Abstract
Viruses are successful and omnipresent. Influenza A is a particularly important virus of humans. The article reviews the 2009 emergence of the pandemic influenza A virus, focusing on the potential origin of the virus and the distinctive clinical and epidemiological impact of the 2009 pandemic.
INTRODUCTION
Viruses are extremely successful. They parasitise all forms of life both prokaryotic (bacteria) and eukaryotic (plants, fungi and animals). There is more genetic diversity within viruses than in all eukaryotic and prokaryotic genomes put together. Viruses are, by far, the most abundant life forms on the planet. It has been estimated that there are >1030 viable virus particles on our planet, most of which are in the oceans. If we were able to line up all of these end to end it would reach to a radius of 5 million light years, which would include not only the Milky Way but also our nearest 50 galaxies.
At an evolutionary level we still do not understand how viruses originated. It is probable that there are multiple origins relating to different groups of viruses - some ancient, relating to elements of the primordial soup that predate the development of life as we know it, and some more recent in evolutionary terms originating from ‘escaped’ cellular genetic elements.
For all their ubiquity and success, viruses are startlingly simple. They are tiny, on average about 1/5000th the size of the typical bacterium. Most have fewer than 10 genes coding for proteins specific to the virus and no metabolic system apart from that of the host cell that they parasitise. Yet these simple biological systems are capable of harnessing and subverting the host's complexity to produce viral components, assemble them into new virus particles and eject them from the host in such a way that they can find a next host. The survival of viruses is entirely dependent on a continuous chain of transmission being available, as most viruses cannot survive for prolonged periods outside the host.
At a fundamental level viruses have shaped us. At least 10% of the human genome is made up of elements that originate from viruses and there is increasing evidence that such sequences have had profound effects on the evolution of complex organisms including humans.
The question is often posed; are viruses alive? Essentially the answer simply depends on how we chose to define life. Using standard definitions, viruses cannot be considered to be living organisms. Rather they are biological entities entirely dependent on the cellular functions of the organism that they parasitise.
There are about 200 viral species known to infect humans. In this article I will focus on one of them; influenza A virus, whose profile rose considerably in 2009 with the emergence of a new pandemic virus: Influenza A H1N1 2009.
INFLUENZA VIRUSES
Influenza viruses circulate in humans every year in every part of the world. Intermittently and unpredictably new viruses arise that are capable of causing pandemics (Table 1). The term ‘pandemic’ refers to the rapid emergence of a new influenza virus that causes an epidemic that covers the whole world.
Table 1.
Key examples of past Influenza A Pandemics and antigenic shift events resulting in generalised human circulation
| Year of emergence | Influenza A Virus type | Comments |
|---|---|---|
| 1889 | H2N2 | Pandemic. |
| Estimated deaths worldwide: 1 million | ||
| Antigenic type deduced by retrospective serological testing of stored sera. | ||
| 1900 | H3N2 | Pandemic. |
| Estimated deaths worldwide: uncertain, <1 million | ||
| Antigenic type deduced by retrospective serological testing of stored sera. | ||
| 1918 | H1N1 ‘Spanish’ | Pandemic. |
| Estimated deaths worldwide: 20 to 100 million | ||
| 1957 | H2N2 ‘Asian’ | Pandemic. |
| Estimated deaths worldwide: 1 to 1.5 million | ||
| 1968 | H3N2 ‘Hong Kong' | Pandemic. |
| Estimated deaths worldwide: 0.75 to 1 million | ||
| 1977 | H1N1 ‘Russian’ | Not considered to be a pandemic in the sense that severity and attack rate were lower than with previous shift events. |
| Estimated deaths worldwide: <100 000 | ||
| Widely conjectured to be a virus of laboratory origin. | ||
| 2002 | H1N2 | No pandemic, few documented deaths. Reassortment between the 2 seasonal viruses (H3N2 and H1N1) so not new antigenically. |
| 2009 | H1N1 2009 ‘Mexican’ ‘swine’ | Pandemic |
| Estimated deaths worldwide: <100 000 |
There are two main types of viruses that cause seasonal influenza –types A and B. Influenza B is an important cause of seasonal influenza but unlike influenza A it does not cause pandemics. Influenza A viruses are further typed into subtypes according to different kinds and combinations of virus surface proteins Haemagglutinin (H) and Neuraminidase (N).
