Abstract
Induced pluripotent Huntington's disease monkey stem cells (rHD-iPSCs) were established by the overexpression of rhesus macaque transcription factors (Oct4, Sox2, and Klf4) in transgenic Huntington's monkey skin fibroblasts. The rHD-iPSCs were pluripotent and capable of differentiating into neuronal cell types in vitro and developed teratoma in immune compromised mice. We also demonstrated the upregulation of endogenous Oct4 and Sox2 after successful reprogramming to pluripotency in rHD-iPSCs, which was not expressed in skin fibroblasts. rHD-iPSCs also developed cellular features comparable to Huntington's disease (HD), including the accumulation of mutant huntingtin (htt) aggregate and the formation of intranuclear inclusions (NIs) paralleling neural differentiation in vitro. Induced pluripotent stem cells from transgenic HD monkeys open a new era of nonhuman primate modeling of human diseases. rHD-iPSCs that develop key HD cellular features and parallel neural differentiation can be a powerful platform for investigating the developmental impact on HD pathogenesis and developing new therapies, which can be evaluated in HD monkeys from whom the rHD-iPSCs were derived.
Introduction
Personal stem cells hold great promise in the development of cell therapies for Parkinson and Huntington's diseases. In addition to its therapeutic potential, a patient's personal stem cells are unique models for investigating disease pathogenesis and the development of personal medicine (Daley, 2010; Kiskinis and Eggan, 2010). Despite significant advancement in iPSC technology that has led to the derivation of human, nonhuman primate, and rodent iPSCs (Liu et al., 2008; Takahashi and Yamanaka, 2006; Takahashi et al., 2007), therapeutic efficacy and safety of iPSCs remain major concerns in clinical applications. Although cell replacement therapy holds great promise in curing diseases, clinical applications remain difficult and unpredictable because a comprehensive evaluation mechanism in appropriate animal models is not yet available, which is crucial for determining not only the safety of stem cell replacement therapy but also for determining therapeutic efficacy that may lead to a better prediction model for clinical outcomes in human patients (Laowtammathron et al., 2010; Thomson and Marshall, 1998; Yang et al., 2008). Although treatment of sickle cell anemia and Parkinson's disease using rodent iPSCs has resulted in clinical improvement (Hanna et al., 2007; Wernig et al., 2008), an animal model with high genomic and physiological similarity to humans will further validate the potential of the iPS technology in future clinical applications. Nonhuman primates share high degrees of physiologic, neurologic and genetic similarity with humans, which make them one of the best models for investigating human physiology, disease pathogenesis, and the development of novel therapies (Chan, 2004; Liu et al., 2008).
Our studies evolved because of our most recent success in developing transgenic Huntington's disease (HD) monkeys (Yang et al., 2008). HD is an autosomal dominant neurological disorder and currently has no cure. The expansion of the ployglutamine (polyQ; >37) stretches at the N-terminus of the huntingtin (htt) protein (Huntington's Disease Collaborative Research Group, 1993) is the primary cause of HD. The accumulation of oligomeric mutant htt, the formation of intranuclear inclusions, and progressive neuronal death are the hallmark pathological features in HD. In addition to revealing unique cellular changes and HD pathogenesis, HD monkeys also developed clinical features including dystonia, chorea, and seizure, which were comparable to human HD patients that no other animal model has yet reported (Yang et al., 2008). Due to the distinctive clinical features of the HD monkeys, iPSCs derived from HD monkeys provide a unique in vitro model for investigating HD pathogenesis and for drug discovery research that could later be validated in HD monkeys.
Although htt is widely expressed in the body with its highest levels of expression in the brain and testis, the primary site of damages in HD are found in the brain (Davies and Ramsden, 2001; Gutekunst et al., 1999; Li and Li, 2006; Rubinsztein, 2002). In order to investigate the mechanism of cell/tissue type-specific degeneration, pluripotent stem cells are a unique model for studying the pathogenic role of mutant htt genes and the impact of development events, because they are capable of differentiating into multiple cell lineages that mimic early embryogenesis.
Although human HD embryonic stem (hHD-ES) cells have been generated by using different methods including the use of human embryos diagnosed with HD (Mateizel et al., 2006) or by induced pluripotency using HD patients skin cells (Mateizel et al., 2006; Park et al., 2008a), follow-up study has been limited. Currently, no HD cell model has been reported that developed hallmark pathological features of HD paralleling neural development. Our recent report on hybrid HD monkey stem cells suggested that pluripotent stem cells with small htt fragments and expanded polyQ develop robust HD cellular features paralleling neural development, which can be a useful model for investigating the developmental impact on HD pathogenesis (Laowtammathron et al., 2010).
In this study, we demonstrate the reprogramming of HD monkey skin fibroblasts to pluripotency by the overexpression of three transcription factors and the development of HD cellular features during the course of in vitro neural development.
