SUMMARY
Many neuropeptides and peptide hormones require amidation of their carboxy terminal for full biological activity. The enzyme Peptidyl-α-hydroxyglycine α-amidating lyase (PAL; EC 4.3.2.5) catalyzes the second and last step of this reaction – N-dealkylation of the peptidyl-α-hydroxyglycine to generate the α-amidated peptide and glyoxylate. Here we report the X-ray crystal structure of the PAL catalytic core (PALcc) alone and in complex with the non-peptidic substrate α-hydroxyhippuric acid. The structures show that PAL folds as a six-bladed β-propeller. The active site is formed by a Zn(II) ion coordinated by three histidine residues; the substrate binds to this site with its α-hydroxyl group coordinated to the Zn(II) ion. The structures also reveal a tyrosine residue (Tyr654) at the active site as the catalytic base for hydroxyl deprotonation, an unusual role for tyrosine. A reaction mechanism is proposed based on this structural data and validated by biochemical analysis of site-directed PALcc mutants.
Keywords: Hg-MAD, peptide activation, Peptidyl-α-hydroxyglycine α-amidating lyase, X-ray structure, Zn-MAD
INTRODUCTION
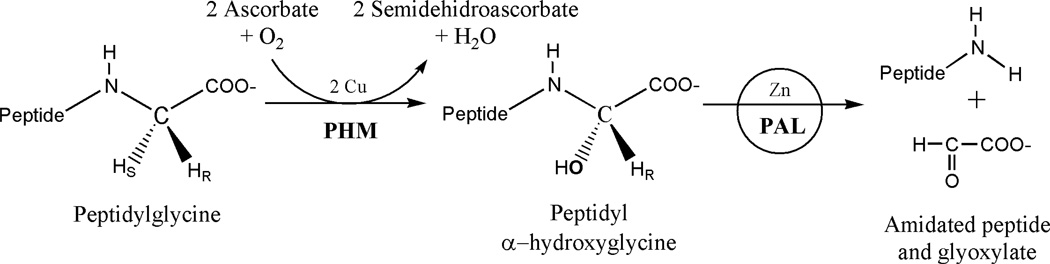
Alpha-amidation of neuropeptides and peptide hormones is a post-translational modification in most cases essential for their full biological activity - receptor recognition and signal transduction (Eipper et al., 1992; Merkler, 1994; Prigge et al., 2000). Active α-amidated peptides are produced by N-oxidative cleavage of glycine-extended substrates generated from precursor proteins by sequential endo- and exoproteolysis. The oxidative cleavage is catalyzed by peptidylglycine a-amidating monooxygenase (PAM), an enzyme localized to the trans-Golgi network and secretory granules of neural and endocrine tissues. PAM is a bifunctional enzyme with two domains: (i) Peptidylglycine α-hydroxylating monooxygenase (PHM; EC 1.14.17.3) and (ii) Peptidyl-α-hydroxyglycine α-amidating lyase (PAL; EC 4.3.2.5). These two domains act sequentially to generate an active α-amidated peptide product and glyoxylate (Figure 1). Although PHM and PAL are encoded by the same transcript in many species (e.g., human, rat, Xenopus), the domains may be encoded by separate genes (Drosophila, Cnidaria and Schistosomes) (Kolhekar et al., 1997).
Figure 1. The sequential stereospecific reactions of PAM catalyzed by PHM and PAL.
PHM is a dicopper, ascorbate-dependent monooxygenase that catalyzes the stereospecific α-hydroxylation of glycine-extended precursor peptides by molecular oxygen (O2). The relevance of PHM in physiology and medicine has been recently highlighted by investigations in rat brain stem that have revealed that intermittent hypoxia (associated with sleep aneas) activates PHM (Sharma et al., 2009). Also, studies on Drosophila have shown that reduced PHM activity leads to defects in memory retention and learning (Iliadi et al., 2008). In the mechanism of PHM, the two electrons required for dioxygen reduction are provided by ascorbate via one-electron reductions of the two copper centers, CuM and CuH. The X-ray structure of PHM (Prigge et al., 1997) revealed that it was organized into N- and C-terminal domains of about 150 residues connected by a linker peptide. Each domain contains nine β-strands and binds one copper ion. The copper centers are separated 11 Å by a solvent- and substrate-accessible cleft. Subsequent studies showed that the substrate binds close to the CuM site, where hydroxylation takes place, while the CuH site has been identified as an electron transfer site (Prigge et al., 1999). Kinetic investigations, kinetic isotope effect measurements, theoretical calculations and structural studies point to a CuM bound reactive oxygen species as responsible for hydrogen-atom abstraction (Chen and Solomon, 2004b; Crespo et al., 2006; Francisco et al., 1998; Prigge et al., 2004).