It can be reasonably argued that influenza is the single most significant infection of humans. Typically, each year about 10% of the population will contract an influenza infection. Worldwide, on average, influenza contributes to the deaths of 250 000 people annually. Influenza is the only common infectious agent in the developed world where one can simply look at weekly gross mortality statistics for a country and determine when the virus circulated in a particular year and whether it was a particularly bad year. In a pandemic, mortality can be much higher than for seasonal influenza.
Pandemics occur when a brand new influenza A emerges, to which the human population does not have any pre-existing immune protection. This results in rapid transmission and high attack rates. The definition of a pandemic is to some extent arbitrary and is not well standardised over time; for example the 1977 H1N1 emergence was not then considered to be a pandemic, but using the modern World Health Organisation (WHO) definitions it undoubtedly would be now.
Pandemics are a regular occurrence. Historical accounts suggest that there have been more than 20 since the 16th century, typically occurring every 10 to 40 years. The 1918 pandemic (H1N1) is widely regarded as the single biggest natural disaster documented to have affected our species; estimates of mortality range from 10 to 100 million deaths. Notably, over 50% of these deaths were in people under the age of 40. However only the 3 pandemics that happened in the last century and last year's H1N1 2009 pandemic are well characterised in virological and epidemiological terms.
THE ANATOMY OF THE VIRUS (FIGURE 1)
Fig 1.
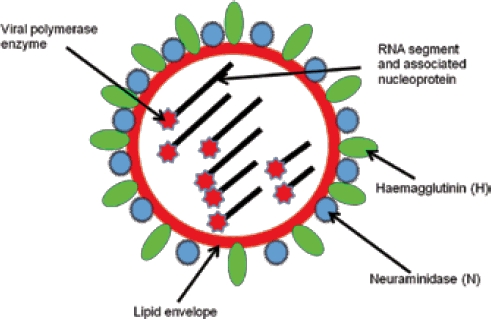
The structure of an influenza virus
The outer surface of the virus is made of a lipid envelope derived from the cytoplasmic membrane of the host respiratory cell from which the virus budded. The envelope is studded with the H and N glycoproteins (proteins with chains of sugar residues) that extend right through the lipid. This lipid envelope is essential to the viability of the virus. Viruses with a lipid envelope are inherently more delicate than viruses without one, as the lipid is very susceptible to environmental influences such as desiccation. This is the reason that influenza viruses will not survive in the environment for long periods of time whereas, for example, hepatitis A virus (non-enveloped) survives much longer. Most human cases of influenza will be contracted by direct inhalation of fresh wet droplets expelled by a cough or sneeze.
The biology textbook teaches that the perfect parasite is one that causes the host no harm. This may be true of some virus infections, for example those that spread by saliva contact. However the production of a disease process itself may be a key survival advantage for the virus - ‘Coughs and sneezes spread diseases’. In animal models, an influenza virus that does not cause disease typically does not spread as well as one that does. Some have conjectured that this evolutionary advantage for a nastier virus is a reason why sometimes a second pandemic wave is of greater disease severity than the first.
‘Influenza A H1N1 2009’ is the name of the current pandemic virus. ‘H’ and ‘N’ are the standard nomenclature for naming influenza A viruses and refer to the type of haemagglutinin (H) and neuraminidase (N) on the surface of the virus. It is worth considering the biological functions of H and N and the resultant opportunities these confer in prophylactic and therapeutic approaches to combating influenza A.
Haemagglutinin specifically attaches to the surface of a respiratory cell. Without this specific attachment the infection of the cell and hence host cannot be initiated. Specific antibody to haemagglutinin can block this attachment and confer immunity. For this reason haemagglutinin is the most important component of an influenza vaccine. The closeness of match between the haemagglutinin in the vaccine and the haemagglutinin in the circulating virus is the most important determinant of vaccine efficacy.
