in the supplemental material). TN/560 and G1 did not react with the ferret postinfection antisera or MAbs raised against contemporary human or swine H1 viruses, which was in good agreement with earlier findings (11). G1 exhibited a slight change in reactivity with H5, E2, and H7 MAbs (Table S2), suggesting that the HA mutation at residue 154 may play a role in antigenicity.
DISCUSSION
In our study, coinfection of human airway cells with two A/H1N1 strains allowed us to study the possible interactions between viruses, which are difficult to assess on the basis of surveillance data alone. Although the NHBE model lacks a functional adaptive immune system, which would obviously play a role in virus adaptation, it is worth mentioning that our model correlated well with what has been seen in nature. Therefore, this work could be useful in identifying new possible A/H1N1 influenza virus evolution scenarios. Our findings suggest that generation of viable intrasubtype reassortment between currently circulating oseltamivir-resistant seasonal and pandemic viruses is possible but requires initial dominance of the seasonal A/H1N1 strain. Although we did not observe the emergence of a drug-resistant reassortant, the G2 variant was more fit than its parental strains for replication in ferrets. The lack of detection of such reassortants before now may be explained by a too-low ratio of seasonal to pandemic A/H1N1 strains.
We anticipate that the more favorable adaptation scenario for the A/H1N1 pandemic virus seems to tend toward an antigenic drift than a shift. Indeed, in two out of three coinfection groups studied, the pandemic genotype dominated. Our data showed that the acquisition of two nonsynonymous mutations in the HA (K154Q) and PA (L295P) genes resulted in rapid selection of the pandemic virus with improved replication and virulence. All pandemic A/H1N1 viruses except TN/560 possessed a proline at residue 295 in the PA protein, indicating a selective advantage for this mutation in the mammalian host, such as better replication in the upper respiratory tract, as confirmed by minigenome data. In contrast, the HA K154Q substitution identified in G1 was not present in any contemporary pandemic A/H1N1 influenza virus examined. Located next to the HA receptor-binding site, the K154Q mutation affected the protein structure such that binding to the 6′-SL receptor became impossible, probably because of an increase of the negative charge of the HA molecule. Thus, this HA mutation, resulting in a typical human virus-like preference due to strong recognition of only 6′-SLN sialosides, could contribute to the increased initial ability of the virus to infect and spread in the human airway and lung epithelium. In addition, it was determined previously that adaptation of the PB2 protein is also crucial for efficient transmission of A/H1N1 influenza virus (12). In this study, a single mutation, PB2 D87N, was responsible for increased G2 RNP polymerase activity at the lower temperature (33°C) found in the mammalian airway and, therefore, probably contributed to better replication in the nose, significantly elevated inflammation, or more efficient transmission. It is noteworthy that alignment of full PB2 proteins showed the D87N mutation only in 0.06% of sequenced isolates. We believe that surveillance should involve careful examination of the adaptive changes identified here as possible markers for enhanced morbidity of the pandemic viruses. In conclusion, our study demonstrates that the emergence of a more virulent A/H1N1 pandemic strain is possible.
MATERIALS AND METHODS
Cell lines and viruses.
MDCK and human embryonic kidney (293T) cells were obtained from the American Type Culture Collection. NHBE cells were obtained from Cambrex Bio Science. All cell cultures were maintained as previously described (13, 14). For NHBE cell differentiation, culture medium was kept on both the basal and the apical sides of the cells for 10 to 14 days, until the cells formed a monolayer. The cells were then left at an air-liquid interface (with medium on the basal side only) for another 2 to 3 weeks so that they could differentiate into polarized cultures.
The seasonal NJ/15 and pandemic TN/560 (H1N1) influenza viruses were obtained from the World Health Organization collaborating laboratories. Stock viruses were prepared in MDCK cells at 37°C for 72 h, and aliquots were stored at –70°C until used.
Coinfection studies with NHBE cells.