Materials and Methods
Production of transgenic HD monkeys by lentiviruses
Transgenic HD monkeys were generated by using high titer lentiviruses introduced into the perivitelline space of mature oocytes followed by intracytoplasmic sperm injection and in vitro culture until embryo transfer at the four- to eight-cell stage. Two gene constructs: (1) exon 1 of the human htt gene with 84 CAG repeats and (2) the green fluorescent protein (GFP) gene regulated by the human polyubiquitin C promoter were inserted into lentiviruses. Successful transgenesis was determined by polymerase chain areaction (PCR) and Southern blot analysis (Yang et al., 2008).
Establishment and characterization of donor skin cells
A miscarried transgenic HD monkey was recovered during the fourth month of pregnancy. Skin samples were collected and a primary culture was established after enzymatic digestion. The expression of mutant htt was determined by immunocytochemistry and Western blot analysis, whereas transgenesis was determined by PCR and Southern blot analysis (Laowtammathron et al., 2010; Yang et al., 2008).
Construction and generation of high titer retroviruses expressing rhesus macaque Oct4, Sox2, and Klf4 cDNA
Rhesus macaque Oct4, Sox2, and Klf4 cDNA were cloned by PCR amplification of rhesus monkey embryonic stem cell cDNA using specific primer sets. Oct4-F (5′-TTC ACC AGG CCC CCG GCT T-3′); Oct4-R (5′-CCT GTC CCT CAT TCC TAG AAG-3′); Sox2-F (5′- CAC AGC GCC CGC ATG TAC AA-3′); Sox2-R (5′-AGT TCG CTG TCC TGC CCT CA-3′); Klf4-F (5′-CAC CTG GCG AGT CTG ACA TG-3′); Klf4-R (5′-TGG GTC ATA TCC ACT GTC TGG-3′). PCR amplified cDNAs were then subcloned into pGem-T easy cloning vector and subsequently inserted in retroviral vector pMXs (Addgene, Cambridge, MA). The resultant retroviral vectors were named pMX–Oct4, pMX–Sox2, and pMX–Klf4. To produce high titer retroviruses, 293T packaging cells were cotransfected with retroviral vectors carrying the transcription factors cDNA, pUMVC (composed of structural genes for virion assembly; Addgene) and pVSVG (Invitrogen, Carlsbad, CA). Culture medium was collected at 48 h posttransfection for 3 consecutive days at 24-h intervals. The supernatant was centrifuged at 25,000 × g for 90 min. The viral vector pellet was resuspended, aliquoted and kept frozen at −80°C.
Derivation and culture of induced pluripotent Huntington's monkey ES-like cells
Day 1: skin fibroblasts were seeded at 100,000 cells in a 35-mm tissue culture dish. Retroviral gene transfer (days 2 and 3): 5 μL of each concentrated retrovirus (MX–Oct4, MX–SOX2, and MX–Klf4) were added together with 8 μg/mL of ploybrene (Sigma, St. Louis, MO) into skin fibroblast culture. Day 4: skin fibroblasts (1 × 105 cells) were passaged onto a 35-mm gelatin-coated dish, with fresh skin fibroblast culture media Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 1 mM glutamine (Invitrogen) and 1 × penicillin/streptomycin (Invitrogen). Day 5: fresh monkey ES cell culture media [Knockout-Dulbecco's modified Eagle's medium (KO-DMEM; Invitrogen) with 20% Knock-out Serum Replacement (KSR; Invitrogen), 1 mM glutamine, 1 × nonessential amino acids (NEAA), and 4 ng/mL of human basic fibroblast growth factor (bFGF; Chemicon, Inc., Tumecula, CA)] was replaced. Valproic acid (VPA; 1 mM) was then supplemented for 2 weeks. At approximately 2 weeks postretroviral transfection, a monkey ES cell-like colony was selected based on morphology and mechanically passaged onto mouse fetal fibroblast (MFF) feeder cells with monkey ES media. The resulting HD monkey iPS cell was named the rhesus macaque iPS cell line (RiPS).
Characterization of induced pluripotent HD monkey ES-like cells (RiPS3)
The htt-84Q gene was detected by PCR using ubiquitin forward primer (5′-GAGGCGTCAGTTTCTTTGGTC-3′) and htt-84Q-R reverse primer (5′-GCTGGGTCACTCTGTCTCTG-3′) with an expected amplicon of 818-bp. Amplication condition: 35 cycles at 96°C for 5 min, 96°C for 45 sec, 62°C for 45 sec, and 72°C for 150 sec, followed by 72°C for 7 min. The number of polyglutamine repeats were determined by sequencing the PCR product generated by using a primer: HD exon 1-F (5′-GGCGACCCTGGAAAAGCTGA-3′). The GFP gene was detected by PCR using ubiquitin forward primer (5′-GAGGCGTCAGTTTCTTTGGTC-3′) and GFP-R reverse primer (5′-TAGTGGTTGTCGGGCAGCAG-3′) with an expected amplicon of 869 bp. WT-monkeys and plasmid DNA of htt-84Q and GFP were used as the negative and positive control.