Neuraminidase is also embedded in the lipid envelope of the virus. It is essential for the release of virus particles allowing them to be released from one respiratory cell and hence available for infection of other cells in the same host or shed in droplets to infect another host. Neuraminidase is also important in facilitating the virus penetration of mucus to allow attachment to the cell surface. Without a functional neuraminidase, the virus is not shed from an infected cell and infection in the host is terminated. The two main antiviral drugs, Oseltamivir (Tamiflu) and Zanamivir (Relenza), are neuraminidase inhibitors and act on the neuraminidase protein preventing efficient release of the virus from the host cell.
Influenza virus is particularly well equipped to change its H and N antigens to escape immune detection and infect previously infected or vaccinated people. Two particular features of the influenza virus contribute to its rapid evolutionary capacity to change its antigens and escape immune recognition.
The first feature is the high error rate during genomic replication. The enzyme that copies the viral RNA to make copies to be packaged in new viruses makes a very high number of mistakes. This enzyme is much more error prone that the DNA polymerase enzymes in our cells that copy our chromosomal DNA, facilitating cell division. This generates a very high rate of mutations in the virus. Most of these will be neutral or detrimental to the survival of the virus but some will contribute to the evolution of antigenic change that favours the survival of the virus. The evolutionary selection and accumulation of these mutations causing gradual changes in the H and N proteins (‘antigenic drift’) happens continuously. This antigenic drift necessitates annually updated influenza vaccines which are matched to the currently circulating strains to render them effective.
The second important feature of the virus is the segmented genome. The virus genetic material is contained in 8 separate RNA segments, each of which codes for one or two proteins. This segmented genome allows the phenomenon of genetic reassortment to occur. Viruses that simultaneously infect the same cell can swap segments - sexual reproduction in viruses: effectively two viruses mating. We know from surveillance studies that this happens frequently, for example, with influenza viruses circulating in wild birds. Genetic reassortment can result in a new H and/or N emerging. This is the phenomenon of ‘antigenic shift’ where a brand new virus appears.
In nature, there at least 17 different H types. Only H1, H2 and H3 have been recognised in viruses capable of sustained transmission in humans. The rest are seen in a wide range of birds and in some mammals. Occasionally avian viruses have crossed over into the human population causing clusters of cases without sustained human to human transmission (Table 2). The spread of H5N1(‘Avian Flu') in birds throughout Asia, Europe and Africa is a recent example of a virus that has proven capable of causing severe disease in humans but which has not, so far, resulted in significant human to human transmission.
Table 2.
Selected examples of zoonotic transmission of influenza to humans that have not resulted in sustained human to human transmission
| Year | Virus Subtype | Location | No. of Cases |
|---|---|---|---|
| 1997 | H5N1 | Hong Kong | 18 (6 deaths) |
| 2003 | H7N7 | Holland | 89 (1 death) |
| 2003- to present | H5N1 | S&SE Asia, Turkey, Egypt and others | 442 (262 deaths) −60% death rate |
Antigenic shift results from the movement of one of these H types from an influenza virus, affecting another animal, into a virus capable of transmission between humans. Figure 2 illustrates the process of reassortment which happens frequently in animal influenza viruses. The example in the diagram relates to the pandemic emergence of the H3N2 virus in 1968 which still represents one of the viruses that cause seasonal influenza.
Fig 2.
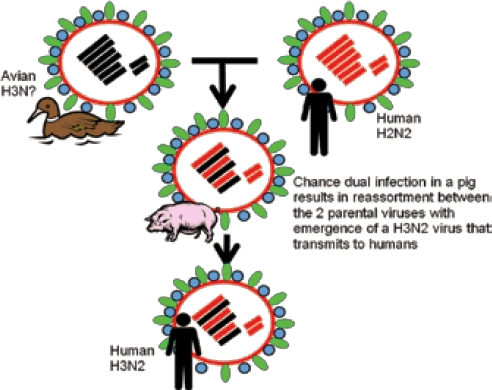
The 1968 pandemic influenza A virus is believed to have originated by reassortment between the H2N2 then currently circulating in humans and an avian H3 virus resulting in a virus that had gene segments from both parents. It is widely postulated that this reassortment event happened in a pig.