Duplicate differentiated NHBE cell cultures were washed extensively with sterile phosphate-buffered saline (PBS) and then were inoculated via the apical side with mixtures of seasonal NJ/15 and pandemic TN/560 virus pairs at different ratios (10:90, 50:50, and 90:10). NHBE cells were inoculated with mixtures of the two viruses at a multiplicity of infection (MOI) of 0.1 in 100 µl of inoculum. After 1 h of incubation, the inoculum was removed, and the cells were then incubated at 37°C for 72 h. Viruses released into the apical compartment of NHBE cells were harvested by the apical addition and collection of 300 µl of medium allowed to equilibrate for 30 min. Then, 100 µl of the sample was used as an inoculum for the next passage.
Genotyping.
To determine if reassortant viruses emerged in the apical medium samples of coinfected NHBE cultures, a genotyping analysis of plaque-purified viruses isolated from the harvested samples was performed after a total of 3 (n = 25) or 10 (n = 30) passages. After completion of genome analysis, two viruses representing G1 and G2 reassortant genotypes were subsequently cloned twice by plaque purification in MDCK cells, and the cloned viruses were passaged once in the MDCK cells for 72 h at 37°C to prepare a virus stock.
One-step reverse transcriptase PCR (RT-PCR; Qiagen) was also used to determine the NJ/15- or TN/560-based origin of each gene in plaque-purified virus populations after a total of 3 (n = 25) or 10 (n = 30) passages in coinfected NHBE cells. Primers were designed to hybridize specifically to each NJ/15- or TN/560-derived gene segment at appropriate annealing temperatures (8). Multiplex reactions using NJ/15- and TN/560-specific primers for each viral gene were performed to simultaneously detect both NJ/15- and TN/560-derived genes of different amplicon sizes.
Real-time RT-PCR.
Copy numbers of HA and NA RNAs of NJ/15 and TN/560 viruses were quantified by multiplex quantitative real-time RT-PCR using primers and probes specific for NJ/15 and TN/560 RNAs (HA-Fwd [5′ CCATTGCCGGTTTCATTG 3′] and HA-Rev [5′ AATGGCATTYTGTGTGC 3′] for H1 HA of both NJ/15 and TN/560 viruses; NJ-HA-PRO [5′ VIC–GCCAGATCCTTGCTCATTC–6-carboxytetramethylrhodamine {TAMRA} 3′] and TN-HA-PRO [5′ 6-carboxyfluorescein {FAM}–GGACAGGGATGGTAGATGGA–TAMRA 3′] for H1 HA of NJ/15 and TN/560, respectively; NA-Fwd [5′ AGTGGATGGGCTATATACA 3′] and NA-Rev [5′ CCATTKGAATGTTTGTCATTTA 3′] for N1 NA of both NJ/15 and TN/560; and NJ-NA-PRO [5′ VIC-AAGAATTGGCTCCAAAGGA-TAMRA 3′] and TN-NA-PRO [5′ FAM-GGAATGCAGAACCTTCTTCT-TAMRA 3′] for N1 NA of NJ/15 and TN/560, respectively). Real-time RT-PCR was performed using a Qiagen one-step RT-PCR kit, 600 nmol/liter of each primer, 500 nmol/liter of the TN/560-specific probe, 100 nmol/liter of the NJ/15-specific probe, and 2 mM MgCl2 (Roche). Quantitative real-time RT-PCR was done using an ABI Fast real-time PCR system 7500 thermocycler and corresponding software (Stratagene), with the following cycling conditions: 50°C for 30 minutes and 95°C for 15 minutes, followed by 40 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 40 s. RNA standards were quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific), and real-time RT-PCR efficiencies were similar for HA and NA RNA of NJ/15 and TN/560H1N1 viruses, ranging from 2.5 to 2.7.
Virus sequence analysis.