Immunocytochemistry of stem cell markers
RiPS3 cells cultured in four-well plates were fixed in 4% PFA followed by permeabilization using 1% Triton-X and blocking by using 2% bovine serum albumin (BSA) and 130 mM glycine in phosphate buffer saline (PBS). Primary antibodies (TRA-1-60; SSEA-4 and Oct4) were incubated overnight followed by thorough washes and incubation with a secondary antibody conjugated with appropriate fluorescent tag. Hoechst 33342 (5 μg/mL) was used for DNA staining, and samples were examined by using BX71 (Olympus, Melville, NY) microscope equipped with an epifluorescent device. Alkaline phosphatase assay was performed as described by manufacturer (Vector Lab, Burlingame, CA).
Immunostaining of mutant htt
In vitro differentiated RiPS3 were prepared as described in previous section. Primary antibody “mEM48 (1:50)” was then incubated overnight at 4°C followed by thorough washes. DAB (Vector Laboratories) staining was then performed after avidin–biotin treatment using the Vectastain Elite ABC kit (Vector Laboratories). Images were captured with the use of MetaMorph software (Universal Imaging, Miami Lakes, FL).
Cytogenetic analysis/G-banding analysis
Cytogenetic analysis was performed by Cell Line Genetics LLC (Madison, WI). In brief, RiPS3 at passage 15 was incubated with KaryoMax® colcemid (Invitrogen) for 20 min, trypsinized, centrifuged, and then resuspended in 0.075 M KCl solution for 20 min. Cell samples were fixed by using 3:1 ratio of methanol to glacial acetic acid and stored at 4°C until G-banding. Fixed cell suspension was dropped on wet slides, air dried, and baked at 90°C for 1 h prior to immersion in 0.5× Trypsin-EDTA (Invitrogen) with two drops of 67 mM Na2HPO4 for 20 to 30 sec. Samples were then stained with Leishman Stain (Sigma) after thorough rinse. A total of 20 metaphases were analyzed and images were captured by using the CytoVysion® digital imaging system (Applied Imaging, Grand Rapids, MI).
In vitro differentiation to neuronal lineage
RiPS3 cells were mechanically removed, as small pieces, followed by suspension culture for 7 days. To differentiate into mature neurons, the resulted embryoid bodies (EBs) were then transferred onto a gelatin coated plate in N1 medium (N1 base medium: KO-DMEM (Invitrogen), nonessential amino acid (Invitrogen), and 200 mM of L-glutamine (Invitrogen) supplemented with N2 supplement) for 7 days, in N2 medium (N1 base medium supplemented with 20 ng/mL bFGF instead of N2 supplement) for 14 days and in N3 medium [KO-DMEM supplemented with 1% FBS (Hyclone, Logan, UT) and B27 supplement (Invitrogen)] for 7 days. In order to mimic postmitotic condition, an extended culture (4 weeks) at N3 stage was performed. Successful differentiation to NPCs and neurons were determined by the expression of nestin, βIII tubulin, and MAP2 (Laowtammathron et al., 2010).
Quantitative RT-PCR (Q-PCR)
Total RNA was extracted by using a RNeasy Mini Kit (Qiagen, Chatsworth, CA) and reverse transcribed as instructed by manufacturer (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems, Norwalk, CT). Q-PCR was performed by using 2× Power SYBR® Green PCR Master Mix (Applied Biosystems), with specific primers and cDNA samples. iQ5 Optical System (BioRad, Hercules, CA) was used, and cDNA samples were first incubated at 96°C for 3 min followed by 45 cycles; at 95°C for 10 sec and 62°C for 30 sec. Mutant htt specific primers were: HD Exon 1-F (5′-ATG GCG ACC CTG GAA AAG CT-3′) and HD Exon 1-R (5′-TGC TGC TGG AAG GAC TTG AG-3′). The specific primer for 18S: 18S F (5′-CGG CTA CCA CAT CCA AGG AA-3′) and 18S R (5′-CCT GTA TTG TTA TTT TTC GTC ACT ACC T-3′). Primers for endogenous rhesus monkey transcription factors are: Oct4: Monkey Oct4-F (5′-GGA ATG AGG GAC AGG GGG AG-3′) and Monkey Oct4-R (5′-ACT CCC CTG CCC CCA CCC T-3′); Sox2: Monkey Sox2-F (5′-GCA GGT TGA CAT CGT TGG TAA T-3′) and Monkey Sox2-R (5′-CAA CTA CGG AAA ATA AAG GGG G-3′); and Klf4: Monkey Klf4-QPCR-F (5′-GAA GGA GCC CAG CCA GAA AG-3′) and Monkey Klf4-QPCR-R (5′-TCA CCC CCT TGG CAT TTT GTA A-3′). Primers for exogenous rhesus monkey transcription factors are: Oct4: Monkey OCT4(iPS)-F (5′-CTG TCT CCG TCA CCA CTC TG-3′) and pMXs vestor-R (5′-ACA GGT GGG GTC TTT CAT TCC-3′); Sox2: pMX-1811F (5′-GAC GGC ATC GCA GCT TGG ATA CAC-3′) and Monkey Sox2-R (5′-CCC CCC GAA GTT TGC TGC G-3′); and Klf4: pMX-1811F (5′-GAC GGC ATC GCA GCT TGG ATA CAC-3′) and Monkey Klf4(iPS)-R (5′-GTG GAG AAA GAT GGG AGC AGC-3′).