Most influenza genetic diversity is in birds. There are very significant difficulties for an avian-adapted virus to infect a human respiratory cell. This species barrier results from the sugar residue (sialic acid chains) arrangements of that the viral H protein that attaches to onto respiratory cells in mammals and birds. The virus H protein will be adapted to attach either to the mammal or the avian sialic acid chain arrangement. Hence it will be either a mammalian or an avian-adapted virus.
Pigs however, are unusual among mammals in that their tracheal respiratory cell surfaces have sialic acid chains with both the avian and mammalian arrangements. This means that pigs are uniquely susceptible to viruses of both avian and mammalian origin. Pigs have been well documented to contract productive infections with swine, avian and human influenza viruses and have been strongly suspected to be the “mixing vessel” responsible for emergence of reassortant strains that cause pandemics in humans.
H1N1 2009 (SWINE FLU)
Until now, there have been 3 seasonal influenza A viruses circulating in humans: H1N1, H1N2 and H3N2. Perhaps naïvely, based on our short series of three well-documented pandemics, it had generally been assumed that the awaited pandemic virus would require a brand new haemagglutinin type, perhaps an H2 or an H5. Instead, the pandemic virus was another H1 virus, but significantly different from the seasonal H1 virus such that immunity resulting from past infection with the seasonal virus or vaccination did not prevent infection.
The virus was first identified in humans in April 2009 in the southern USA and it was quickly established that it was spreading rapidly from person to person and had been causing widespread disease in Mexico since early March. From the beginning it was recognised that this was a swine origin virus. On June 11th, 2009, with virus transmission documented in more than 70 countries, WHO declared that a global pandemic of novel influenza A (H1N1 2009) was underway. This very rapid spread from North America to the rest of the world was notable as the first pandemic to unfold in the context of post-globalisation modern human travel patterns.
WHERE DID H1N1 2009 COME FROM?
There are many unanswered questions. H1N1 2009 is essentially a reassortment of 3 viruses that have been endemic in farmed pigs over the past 15 years. Where, when and how these 3 viruses reassorted is unclear.
Six of the 8 segments are directly descended from swine H1N2 virus ‘triple-reassortant’ influenza viruses isolated from pigs in North America around 1999–2000. These triple reassortant viruses emerged in 1999 and had segments of pig, human and avian origin. However the other two gene segments are from 2 different Eurasian viruses of pigs; the N gene is closest to European H1N1 viruses from 1991–1993, and the MP gene is closest to H3N2 viruses isolated in Asia in 1999–2000. Hence the 8 genetic segments have an origin in 3 different known pig-associated viruses.
There are two difficult aspects to explain. Firstly, the closest relatives for all 8 segments are virus isolates collected more than a decade before the human pandemic started. Where have these segments been in the meantime?
Secondly, how did European and Asian viruses reassort with an American virus, as there is normally little mixing of swine viruses across the Atlantic/Pacific? These two key unanswered questions have given rise to some speculation about the evolutionary origin of the virus and in particular the role of human activity in this. It is possible that the reassortant event occurred 10 years ago and that the virus lineage has been in pigs as a minor unrecognised virus. Lack of surveillance may have resulted in it going undetected until it emerged in humans. However a possible research or diagnostic laboratory origin for the virus has to be considered, possibly related to swine vaccine manufacture. The three parental lines for H1N1 2009 may have been assembled in one place by natural means, such as by migrating birds; however the consistent link for all 8 segments with pig viruses suggests that human activity could be implicated. It is difficult to entirely discount human activity as the possible origin of the virus on current evidence.