Viral RNAs (vRNAs) were isolated from virus-containing cell culture fluid by using an RNeasy minikit (Qiagen). Samples were reverse transcribed and analyzed by PCR using universal primers, as described previously (15). Sequencing was performed by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital and edited by using the Lasergene sequence analysis software package (DNASTAR).
Infectivity and replication kinetics of A/H1N1 influenza viruses.
The infectivity of A/H1N1 viruses was determined by 50% tissue culture infective dose (TCID50), PFU count, and 50% egg infectious dose (EID50) as described previously (13, 16).
To determine multistep growth curves for NHBE cells, triplicate cell cultures growing in 12-mm-diameter inserts were inoculated via the apical side with each virus at an MOI of 0.1 at 33°C or 37°C. After 1 h of incubation, the inoculum was removed. Viruses released into the apical compartment of NHBE cells were harvested at the indicated time points by the apical addition and collection of 300 µl of medium allowed to equilibrate for 30 min at 33°C or 37°C. The virus titers were determined as log10 numbers of PFU/ml in MDCK cells.
Assessment of virus pathogenicity and transmission in ferrets.
Pathogenicity in 4- to 5-month-old male ferrets obtained from Marshall Farms (North Rose, NY) was tested. All ferrets were seronegative for influenza A H1N1, H3N2, and H5N1 and for influenza B viruses. Donor ferrets were initially housed separately from contact ferrets. The donor ferrets (two for each group) were inoculated intranasally under light isoflurane anesthesia with 106 PFU of NJ/15, TN/560, G1, or G2 virus in 1 ml of sterile PBS. For the direct-contact transmission experiment, 24 h after inoculation (day 1 postinoculation [p.i.] for the inoculated ferrets and day 0 postcontact [p.c.] for the contact ferrets), two naïve direct-contact ferrets were housed in the cage with two donor ferrets. Clinical signs of infection, relative inactivity index (17), weight, and temperature were recorded daily.
To monitor virus shedding, nasal washes were collected from ferrets on days 1, 2, 4, 6, 8, and 10 p.i. (or days 1, 3, 5, 7, and 9 p.c.). The virus titers were determined as log10 TCID50/ml in MDCK cells. The limit of virus detection was 0.5 log10 TCID50/ml. For calculation of the mean, samples with virus titers of <0.5 log10 TCID50/ml were assigned a value of 0. Inflammatory cell counts and protein concentrations in nasal washes were also determined.
To determine inflammatory-cell counts, the nasal washes were centrifuged at 2,000 rpm for 10 min. The cell pellet was resuspended in PBS, and the cells were counted microscopically with a Countess automated cell counter (Invitrogen). The total number of inflammatory cells was calculated on the basis of the initial volume of nasal wash. The protein concentration in cell-free nasal washes was measured by using Bio-Rad protein assay dye reagent (Hercules).
All animal experiments with H1N1 influenza A viruses were performed in biosafety level 2+ facilities at St. Jude Children’s Research Hospital (Memphis, TN). All animal studies were approved by the St. Jude Children’s Research Hospital Animal Care and Use Committee and were conducted according to applicable laws and guidelines.
Titration of virus in ferret organs.
One donor animal inoculated with NJ/15, TN/560, G1, or G2 virus was euthanized by intracardiac injection of euthanasia V solution on day 3 p.i., and tissue samples (~0.5 g each) were collected from lungs (4 lobes tested separately), nasal turbinates, tracheae, spleens, livers, and small intestines. Samples were homogenized in 1 ml of sterile PBS with antibiotics, and the virus titer (log10 TCID50/gram tissue) in MDCK cells was determined.
Histopathologic analysis.
Tissues (lung, nasal turbinate, trachea, spleen, liver, and small intestine) of donor ferrets were collected at the time of necropsy, fixed in 10% neutral buffered formalin, and embedded in paraffin. Sections 5 µm thick were stained with hematoxylin and eosin and studied by light microscopy.
Serologic tests.