Western blot analysis
All proteins were extracted from undifferentiated and differentiated RiPS3 cells, and their concentration was determined by Bradford assay (Pierce, Rockford, IL). Equal amounts (20–30 μg) of protein extract with loading dye were boiled prior to loading into a 9% polyacrylamide gel (BioRad). Proteins were transferred to a PVDF membrane, after electrophoresis, followed by blocking and incubation with primary antibodies (mEM48 and γ-tubulin at 1:50 and 1:2000, respectively). After thorough washes, the membrane was incubated with secondary antibody conjugated with peroxidase followed by detection using Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer, Norwalk, CT) (Yang et al., 2008)
Formation of teratoma in severe compromised immune deficient (SCID) mice
RiPS3 cells were surgically grafted under the kidney capsule of SCID mice. Animals were euthanized at 2–3 months after transplantation. Teratoma was recovered for histological analysis. All animal procedures were approved by the IACUC at the Emory University.
Statistical analysis
A Student's t-test was used for statistical analysis. Differences of p < 0.05 were considered statistically significant.
Results
Generation of HD monkey ES-like cells by induced pluripotency
A primary culture of skin fibroblasts was established from transgenic male HD monkey at 4 months of gestation. Transgenic status of the mutant htt and GFP gene was determined by PCR and Southern blot analysis (data not shown). The mutant htt gene contained 72 CAG repeats. The expression of transgenic mutant htt was confirmed by Western blot and immunohistochemistry with mEM48, a monoclonal antibody whose reaction with human htt is enhanced by polyQ expansion (Laowtammathron et al., 2010; Yang et al., 2008).
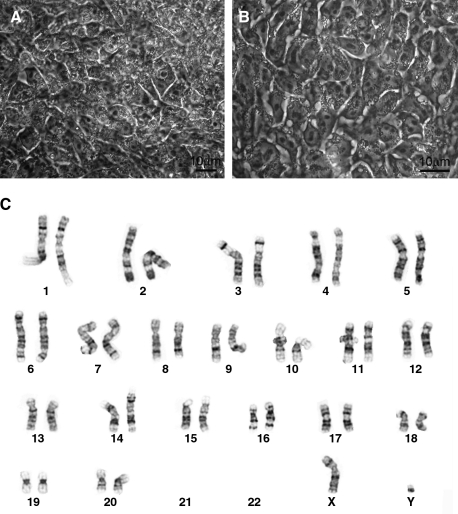
HD monkey skin fibroblasts were coinfected with retroviruses expressing rhesus monkey Oct4, Sox2, and Klf4. A second infection was performed on the following day and transformed skin cells were replated on the next day. VPA was then supplemented for up to 2 weeks prior the pickup of ES cell-like colonies. ES cell-like cells were identified based on a distinctive ES cell morphology of a high nuclear to cytoplasmic ratio (Fig. 1A). ES cell-like colonies were identified and mechanically passaged onto freshly prepared feeder cells. A total of four rHD-iPS cell lines were established. Among the four cell lines, RiPS-3 was randomly selected for further characterization. Similar to the other rHD-iPS cell lines, RiPS-3 retains monkey ES cell morphology at passage 15 (Fig. 1A and B) and is pluripotent as shown in the following sections. Cytogenetic analysis verified that RiPS-3 is a male diploid cell line (42 XY; Fig. 1C), which is identical to the skin fibroblast monkey donor.
FIG. 1.
Induced pluripotent Huntington monkey ES-like cells. (A) RiPS-3 before first passage. (B) RiPS-3 at Passage 13. (C) G-banding analysis of RiPS-3. Cytogenetic analysis of RiPS-3 demonstrated diploid chromosome (42; XY).
Stem cell properties and multipluripotent differentiation capability of RiPS3
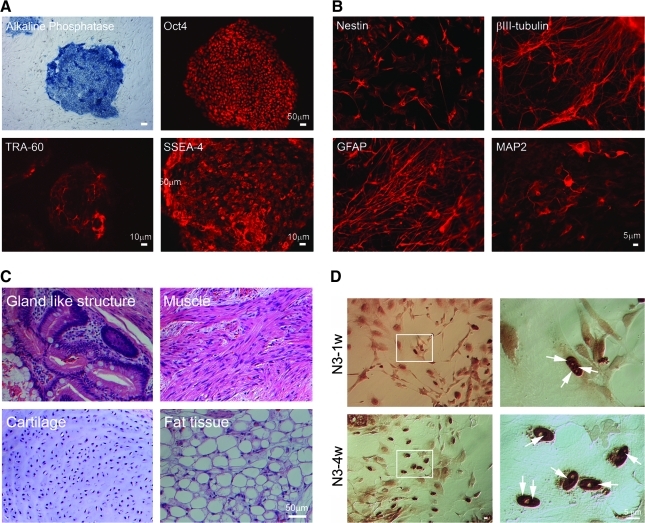
The expression of stem cell markers including Oct4 and stem cell specific surface antigens (SSEA4 and TRA-1-60) in RiPS3 was determined by immunocytochemistry and the expression of Alkaline phophatase (AP) (Fig. 2A).
FIG. 2.