While a large amount of data is now available for the interpretation of the reaction mechanism of PHM (Chen and Solomon, 2004a; Crespo et al., 2006; Klinman, 2006), much less is known about the PAL domain of PAM, which catalyzes the N-dealkylation of the peptidyl-α-hydroxyglycine intermediate generated by PHM (Kolhekar et al., 2002). The PAL reaction resembles the reaction catalyzed by ureidoglycolate lyase (UGL; EC 4.3.2.3), which cleaves ureidoglycolate to produce urea and glyoxylate (Trijbels and Vogels, 1966). Although PAL can function as an ureidoglycolate lyase (De et al., 2006), no sequence homology is apparent between PAL and B. cepacia UGL (McIninch et al., 2003) and the lack of structural data for UGL precludes further comparison. Previous investigations defined the PAL catalytic core (PALcc; residues 498 to 820), revealed that the protein contains equimolar amounts of Zn(II) and Ca(II), and showed that a conserved tyrosine residue (Tyr654) plays an essential role in catalysis (De et al., 2006; Kolhekar et al., 2002). Both divalent cations were shown to play an important role in maintaining the stability of PAL (Kolhekar et al., 2002). At pH > 7 peptidyl-α-hydroxyglycine substrates do not require an enzyme to form the amidated peptide (Katopodis et al., 1991), suggesting that deprotonation by itself may trigger N-C(α) cleavage. However, the physiological pH of secretory granules, where the N-dealkylation reaction takes place, is acidic (pH = 5.0–6.0) (Prigge et al., 2000). It was proposed that in the PAL-catalyzed reaction zinc is part of the active site but it was not possible to determine whether the metal binds to the substrate directly or whether a zinc-water/hydroxyl group serves as the deprotonating agent.
Here we report the X-ray crystal structure of the rat (Rattus norvegicus) Peptidyl-α-hydroxyglycine α-Amidating Lyase catalytic core (PALcc) alone and in complex with the non-peptidic substrate α-hydroxyhippuric acid. PAL folds as a six-bladed β-propeller with a zinc ion at the active site coordinated by three histidine residues. The previously identified essential tyrosine (Tyr654) (De et al., 2006) and a key arginine (Arg706) are positioned in the active-site. A mechanism for the PAL reaction with essential roles for both of these residues is proposed and validated by biochemical analysis of PALcc mutants. The calcium binds to three β-strands in a fashion that is consistent with its important structural role in enhancing the stability of the protein (Kolhekar et al., 2002).
RESULTS
Structure determination and refinement
PALcc, consisting of residues 498–820 of rat PAM and an N-terminal His6 tag, was expressed in stably transfected CHO cells and purified as described in Materials and methods. The protein crystallizes in the orthorhombic space group (P212121) in the presence of equimolar amounts of Zn(II) and Fe(III). Initial phases were determined by multiwavelength anomalous diffraction (MAD) of an Hg derivative. These initial phases were used to locate the catalytic Zn2+ site using Zn-MAD data from a native crystal. Combination of the Hg and the Zn MAD data using the program autoSHARP (Vonrhein et al., 2007) resulted in a clear electron density map into which the structure was built. Cycles of manual rebuilding and refinement gave an R value of 20% (R-free 26%) (Table II). The His6 tag is ordered in the structure, with the first 2 histidines coordinating an iron(III) ion present in the mother liquor (ML) (or a mercury(II) ion when the protein was crystallized in the presence of HgAc2 instead of ZnAc2/FeCl3). Interestingly, the His6 tag remained ordered even when the protein was crystallized in the absence of metal (0.1 mM NaAc pH 4.6; 9–12% PEG8000).
Table II.
Statistics for crystallographic data collection and refinement
| PALcc | PALcc-substrate† | |
|---|---|---|
| Space group | P212121 | P212121 |
| a (Å) | 52.2 | 51.9 |
| b (Å) | 75.1 | 75.1 |
| c (Å) | 97.5 | 97.0 |
| Data collection | ||
| Source | X4A | APS |
| Wavelength (Å) | 1.28241 | 1.00724 |
| Resolution (Å) | 39 – 2.35 | 59 – 2.50 |
| Observed reflections | 187,201 | 90,551 |
| Unique reflections | 16,524 | 13,347 |
| Redundancy | 11.3 (7.9) | 6.8 (5.7) |
| Completeness (%) | 99.7 (96.9) | 99.6 (98.9) |
| Signal [I/σ(I)] | 60.9 (4.2) | 23.5 (2.7) |
| Rsym | 0.06 (0.49) | 0.075 (0.48) |
| Refinement | ||
| Rwork | 0.20 | 0.21 |
| Rfree | 0.26 | 0.26 |
| Stereochemistry | ||
| R.M.S. bond lengths (Å) | 0.008 | 0.008 |
| R.M.S. angles (°) | 1.25 | 1.25 |
| B-factor | ||
| Protein | 54.7 | 44.9 |
| Zn | 61.7 | - |
| Hg (active site) | - | 69.8 |
| Ca | 49.8 | 43.0 |
| Fe (contact site) | 49.0* | - |
| Hg (contact site) | - | 62.2* |
| Acetate | 60.0 | - |
| Substrate†† | - | 48.7 |
| Glycerol | 57.4 | - |
| Water | 56.6 | 43.9 |
| Model composition | ||
| Amino acids (atoms) | 329 (2,660) | 329 (2,660) |
| Metals | 3 | 3 |
| Acetate | 1 (4) | - |
| Substrate† | - | 1 (14) |
| Glycerol | 2 (12) | - |
| Water | 149 | 118 |
Data collection statistics given in parentheses are for the highest resolution shell.