If this is the case it would not be the first time that a laboratory source for an influenza virus has been postulated. The 1977 H1 virus (Table 1) is widely regarded as having a likely laboratory origin as it is so similar to viruses from the 1950's that this seems the most biologically feasible source. It is important that further work on the evolutionary origin of the H1N1 2009 virus is carried out and that the origin is eventually understood as this may shed important light on future prevention.
H1N1 2009 – DISTINCTIVE FEATURES
All pandemics are different in their clinical impact and epidemiology. The H1N1 2009 pandemic has displayed a number of interesting features that are worth considering.
AGE PROFILE
This was unusually young compared to seasonal influenza. It is estimated that only 2% of total cases were over the age of 65. This was undoubtedly a major factor in the clinico-epidemiological impact of the pandemic as older people are, typically, the largest vulnerable group for influenza complications. Serological studies have shown that older people have some cross protective antibody arising from either repeated past vaccination or infection with H1 viruses.
ASYMPTOMATIC INFECTION
The vast majority of pandemic infections were mild. Asymptomatic and very mild infection, especially in children, was much higher than expected. In London and Birmingham, during the first wave of infection in summer 2009, serological testing showed that over 30% of children were infected by the pandemic virus. This was ten times more than was estimated from clinical surveillance.
NEW RISK FACTORS
Particular risk factors emerged as more prominent in this pandemic than in previous ones, Neurodevelopmental delay, pregnancy and obesity were all recognised as potential co-factors in increased mortality and morbidity in flu infections. However all 3 of these risk factors, previously viewed as of relatively minor importance, came to a prominence in this pandemic as major predictors of severe morbidity and mortality.
HIV INFECTION
This is the first pandemic of the HIV era and there had been concern that HIV would be a significant risk factor. Reassuringly this was not the case and several studies found that HIV status had little impact on the severity or outcome of H1N1 2009 infection, although very low CD4 counts may increase the risk of worse outcomes and opportunistic infections may complicate diagnosis.
INDIGENOUS PEOPLES
The attack rates and severity in indigenous populations were higher than for the general population. They had a three to six-fold higher risk of admission to hospital, severe disease and death. This was noted in Native Americans, Inuit, Australian aborigines, Maori and Pacific island people. The mechanism for this is still unclear. It has been suggested that this may be related to the generally higher prevalence of obesity, diabetes, asthma, chronic obstructive pulmonary disease (COPD) or social factors such as crowding, poverty and difficulty with access to health provision. However, it is likely that there are genetic factors involved.
PARADOXICAL INCREASED SEVERITY OF HOSPITALISED CASES
Although the severity of cases was generally mild with hospitalisation rates of 0.3%, comparing very favourably with previous pandemics and even with seasonal influenza, paradoxically a much higher than expected proportion (20%) of hospitalised cases required admission to intensive care units (ICU), placing disproportionate pressure on them. ICU admissions reflected the ‘new’ risk factors of pregnancy, obesity and neurodevelopmental delay, but over 20% were in patients without clear risk factors. Admission to ICU was typically for rapidly progressive respiratory failure with hypoxaemia. Secondary bacterial infection was less common that had been anticipated.
PCR DIAGNOSIS
This was the first pandemic to occur since the advent of molecular techniques. These were used extensively for surveillance and diagnosis, having many advantages over traditional diagnostic methods. Widespread use of combined nose and throat swab sampling proved very successful.
However in severe cases requiring ICU admission, approximately 20% were found to have a negative nose & throat swab result ,despite having detectable virus in the lower respiratory tract (tracheal secretions or lower). The mechanism of this phenomenon is unclear but may reflect some altered tropism of the virus for the lower respiratory tract in more severe cases.
H1N1 2009 INFECTION IN NON-HUMAN ANIMALS
Many countries have reported H1N1 2009 in animals. The source has been regarded as human, with subsequent transmission between farm animals but limited evidence of farm to farm transmission. It is noteworthy that the first European detections of H1N1 2009 in animals occurred in Northern Ireland, affecting pigs on 4 separate farms. Farmed pigs are the animals most recognised to be infected but outbreaks in farmed turkeys and isolated cases in pet cats and dogs have been recorded.