Serum samples were collected from ferrets 3 weeks after inoculation, treated with receptor-destroying enzyme, heat inactivated at 56°C for 30 minutes, and tested by a hemagglutination inhibition (HI) assay with 0.5% packed turkey red blood cells by a standard method as described previously in the World Health Organization manual on animal influenza diagnosis and surveillance (18). Virus-neutralizing titers were determined by infection of MDCK cells and expressed as the reciprocal of the highest serum dilution that neutralized 50% of 100 TCID50 of virus after incubation at 37°C for 72 h.
Minigenome assay for polymerase activity.
Subconfluent monolayers of 293T cells (7.5 × 105 cells in 35-mm dishes) were transfected with the luciferase reporter plasmid (enhanced green fluorescent protein [EGFP] open reading frame in pHW72-EGFP replaced with the firefly luciferase gene) (19) and a mix of PB2, PB1, PA, and NP plasmids (TN/560 or mutated) in quantities of 1, 1, 1, and 2 µg, respectively. The plasmid pGL4.75[hRluc/CMV] vector, which expresses Renilla luciferase (Promega), was used as an internal control for the dual-luciferase assay. As a negative control, 293T cells were transfected with the same plasmids, with the exception of the NP expression plasmid. After 24 h of incubation at 33°C, 37°C, or 39°C, cell extracts were harvested and lysed, and luciferase levels were assayed with the dual-luciferase assay system (Promega) and a BD Monolight 3010 luminometer (BD Biosciences). Experiments were performed in triplicate.
Receptor-binding assay.
The binding of H1N1 influenza viruses to fetuin (containing α2,3- and α2,6-linked sialyl receptors) was measured in a direct solid-phase assay using the immobilized virus and horseradish peroxidase-conjugated fetuin, as described previously (9). The affinity of viruses for synthetic 3′- and 6′-sialylglycopolymers obtained by conjugation of a 1-N-glycyl derivative of 3′- or 6′-sialyllactose (3′-SL or 6′-SL, respectively) or 3-aminopropylglycoside of 6′-sialyllactosamine (6′-SLN) with poly(4-nitrophenylacrylate) was measured in a competitive assay based on the inhibition of binding to peroxidase-labeled fetuin (9). Association constant (Kass) values were determined as the sialic acid (N-acetylneuraminic acid [Neu5Ac]) concentration at the point represented by maximum binding site value (Amax)/2 on Scatchard plots.
Statistical analysis.
The virus yields, plaque sizes, virus titers in nasal wash samples, and differences in fevers, weights, total numbers of cells, and protein concentrations in nasal wash samples observed for ferrets inoculated with H1N1 viruses were compared by analysis of variance (ANOVA). A probability value of 0.05 was prospectively chosen to indicate that the findings were not the result of chance alone.
ACKNOWLEDGMENTS
We thank Elena A. Govorkova for helpful discussions, Jon P. Seiler and Alexey M. Khalenkov for excellent technical assistance, Sharon Lokey for the assistance with animal work in the ABSL2+ laboratory, Julie Groff for illustrations, and David Galloway for editorial assistance.
This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN266200700005C, by RAS Presidium grant “Molecular and Cell Biology,” by F. Hoffmann-La Roche, Ltd., and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Citation Ilyushina, N. A., M. F. Ducatez, J. E. Rehg, B. M. Marathe, H. Marjuki, et al. 2010. Does pandemic A/H1N1 virus have the potential to become more pathogenic? mBio 1(5):e00249-10. doi:10.1128/mBio.00249-10.