Occupancy: Fe=0.7, Hg=0.35;
Substrate: α-hydroxyhippuric acid (HydHipA)
Overall structure
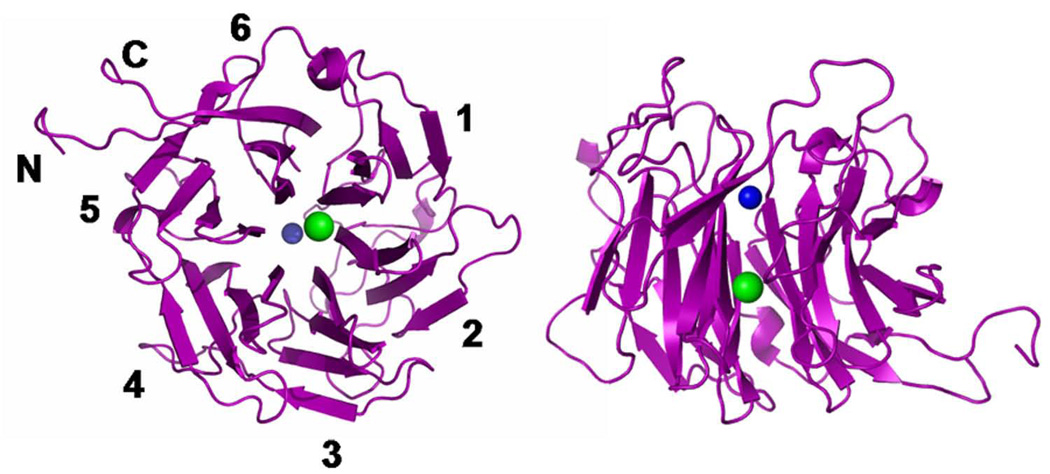
PAL folds as a β-propeller, with six blades positioned around a central cavity (central pore) (Figure 2). Each blade contains four antiparallel β-strands, with the first strand at the center of the propeller and the last at the edge. The loops connecting strand 4 of one blade to strand 1 of the next blade (loops 4–1) and the loops connecting strands 2 and 3 of each blade (loops 2–3) form the “cup” of the β-propeller. As in other β-propeller-enzymes, the cup encompasses the active site. Loop 2–3 of the first blade contains 20 residues, significantly longer than the equivalent loop in the other blades, which are 4–8 residues long. This long first blade loop provides a tryptophan (Trp538) that creates a hydrophobic pocket for the substrate and may also provide a surface for interacting with the PHM domain in the complete PAM enzyme. The N-terminal His-tag starts at the periphery of the propeller, preceding the N-terminal sequence of PALcc that forms β-strand 4 of the sixth blade. β-Strands 1 to 3 of the sixth blade are formed by the C-terminus of PALcc, which ends just after completing β-strand 3. In the construct under study, the N-terminal His-tag and the C-termini of PALcc form a “latch” that ties the structure together. In the bifunctional PAM protein, the linker that joins PHMcc to PALcc would replace this His-tag (Prigge et al., 2000). In membrane-bound PAM, PALcc is followed by a transmembrane helix that anchors the enzyme to the membrane (Prigge et al., 2000). This “latch”-like arrangement of PALcc places PHMcc close to the membrane, where PHMcc is believed to receive electron equivalents from cytochrome b561 via regeneration of intravesicular ascorbate (Iliadi et al., 2008; Kent and Fleming, 1987; Tsubaki et al., 2005).
Figure 2. Crystal structure of His6-PALcc.
The PAL enzyme folds as a six-bladed β-propeller; the figure shows two views: oriented with the central cavity normal to the page, left, and with the central cavity in plane of the page, right. Four strands form each of the blades, numbered 1–6 around the periphery. The Zn(II) and Ca(II) ions are indicated as blue and green balls, respectively (Zn-Ca distance = 11 Å).
PAL contains four NHL repeats (581–608; 633–662; 686–714; and 782–809). NHL repeats, first identified in NCL-1, HT2A and LIN-41 proteins, consist of sequences rich in glycine, proline, hydrophobic residues and a cluster of charged residues, that in the case of PAL are aspartates (Slack and Ruvkun, 1998). The NHL repeats correspond to the first, second and third strands of blades 2, 3, 4 and 6, where they contribute to intramolecular β-strand to β-strand interactions.