Characterization of RiPS3. (A) Expression of ES-cell specific markers: alkaline phosphatase, Oct4, TRA-1-60, and SSEA4. (B) RiPS-3 was differentiated toward neuronal lineage in vitro using a step-wise differentiation protocol. Antibodies specific for neural progenitor cells (nestin), βIII-tubulin, glial fibrillary acidic protein (GFAP), and MAP2 were used for immunostaining. (C) Hematoxylin and eosin staining of teratoma derived from undifferentiated RiPS-3 grafted under the kidney capsule. (D) Immunostaining of RiPS-3 at the N3-1w and N3-4w stage. Arrow heads indicate intranuclear inclusions.
In vitro differentiation of RiPS3 to neural cell types (Fig. 2B) and the formation of teratoma (Fig. 2C) in immune compromised mice was performed in order to demonstrate the multipotent differentiation capability of RiPS3. Spontaneous differentiation to contracting myocardiac tissue was also observed (data not shown). For in vitro neural differentiation, a step-wise protocol was used in this study (Laowtammathron et al., 2010). The expression of nestin (a progenitor cell marker), Glial fibrillary acidic protein (GFAP), neural specific βIII tubulin, and microtubule-associated protein (MAP2) were detected by immunostaining of differentiated RiPS-3 (Fig. 2B), which suggested neural differentiation capability of RiPS-3.
To determine pluripotency of RiPS-3 in vivo, undifferentiated RiPS-3 were implanted under the kidney capsule of SCID mice for 3 months before the recovery of teratoma for histological analysis (Fig. 2C). Histological study of RiPS-3 derived teratoma revealed gland-like epithelium, muscle, cartilage, and fat tissues (Fig. 2C).
Expression of endogenous and exogenous transcription factors in RiPS-3 cells during in vitro neural differentiation
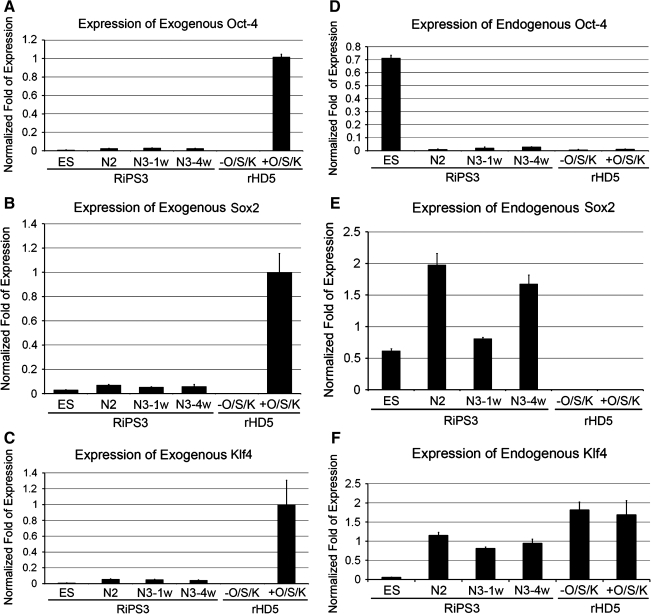
One of the fundamental phenomenon of iPS technology is to reprogram differentiated skin fibroblasts to regain pluripotency by overexpressing a combination of transcription factors including core stemness factors Oct4 and Sox2 that were downregulated in somatic cells, while Klf4 is expected to modify chromatin structure for efficient binding of Oct4 and Sox2 to the target sites (Takahashi and Yamanaka, 2006). By overexpressing a cocktail of transcription factors, the cascade of reprogramming events is elicited and results in the upregulation of endogenous stemness transcription factors to maintain pluripotency. We have measured the expression level of exogenous transcription factors that were introduced by retroviruses in skin fibroblasts and the expression level of endogenous transcription factors in skin fibroblasts transfected by retroviruses, and undifferentiated and differentiated RiPS-3 cells during in vitro neural differentiation. Neuronal maturation is commonly cultured for 1 week at N3 stage (N3-1w). However, we observed an enhanced accumulation of mutant htt aggregate in hybrid HD monkey stem cells when the N3 stage culture is extended to 4 weeks (N3-4w; Laowtammathron et al., 2010).
Exogenous transcription factors were all highly expressed in retroviruses transfected skin fibroblasts, but not in the primary culture of skin fibroblasts, while a relatively low level was observed in RiPS-3 cells at undifferentiated and differentiated stages (Fig. 3A–C). On the other hand, endogenous Oct4 and Sox2 were not detected or at extrememely low levels for Oct4 in skin fibroblasts with or without being transfected by retroviruses (Fig. 3D and E). Endogenous Oct4 was highly expressed only in undifferentiated RiPS-3 cells and was down regulated as RiPS-3 differentiated toward neuronal lineage (Fig. 3D). Endogenous Sox2 was detected in RiPS-3 at both undifferentiated and differentiated stages (Fig. 3E). Unlike Oct4 and Sox2, Klf4 expressed in cells at all differentiated stages and was expressed at relatively low levels in undifferentiated RiPS-3 cells (Fig. 3F) while skin fibroblasts expressed at similar levels with or without retroviral transfection of Klf4 (Fig. 3F).