SUPPLEMENTAL MATERIAL
REFERENCES
- 1. Kilbourne E. D. 2006. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12:9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization 2009. Global alert and response: pandemic (H1N1)—update 58. http://www.who.int/csr/don/2009_07_06/en/index.html Accessed 6 July 2010
- 3. Hurt A. C., Holien J. K., Parker M. W., Barr I. G. 2009. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs 69:2523–2531 [DOI] [PubMed] [Google Scholar]
- 4. Janies D. A., Voronkin I. O., Studer J., Hardman J., Alexandrov B. B., Treseder T. W., Valson C. 2010. Selection for resistance to oseltamivir in seasonal and pandemic H1N1 influenza and widespread co-circulation of the lineages. Int. J. Health Geogr. 9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peacey M., Hall R. J., Sonnberg S., Ducatez M., Paine S., Nicol M., Ralston J. C., Bandaranayake D., Hope V., Webby R. J., Huang Q. S. 2010. Pandemic (H1N1) 2009 and seasonal influenza A (H1N1) co-infection, New Zealand, 2009. Emerg. Infect. Dis. 16(10):1618–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perez D. R., Sorrell E., Angel M., Ye J., Hickman D., Pena L., Ramirez-Nieto G., Kimble B., Araya Y. 2010. Fitness of pandemic H1N1 and seasonal influenza A viruses during co-infection: evidence of competitive advantage of pandemic H1N1 influenza versus seasonal influenza. PLoS Curr. 1:RRN1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization 2009. Global alert and response: pandemic (H1N1)—update 103. http://www.who.int/csr/don/2010_06_04/en/index.html Accessed 6 July 2010
- 8. Ducatez M. F., Sonnberg S., Hall R. J., Peacey M., Ralston J., Webby R. J., Huang Q. S. 2010. Genotyping assay for the identification of 2009-2010 pandemic and seasonal H1N1 influenza virus reassortants. J. Virol. Methods 168:78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matrosovich M. N., Gambaryan A. S., Tuzikov A. B., Byramova N. E., Mochalova L. V., Golbraikh A. A., Shenderovich M. D., Finne J., Bovin N. V. 1993. Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology 196:111–121 [DOI] [PubMed] [Google Scholar]
- 10. Mochalova L., Gambaryan A., Romanova J., Tuzikov A., Chinarev A., Katinger D., Katinger H., Egorov A., Bovin N. 2003. Receptor-binding properties of modern human influenza viruses primarily isolated in Vero and MDCK cells and chicken embryonated eggs. Virology 313:473–480 [DOI] [PubMed] [Google Scholar]
- 11. Garten R. J., Davis C. T., Russell C. A., et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 (A)H1N1 influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tumpey T. M., Maines T. R., Van Hoeven N., Glaser L., Solorzano A., Pappas C., Cox N. J., Swayne D. E., Palese P., Katz J. M., García-Sastre A. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655–659 [DOI] [PubMed] [Google Scholar]
- 13. Ilyushina N. A., Govorkova E. A., Gray T. E., Bovin N. V., Webster R. G. 2008. Human-like receptor specificity does not affect the neuraminidase-inhibitor susceptibility of H5N1 influenza viruses. PLoS Pathog. 4:e10000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson C. I., Barclay W. S., Zambon M. C., Pickles R. J. 2006. Infection of human airway epithelium by human and avian strains of influenza A virus. J. Virol. 80:8060–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 16. Reed L. J., Muench H. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 17. Reuman P. D., Keely S., Schiff G. M. 1989. Assessment of signs of influenza illness in the ferret model. J. Virol. Methods 24:27–34 [DOI] [PubMed] [Google Scholar]
- 18. Palmer D. F., Dowdle W. R., Coleman M. T., Schild G. C. 1975. Advanced laboratory techniques for influenza diagnosis. Immunology series no. 6 U.S. Department of Health, Education, and Welfare, Center for Disease Control, Atlanta, GA [Google Scholar]
- 19. Salomon R., Franks J., Govorkova E. A., Ilyushina N. A., Yen H. L., Hulse-Post D. J., Humberd J., Trichet M., Rehg J. E., Webby R. J., Webster R. G., Hoffmann E. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russell R. J., Gamblin S. J., Haire L. F., Stevens D. J., Xiao B., Ha Y., Skehel J. J. 2004. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287–296 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.