Structure of the active site and Zinc(II) coordination
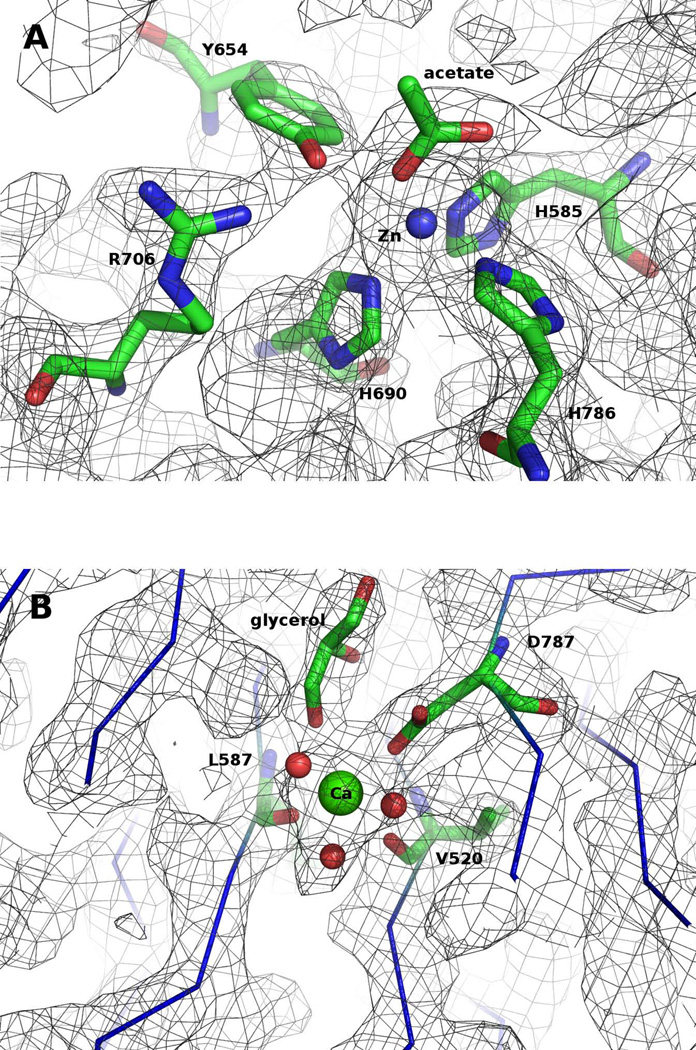
Zinc(II) and calcium(II) are present in the PAL structure in agreement with the results of inductively coupled plasma (ICP) optical emission spectrometry (De et al., 2006). The zinc ion, accurately located using anomalous scattering data collected at the Zn absorption edge wavelength (1.28 Å), is on the central axis of the propeller, at the “bottom” of the cup (Figure 2). It is coordinated by the Nε atoms of three conserved histidine residues (His585, His690 and His786) and an acetate ion bound as a monodentate ligand (Figure 3A). Proline residues (Pro584, Pro689 and Pro785) precede each of the three histidines that coordinate the Zn(II) ion. Since the substrate also binds at this site (see below), it is clear that this is the catalytic site of PAL. The Zn-Nε bond distances (2.1 Å, 2.1 Å and 2.1 Å, Figure 3A) are within expected values (Harding, 2006) but the Nε-Zn2+-Nε angles (97.9°; 95.0° and 103.7°) are intermediate between octahedral and tetrahedral geometries. A Zn(His)3 motif has been found in the catalytic site of enzymes such as carbonic anhydrase (Krishnamurthy et al., 2008), β-lactamase (Toney et al., 2001), epimerase (Luo et al., 2001), protease (Hege and Baumann, 2001), proteinase (Maskos et al., 1998; Schlagenhauf et al., 1998), macrophage elastase (Lang et al., 2001), and aldolase (Hall et al., 2002), and mediates dimerization in human interferon β (Karpusas et al., 1997).
Figure 3. (A) Active site of PALcc.
The zinc(II) (blue ball) ion is in close proximity to the essential tyrosine, Y654, and a key arginine, R706, and is coordinated in a very distorted tetrahedral polyhedron liganded by three histidines and one acetate bound in a monodentate fashion. Relevant distances include: Zn-NεH585 = 2.1 Å; Zn-NεH690 = 2.1 Å; Zn-NεH786 = 2.1 Å; Zn-O1acetate = 2.0 Å; Zn-O2acetate = 3.3 Å; Zn-OHY654 = 3.2 Å. Carbons are colored green, nitrogens blue, and oxygens red. The gray mesh represents the 2Fo-Fc electron density contoured at 1σ. (B) Calcium site of PALcc. The calcium(II) ion (green ball) is bound in the middle of the central cavity by three residues that belong to three different β-strands: the carboxylate of D787 (as a monodentate ligand), and the main-chain carbonyl groups of V520 and L587. Three water molecules and a glycerol complete the coordination sphere (Ca-OD787 = 2.45 Å; Ca-OV520 = 2.5 Å; Ca-OL587 = 2.3 Å; Ca-OW= 2.5 Å; Ca-OW= 2.6 Å; Ca-OW= 2.7 Å; Ca-Oglycerol = 2.75 Å). Carbons are colored green, nitrogens blue, and oxygens red. The gray mesh represents the 2Fo-Fc electron density contoured at 1σ.
As revealed during the Hg-MAD experiments, Hg(II) replaces Zn(II) at the active site when the enzyme is exposed at 1 mM mercury(II) acetate, rendering the enzyme inactive (Figure S1 in Supplemental data). Interestingly, the activity of PAL increases in the presence of rising concentrations of Cd(II) suggesting that Cd(II) can replace Zn(II) and makes the enzyme more active (Figure S1 in Supplemental data).