FIG. 3.
Expression of exogenous and endogenous transcription factors. Expression of exogenous and endogenous transcription factors (Oct4, Sox2, and Klf4). (A) Exogenous Oxt4, (B) Exogenous Sox2, (C) Exogenous Klf4, (D) Endogenous Oct4, (E) Endogenous Sox2, and (F) Endogenous Klf4. Error bars indicate standard deviation.
Expression of mutant htt in RiPS-3 during in vitro neural differentiation
In order to determine if the development of HD cellular characteristics is influenced by the expression of mutant htt and is related to the advancement of neural development, quantitative real-time PCR (Q-PCR), Western blot, and immunostaining were used to determine the expression pattern and the accumulation of mutant htt aggregates as well as the formation of NIs during in vitro neural development.
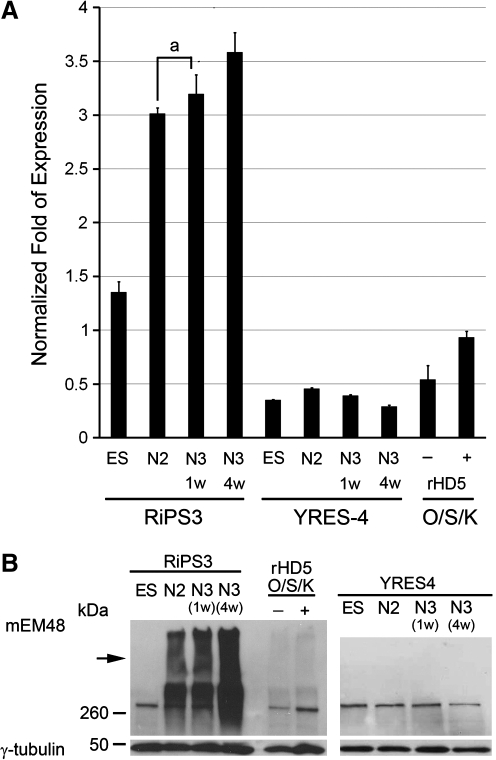
Quantitative measurement of htt transcripts (endogenous and mutant htt) revealed an elevated expression of total htt transcripts in undifferentiated RiPS-3 compared to undifferentiated and differentiated wild-type monkey ES cells, YRES4 (Fig. 4A). Similar expression levels of the htt gene in YRES4 at all stages suggested the basal expression level of endogenous monkey htt gene in monkey ES cells. The expression of mutant htt was significantly increased in RiPS-3 at N2, N3-1w, and N3-4w when compared to undifferentiated RiPS-3. The expression level of htt gene in YRES4 remained relatively low when compared with the respective stages of RiPS3 (Fig. 4A). Higher expression of the mutant htt was revealed in N3-4w when compared to RiPS3 at N2 and N3-1w. However, there is no difference between N2 and N3-1w (Fig. 4A). RiPS-3 and YRES4 cells at all stages, and skin fibroblast samples were also used for Western blot analysis. Oligomeric mutant htt was observed in RiPS-3 at all differentiation stages, YRES4 at all differentiation stages, and skin fibroblasts with or without being transfected with transcription factors, but was not observed in undifferentiated RiPS-3 (Fig. 4B). We have demonstrated a consistent finding on the gradual increase in the expression of mutant htt and the increased amount of oligomeric mutant htt as RiPS3 progresses during in vitro neural differentiation (Fig. 4B). Extended culture at the N3 stage also revealed a significant increase in the accumulation of oligomeric mutant htt compared to N2 and M3-1w, which suggests the possible impact of postmitotic neural cell condition in HD pathogenesis (Fig. 4B). Immunostaining using mEM48 (Gutekunst et al., 1999) revealed mutant htt aggregates and intranuclear inclusions in differentiated RiPS-3 at N3 stages (Fig. 2D). No positive immunostaining was observed in undifferentiated RiPS-3 cells (data not shown), which was consistent with our findings in the quantitative measurement of htt transcripts and Western blotting analysis (Fig. 4)
FIG. 4.
Expression of mutant htt in neural differentiated RiPS-3. (A) Expression levels of mutant htt at various developmental stages during in vitro neural differentiation were determined by Q-PCR. YRES4 is a nontransgenic wild-type monkey ES cell line and was used as a control. The expression levels of mutant htt in differentiated RiPS-3 were significantly increased at N2, N3-1w, and N3-4w compared to undifferentiated RiPS-3 (ES), YRES4 at all stages and skin fibroblasts with or without being transfected with transcription factors. Columns with the same letter indicate no significant difference (p > 0.05). (B) Western blot analysis using mEM48 revealed a gradual increase of oligomeric transgenic mutant htt as RiPS-3 progresses during neural differentiation (N3-4w > N3-1w > N2), whereas no high molecular weight mutant htt aggregates were detected in undifferentiated ES cells, YRES4 at all stages and skin fibroblasts with or without viral transfection. Arrow indicates mutant htt aggregate at the stacking gel.