The viruses from these animal outbreaks that have been studied and sequenced, so far have been found to be essentially identical to the pandemic virus circulating in humans. Infections in pigs are probably even more common than the reported detections suggest as the symptoms in the pigs are very mild. Much of the resulting public health message has been reassuring, stressing the obvious safety of eating cooked pork. However the current unique situation of a widespread virus of swine that can transmit equally efficiently from human to human and from pig to pig is worrying. This is potentially the perfect shuttle vector capable of introducing pig virus segments to viruses that can transmit efficiently in humans.
Industrial pig farming has undoubtedly accelerated the evolution of influenza viruses in farmed pigs. There is considerable influenza genetic variability in currently circulating pig viruses. There is also significant evidence of frequent reassortment events in pig viruses. The risk of such reassortment resulting in nastier viruses emerging into humans is one that we should take seriously in the immediate future.
H1N1 2009 RATE OF GENETIC VARIATION
As discussed, influenza viruses are expected to evolve rapidly and to change antigenicity over time such that their surface proteins are not recognised by antibody resulting from a previous infection or vaccination. However, so far, the H1N1 2009 virus has so far shown very little genetic change. At an antigenic level the virus has essentially remained unchanged from the viruses that emerged in April 2009.
The vaccine for the southern hemisphere winter currently being produced will continue to be made using virus from California (at the start of the pandemic) rather than a more recent isolate. Whether this low mutation rate simply reflects lack of selection pressure in initial pandemic waves, or is some inherent property of this virus, remains to be seen.
ANTIVIRAL RESISTANCE TO TAMIFLU IN H1N1 2009
The H1 seasonal virus circulating up until 2010 was almost entirely resistant to Oseltamivir, and this resistance had emerged over the previous 2 flu seasons. A single aminoacid change in the active site of the N protein can confer resistance.
So far (March 2010), 40 of the 5,462 pandemic viruses tested have been confirmed to carry the mutation which confers resistance to Oseltamivir. The biggest risk for the emergence of Oseltamivir resistant virus appears to be in immunocompromised patients; particularly where lower prophylactic dosages are used after a patient has been exposed to the virus. For example, there have been two recognised outbreaks of drug-resistant virus in haematology units following prophylactic use of Tamiflu in Wales and the USA.
THE FUTURE
It is generally considered that there will not be any further significant pandemic waves unless we see a very substantial change in the virus. It is likely, based on previous pandemics, that the currently circulating seasonal H1 viruses will disappear to be replaced by the H1N1 2009 virus as a cause of seasonal flu each winter. A key concern is that we are now in an inherently less stable situation with a circulating virus that can transmit to and from pigs and that this has the possibility of allowing segments from pig viruses get into humans with unpredictable consequences.
Future pandemics will occur. As pandemics go, H1N1 2009 has certainly been at the benign end of the spectrum. By March 2010, the documented death toll was 457 in the UK, much less than typically seen in seasonal influenza. Probably ten million people in the UK have had swine flu and most will have had either no symptoms or a mild illness.
We do not know if a ‘1918 type’ event could happen again. The 1918 virus has been reconstructed and studied. Undoubtedly it had distinctive biological properties that made it considerably more pathogenic than any other influenza viruses about which we are historically aware and which are established in humans. However the availability of antibiotics, antivirals, vaccines and intensive care support is likely to have substantial impact on the morbidity and mortality rates even in the face of a very nasty virus.
CONCLUSION
In comparative historical terms, the 2009 emergence of a new pandemic influenza represented an overdue event. In terms of severity and impact on society, it was a best case scenario and an opportunity to rehearse and hone preparations for the inevitable future pandemics that may be less benign. The new dynamic of a circulating human influenza virus that can slip in and out of pigs is concerning as it could lead to future reassortant events between pig and human viruses.
The author has no conflict of interest.