Calcium(II) site
A calcium ion is present in the middle of the central pore, 11 Å from the zinc (Figure 2). The identity of this Ca2+ was determined by anomalous scattering data and confirmed by its typical heptacoordination and metal-ligand distances (Ca-O = 2.3–2.5 Å) (Harding, 2006). It is coordinated by the carboxylate group (as monodentate ligand) of Asp787 and by main-chain carbonyls of Val520 and Leu587 (Figure 3B). These residues belong to three different β-strands, consistent with the structural role of the calcium demonstrated before (Kolhekar et al., 2002) (see discussion below).
Tyrosine 654 and Arginine 706 at the active site
Tyr654, the only tyrosine residue conserved in every PAL known to be catalytically active, has an essential role in catalysis: mutation of Tyr654 to Phe results in complete elimination of enzymatic activity (De et al., 2006). In the structure of PAL this tyrosine is located in the active site with its OH oxygen at 3.2 Å from the catalytic Zn2+ ion (Figure 3A). The adjoining residue, Cys655, forms a disulfide bridge with Cys634, positioning the tyrosine accurately for catalysis. Reduction of this disulfide bridge may explain why reducing agents such as ferrocyanide, ascorbate and dithionite lead to inhibition of PAL (De et al., 2006). Also at the active site, a highly conserved arginine residue (Arg706) is located in close proximity to Tyr654, with its guanidinium group at H-bond distance (2.7 Å) from the hydroxyl of the tyrosine (Figure 3A); the Glu707 helps position of this arginine. As with Tyr654, Arg706 is essential for catalysis, as revealed by site-directed mutagenesis studies (see below).
Structure of the PAL-hydroxyhippuric acid complex
To generate stable crystals of PAL in complex with its substrate, the protein was rendered inactive by crystallizing it with Hg(II) instead of Zn(II) in the active site (see above). The structure, PAL crystals grown in 0.5 mM Hg(II) and then soaked in mother liquor (ML) supplemented with the non-peptide substrate a-hydroxyhippuric acid (HydHipA) (final substrate concentration ∼ 5 mM) (Katopodis and May, 1990), was determined by molecular replacement using the coordinates of native PALcc as the search molecule. 2Fo-Fc (contoured at 1 σ) and the Fo-Fc (contoured at 2.5 σ) electron density maps showed extra density at the active site compatible with the presence of a complete HydHipA molecule (Figure 4). As expected, there was no reaction in the presence of Hg(II) and the HydHipA was not cleaved.
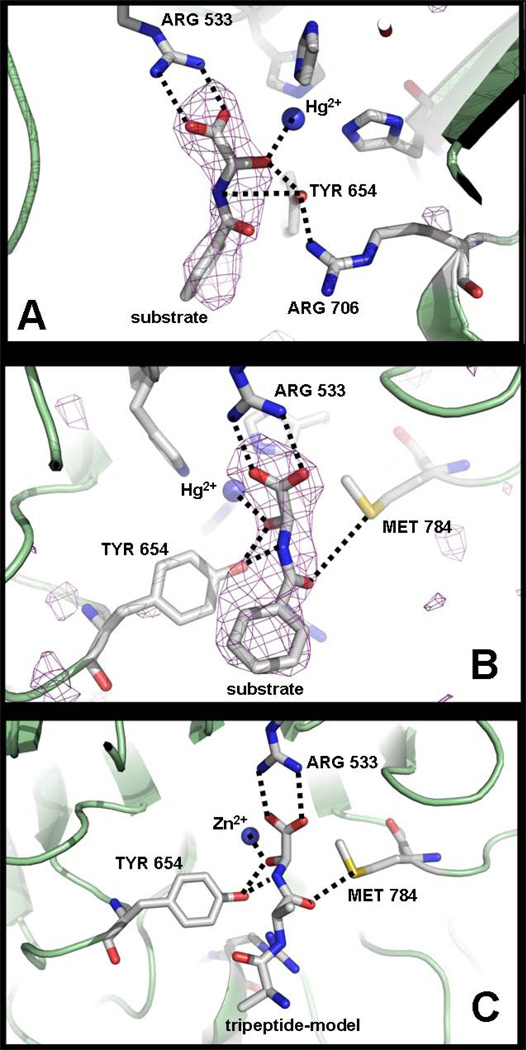
Figure 4. PAL-substrate complex.
A: PALcc-HydHipA (α-hydroxyhippuric acid) complex structure showing the substrate bound to the metal, Arg533 and Tyr654. B: A different view of the PALcc-HydHipA structure showing also the substrate interacting with Met784. C: PAL-tripeptide [Ala-Ala-Gly(OH)] model complex showing the most relevant interactions, as seen in figure B. See text for further discussion. Carbons are colored gray, nitrogens blue, and oxygens red. A purple mesh represents the calculated sigmaA weighted mFo-DFc omit map (Refmac5) of the substrate, contoured at 2σ (Figures A and B). The dashed lines represent relevant interactions between the substrate and the protein (see text).