Discussion
Pluripotent stem cells are unique cell types that are capable of differentiating into multiple cell types upon appropriate induction in vitro or modulated by the microenvironment in vivo. Although mutant htt expresses in all body cells, the primary pathogenic targets are neuronal cell types, especially in the striatal region where peripheral tissues rarely developed comparable cellular pathology (Davies and Ramsden, 2001; Li and Li, 2006; Rubinsztein, 2002). Although the mechanism of cell type-specific pathology may hold the key for understanding HD pathogenesis, it is important to investigate the impact of mutant htt in peripheral tissues and subsequent HD development (Martin et al., 2008; Sathasivam et al., 1999). Because stem cells retain multipotent differentiation capability, they are powerful tools for investigating the underlying mechanism of cell type-specific events and the influence of developmental events that lead to pathologic consequences (Byrne et al., 2007; Dimos et al., 2008; Hanna et al., 2007; Lakshmipathy et al., 2007a, 2007b; Laurent et al., 2008; Muller et al., 2008; Park et al., 2008a, 2008b, 2008c; Pruszak et al., 2007; Rutllant et al., 2003). Thus, pluripotent stem cells that develop HD cellular features paralleling neural development is a useful model for studying HD pathogenesis. Furthermore, pluripotent ES-like cells generated by induced pluripotency using skin cells of HD monkeys can uniquely mimic pathologic events paralleling disease progression in HD monkeys. This could be a unique and powerful model system for the development of novel biomarkers and treatments, which can be validated in HD monkey skin cell donors that share an identical genetic constitution in the future.
One may question why RiPS-3 developed distinct HD cellular features that are rarely reported in mouse and human HD-ES cells. This could result from the overexpression of a small htt fragment driven by ubiquitin promoter in RiPS-3 compared to a full-length htt gene in mouse and human HD-ES cells. We hypothesized that smaller htt fragments are more toxic than full-length htt (Bates et al., 1998; Schilling et al., 1999; Wang et al., 2008); therefore, pluripotent stem cells derived from HD patients' may not develop robust HD cellular pathology because pathological events that lead to signature HD cellular features may take much longer time to develop and accumulate cellular damages. Our recent report on a hybrid HD monkey stem cell derived by using the same skin fibroblast, has further confirmed the notion that Exon 1 of htt with 72Q is sufficient to elicit a pathological cascade of HD and resulted in robust cellular phenotypes (Laowtammathron et al., 2010).
We have also demonstrated the transition from overexpression to downregulation of exogenous transcription factors upon the establishment of pluripotency in RiPS-3 (Fig. 3A–C). A similar transition was also observed in core endogenous stemness transcription factors, Oct4 and Sox2 (Fig. 3D and E), which were not expressed in skin fibroblasts when exogenous Oct4 and Sox2 were highly expressed (Fig. 3A and B). However, upon the establishment of pluripotency, endogenous Oct4 and Sox2 were up regulated (Fig. 3D and E). Although Sox2 is expressed in both stem and neural progenitor cells, the expression of Sox2 in both undifferentiated and differentiated RiPS-3 cells were not unexpected (Fig. 3E). In contrast, Oct4 is a stem cell specific transcription factors that is highly expressed in pluripotent stem cells. A distinct transition in the expression pattern of endogenous Oct4 was observed as skin fibroblasts were reprogrammed to pluripotency (Fig. 3A and D). One possible explanation of the unique expression pattern of exogenous transcription factors under the control of retroviral promoter is the presence of stem cell factors that bind to the retrovirus primer binding site at the 5′ untranslated region and inactivate transcription after skin fibroblasts regained pluripotency (Petersen et al., 1991a, 1991b; Soriano et al., 1991; Yamauchi et al., 1995). This unique transition in gene regulatory patterns further suggested the successful reprogramming of skin fibroblasts to pluripotent stem cell characteristics.
In this study, we showed that RiPS-3, an induced pluripotent Huntington monkey stem cell, can be derived from HD monkey skin fibroblasts and is pluripotent. RiPS-3 expresses mutant htt and develops distinct HD cellular pathology when neural differentiation progresses. The expression of mutant htt in RiPS-3 progressively increased as neural differentiation progressed in vitro (Fig. 4A). We have also demonstrated the increased accumulation of oligomeric mutant htt aggregates at advance differentiation stages (N3-4w > N3-1w > N2; Fig. 4), while no detectable aggregates were observed in undifferentiated RiPS-3, YRES4, and a primary culture of skin fibroblasts (Fig. 4B). Similar expression patterns and pathological changes have been described in human HD patients and HD animal models. Neuronal tissues are the major targets in HD. The increased accumulation of oligomeric mutant htt aggregates in neuronal tissues parallel disease progression while pathological impact on peripheral tissues is minimal. Our findings suggest that pluripotent stem cells can be derived from HD monkeys by the expression of three key transcription factors (Oc4, Sox2, and Klf4). The resulting rHD-iPS cells also develop key cellular HD features during in vitro neural differentiation; thus, rHD-iPS cells could be a powerful in vitro model for investigating HD pathogenesis, developing potential cures, and validated in HD monkeys.