The structure revealed at least three relevant interactions between the protein and the substrate. The HydHipA carboxylate interacts with the Arg533 guanidinium via a bidentate salt bridge. This guanidinium is in turn accurately positioned through hydrogen-bonds to two adjacent residues (Gln516, 2.8 Å; main-chain carbonyl Pro584, 2.6 Å). The substrate α-OH is coordinating the Hg(II) (Hg-O = 2.4 Å) and is in close proximity to the hydroxyl group of Tyr654 (2.4 Å). One of the substrate carboxylate oxygens is at 3.0 Å from the Hg(II) (O-Hg-His690 = 170.0°; O-Hg-His786 = 87.2°). In addition, the Tyr654 hydroxyl is close to the substrate –NH (3.7 Å), the leaving group of the PAL reaction (Figure 4). Met784 is also part of the substrate binding site: its sulfur atom is at 4.2 Å from the carbonyl group of the substrate peptide bond, providing the only contact to the residue preceding the hydroxylated glycine.
Model of the PAL-[Ala-Ala-Gly(OH)] complex
A PAL-substrate complex was modeled using a (S)-α-hydroxyglycine tripeptide model, Ala-Ala-Gly(OH) (Figure 4C). This peptide was manually built into the PAL active site by matching the atoms of the hydroxyglycine moiety with the equivalent atoms of the α-hydroxyhippuric acid, for which experimental data are available (see above). Also, the diastereomer (R)-α-hydroxyglycine Ala-Ala-Gly(OH) was modeled to investigate whether the structural data here reported account for the stereospecificity exhibited by PAL reaction (Ping et al., 1992). When the stereoisomer (R) is placed in the same conformation as the stereoisomer (S) with only a different Cα–OH configuration, the binding mode is non-productive because the putative –OH is not coordinated to the Zn2+ and placed away from the catalytic Tyr654. When the stereoisomer (R) is placed with the –OH coordinated to the Zn2+ and hydrogen-bonded to the Tyr654, in an apparent productive binding, the substrate amide group and the protein Met784 collide. Thus, the Met784 appears to have a role in determining the stereospecificity of PAL reaction.
Site-Directed Mutagenesis of PALcc
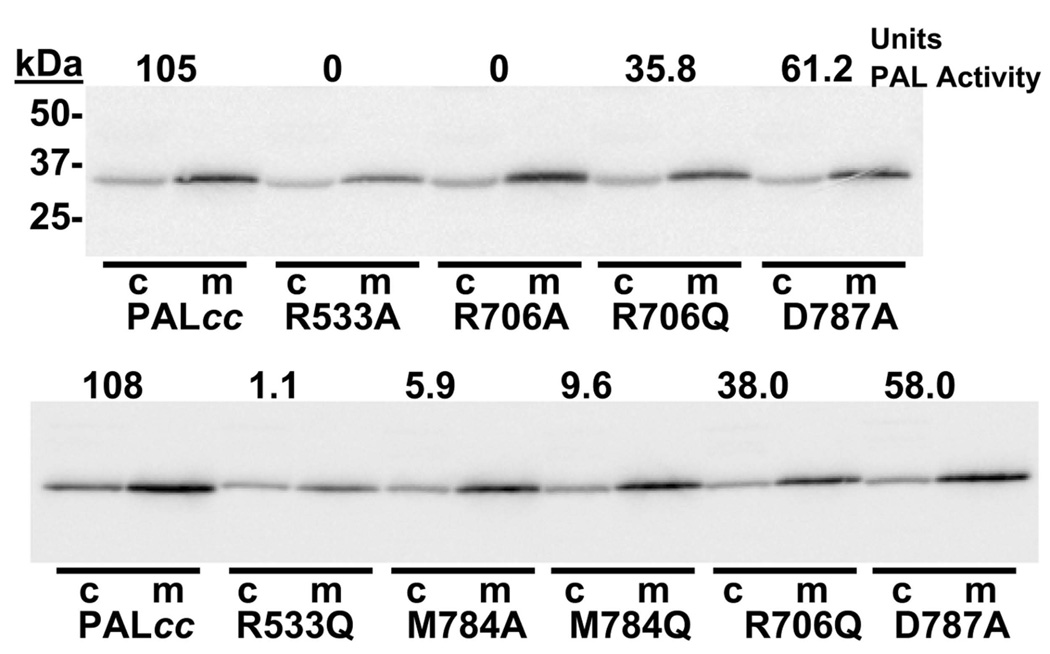
The PAL structures point to Tyr654, Arg706, Arg533 and Met784 as residues that participate in substrate binding and may be expected to play an important role in catalysis. Consistent with these observations, previous mutagenesis studies showed that Tyr654 plays an essential role in catalysis (De et al., 2006). Mutation of PALcc at residues Arg706, Arg533 and Met784 was studied to determine their effect on the catalytic activity. Asp787, one of the residues that binds Ca(II), was also mutated to determine whether perturbation of the calcium site had any effect on PAL activity. The ability of transiently transfected cells to secrete mutant PALcc efficiently was taken as an indication that the protein folded and acquired enough native structure to exit the endoplasmic reticulum (Figure 5). Each of the PALcc mutants examined was secreted as efficiently as wild type PALcc.