Acknowledgments
We thank the veterinary staff and the animal resources at the Yerkes National Primate Research Center (YNPRC), Cell Line Genetics LLC for Cytogenetic analysis, and the critical review provided by Ms. Leslee Sinclair. mEM48 was provided by Dr. Xiao-Jiang Li. All procedures were approved by YNPRC/Emory Animal Care and Biosafety Committees. The YNPRC is supported by the base grant RR-00165 awarded by the Animal Resources Program of the NIH. This study is supported in part by NCRR at NIH (RR018827) and the Atlanta Clinical & Translational Science Institute award to A.W.S.C. A.W.S.C. is supported by NIH (RR018827-05A1, RR018827-05A1S1, and NS064991).
Author Disclosure Statement
The authors have no competing financial interests.
References
- Bates G.P. Mangiarini L. Wanker E.E. Davies S.W. Polyglutamine expansion and Huntington's disease. Biochem. Soc. Trans. 1998;26:471–475. doi: 10.1042/bst0260471. [DOI] [PubMed] [Google Scholar]
- Byrne J.A. Pedersen D.A. Clepper L.L., et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Chan A.W.S. Transgenic nonhuman primates of neurodegenerative diseases. Reprod. Biol. Endocrinol. 2004;2:39. doi: 10.1186/1477-7827-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q. Stem cells: roadmap to the clinic. J. Clin. Invest. 2010;120:8–10. doi: 10.1172/JCI41801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S. Ramsden D.B. Huntington's disease. Mol. Pathol. 2001;54:409–413. doi: 10.1136/mp.54.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimos J.T. Rodolfa K.T. Niakan K.K., et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Gutekunst C.A. Li S.H. Yi H., et al. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J. Wernig M. Markoulaki S., et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Kiskinis E. Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J. Clin. Invest. 2010;120:51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U. Love B. Adams C., et al. Micro RNA profiling: an easy and rapid method to screen and characterize stem cell populations. Methods Mol. Biol. 2007a;407:97–114. doi: 10.1007/978-1-59745-536-7_8. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U. Love B. Goff L.A., et al. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007b;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laowtammathron C. Cheng E.C. Cheng P.H., et al. Monkey hybrid stem cells develop cellular features of Huntington's disease. BMC Cell Biol. 2010;11:12. doi: 10.1186/1471-2121-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L.C. Chen J. Ulitsky I., et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Li S. Li X.J. Multiple pathways contribute to the pathogenesis of Huntington disease. Mol. Neurodegener. 2006;1:19. doi: 10.1186/1750-1326-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Zhu F. Yong J., et al. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Martin B. Golden E. Keselman A., et al. Therapeutic perspectives for the treatment of Huntington's disease: treating the whole body. Histol. Histopathol. 2008;23:237–250. doi: 10.14670/hh-23.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateizel I. De Temmerman N. Ullmann U., et al. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum. Reprod. 2006;21:503–511. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- Muller F.J. Laurent L.C. Kostka D., et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H. Arora N. Huo H., et al. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H. Lerou P.H. Zhao R., et al. Generation of human-induced pluripotent stem cells. Nat. Protoc. 2008b;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Park I.H. Zhao R. West J.A., et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008c;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Petersen R. Kempler G. Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol. Cell. Biol. 1991a;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R. Sobel S. Wang C.T., et al. Cellular transcripts encoded at a locus which permits retrovirus expression in mouse embryonic cells. Gene. 1991b;101:177–183. doi: 10.1016/0378-1119(91)90409-5. [DOI] [PubMed] [Google Scholar]
- Pruszak J. Sonntag K.C. Aung M.H., et al. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25:2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C. Lessons from animal models of Huntington's disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- Rutllant J. Pommer A.C. Meyers S.A. Osmotic tolerance limits and properties of rhesus monkey (Macaca mulatta) spermatozoa. J. Androl. 2003;24:534–541. doi: 10.1002/j.1939-4640.2003.tb02705.x. [DOI] [PubMed] [Google Scholar]
- Sathasivam K. Hobbs C. Turmaine M., et al. Formation of polyglutamine inclusions in non-CNS tissue. Hum. Mol. Genet. 1999;8:813–822. doi: 10.1093/hmg/8.5.813. [DOI] [PubMed] [Google Scholar]
- Schilling G. Becher M.W. Sharp A.H., et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Soriano P. Friedrich G. Lawinger P. Promoter interactions in retrovirus vectors introduced into fibroblasts and embryonic stem cells. J. Virol. 1991;65:2314–2319. doi: 10.1128/jvi.65.5.2314-2319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A. Marshall V.S. Primate embryonic stem cells. Curr. Top. Dev. Biol. 1998;38:133–165. doi: 10.1016/s0070-2153(08)60246-x. [DOI] [PubMed] [Google Scholar]
- Wang C.E. Tydlacka S. Orr A.L., et al. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington's disease. Hum. Mol. Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M. Zhao J.P. Pruszak J., et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi M. Freitag B. Khan C., et al. Stem cell factor binding to retrovirus primer binding site silencers. J. Virol. 1995;69:1142–1149. doi: 10.1128/jvi.69.2.1142-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H. Cheng P.H. Banta H., et al. Towards a transgenic model of Huntington's disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]