Figure 5. Site-directed mutagenesis studies on PALcc.
PEAK-Rapid cells were transiently transfected with vectors encoding wild-type PALcc or each of the mutants indicated. After 24 hours, cells were rinsed and incubated in complete serum-free medium (m) and cells (c) were harvested in TES-mannitol-Triton buffer with protease inhibitors. Samples were analyzed by Western blot (0.8% of the medium; 4% of the cell extract) using an anti-peptide polyclonal antibody and aliquots were assayed for PAL activity. The activity of each sample is shown at the top of each gel lane.
Km and Vmax values for each PALcc mutant were determined by varying the concentration of Ac-Tyr-Val-α-hydroxyglycine from 1 µM to 50 µM (Table I). Relative Vmax values were calculated by quantifying PALcc protein levels based on Western blot analysis. The relative Vmax of PALcc R706A was only 3% that of wildtype PALcc, confirming the essential role of Arg706 in PAL catalysis. PALcc R706Q was 26 % as active as wild-type PALcc; the amide group of Gln706 interacting to Tyr654 in a similar manner to the guanidinium of Arg706 does to Tyr654 in the wild-type enzyme (see discussion below) could explain the 26 % activity exhibited by R706Q mutant. This activity may be aided by Glu707 (mentioned above) that may become a potential proton acceptor. Neither mutation of R706 had a dramatic effect on the Km value. The relative Vmax of PALcc bearing an Arg533 mutation (to Ala or Gln) was reduced about 50-fold compared to wildtype PALcc. With so little activity, Km values were difficult to measure, but appeared to increase slightly; lack of an interaction between the substrate carboxylate and Arg533 may compromise the ability of substrate to adopt the precise orientation required for effective catalysis. The PALcc-HydHipA structure identified Met784 as a key part of the substrate binding site; mutation of this residue to Ala or Gln increased Km approximately 6-fold while reducing relative Vmax 2-fold. Asp787 binds calcium in the crystal structure; PALcc bearing an Asp787 to Ala mutation showed no change in relative Vmax and a 2-fold increase in Km.
Table I.
Kinetic Analysis of PALcc mutants
| PAL tested | Km (µM) | Relative Vmax (pmol/h/OD) |
|---|---|---|
| PALcc | 13.8 ± 2.8 | 1.00 ± 2.8 |
| R533A | 21.3b | 0.03 ± 0.006a |
| R533Q | 24.2b | 0.02 ± 0.007a |
| R706A | 25.5b | 0.03 ± 0.006a |
| R706Q | 15.8 ± 1.5a | 0.26 ± 0.039a |
| M784A | 95.2b | 0.41 ± 0.005a |
| M784Q | 94.6 ± 21.6a | 0.39 ± 0.014a |
| D787A | 33.4 ± 5.5 a | 1.19 ± 0.16 |
significantly different from PALcc, p<0.01
determined once
DISCUSSION
During peptide amidation, following hydroxylation of the terminal glycine residue, the extended peptide disproportionates into the amidated peptide and glyoxylate. This reaction, which occurs spontaneously at neutral pH, requires the action of an enzyme, PAL, because in vivo it takes place in the acid environment of the secretory granules (Katopodis et al., 1991). PAL folds as a six-blade β-propeller with extended loops at the “top” and “bottom” ends of the propeller. A Zn2+ ion is present at the top of the propeller, coordinated by three histidine residues located in three different β-strands. A Ca2+ ion, located in the middle of the propeller 11 Å from the Zn2+, is ideally positioned to confer additional stability to the structure, in agreement with biochemical experiments showing that chelation of the Ca2+ reduces the thermal stability of PAL as well as sensitivity to proteolysis (Kolhekar et al., 2002).
Earlier studies showed that the physiologic metals for PAL are Zn2+ and Ca2+ (De et al., 2006). However, these metals can be removed by chelators such as EDTA or EGTA and subsequent addition of different divalent metal ions (e.g. Mn2+, Co2+, Ni2+, Cd2+, Zn2+) can restore activity (Kolhekar et al., 2002). Interestingly, while the enzyme is inactive with Hg2+, it has enhanced activity with Cd2+. The structure with Hg2+ at the active site and substrate was determined; the geometry around Hg2+ is essentially the same as in the native enzyme with Zn2+ and an acetate ion at the active site (M2+-Nε bonds are larger because of the bigger size of the Hg2+ ion, but the Nε-M2+-Nε angles do not change significantly). This indicates that the lack of activity of PAL with Hg2+ depends upon the electronic properties of the metal. Hg2+ is a soft Lewis acid and its strength towards the substrate –OH (a hard Lewis base) is weak and unable to polarize the O-H bond for catalytic deprotonation. Cd2+ is harder than Hg2+ and exhibits electronic properties more similar as the physiological Zn2+.
The lack of activity of PAL with Hg2+ was exploited to prepare a crystalline complex of PAL with a true substrate, α-hydroxyhippuric acid (HydHypA). The structure of this complex unequivocally identified the catalytic site and the interactions between the substrate and the enzyme. A model of the complex of PAL with a hydroxylated peptide [Ala-Ala-Gly(OH)] was generated based on the experimental structure of PAL-HydHypA. In the PAL-HydHypA structure as well as in the model, Arg533 anchors the terminal carboxylate of the substrate and Met784 helps position the carbonyl of the preceding amino acid.
The most salient feature of the catalytic site, however, is the presence of two residues, Tyr654 and Arg706, which seem to be part of a catalytic dyad. Tyr654, in particular, has multiple interactions that are compatible with an important role in catalysis. Significantly, this residue is absolutely conserved in all PAL enzymes (Kolhekar et al., 2002) and previously reported experiments showed that replacement of this residue by phenylalanine results in total loss of enzymatic activity (De et al., 2006). The substrate α-OH is directly coordinated to the Zn2+ and at the same time makes a H-bond with the –OH of Tyr654. Tyr654, in turn, makes a short H-bond with Arg706, and it is at a close distance from the –NH leaving group.
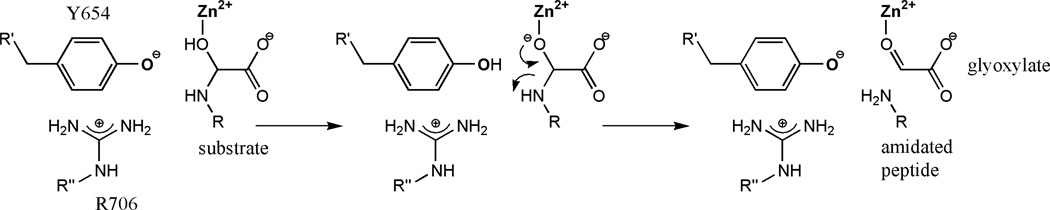
The arrangement of groups in the active site of PAL suggests a mechanism for the lyase reaction (Figure 6). The most likely first step is the deprotonation of the substrate’s α-OH group. Coordination to the Zn2+ must lower significantly the pKa of this hydroxyl group, favoring the release of the proton. This step would be enhanced by the presence of a protein base able to accept the proton. The only group that is in the appropriate location to carry out this task is the –OH group of Tyr654. For this group to accept the proton, it is necessary that its pKa be lowered. The interaction with Arg706 and the proximity to the Zn2+ probably have a significant effect on the pKa of this tyrosine. We propose that this tyrosine is deprotonated prior to substrate binding and that the resulting tyrosinate acts as the catalytic base. For the final bond-breaking step, the tyrosine delivers the proton to the leaving –NH group.
Figure 6. PAL reaction mechanism.
A mechanism for PAL is proposed on the basis of structural and biochemical data. After substrate binding to the active site through coordination of the substrate a-hydoxyl group to the Zn(II) ion, the Tyr654 (as phenolate) deprotonates the substrate α-OH, triggering Cα-N bond cleavage and formation of the amidated peptide and glyoxylate.
A similar role for a tyrosine has been proposed in the mechanism of 3α-Hydroxysteroid Dehydrogenase/Carbonyl Reductase, in which a lysine residue lowers the pKa of the tyrosine allowing it to act as the catalytic base for proton abstraction from the substrate hydroxyl (Penning et al., 1997; Chang et al., 2007). In β-lactam synthetase (Miller et al., 2002) and carbapenam synthetase (Miller et al., 2003), homologous enzymes that catalyze the formation of the four-member β-lactam ring of clavulanic acid and carbapenems, respectively, a tyrosine has also been proposed to act as the catalytic base. However, in these cases a glutamate assists the tyrosine that in turn facilitates deprotonation of the critical α-ammonium ion to initiate the reaction. In PAL, the proton of the catalytic tyrosine is probably transferred to solvent before the start of the reaction, allowing the resulting phenolate to remove the proton from the α–OH, which is also coordinated to the Zn2+. This tyrosine is positioned to transfer a proton to the –NH of the leaving group. The proximity of the positive charges of Zn2+ and Arg706 provide a path to lower the pKa of the tyrosine –OH to values compatible with the proposed mechanism. This arrangement, in which the same tyrosine residue accepts a proton from the substrate and donates a proton to the leaving group, is particularly attractive because, despite its simplicity, it is fully compatible with the structural data. The short distances from the tyrosine –OH to the Cα–OH (2.4 Å) and to the peptide –NH (3.7 Å) are consistent with the proposed dual function for this tyrosine. The absence of any other side chains in the proximity of the scissile bond and the effect on PAL activity of the mutations of the residues identified as participating in the catalysis, argue strongly in favor of this mechanism.
In summary, PAL folds as a six blades β-propeller with a structural Ca2+ located in the middle of the pore and a catalytic Zn2+ at the active site. We propose a mechanism for the PAL reaction, validated by site-directed mutagenesis studies. The substrate binds the enzyme with the a-hydroxyl group directly coordinated to the Zn2+ and a Tyr654-Arg706 dyad at the active site plays an essential role in catalysis. The Arg706 lowers the pKa of the Tyr654 allowing the tyrosine to act as the catalytic base for hydroxyl deprotonation. The β-propeller structure of PAL and the proximity of its N- and C-termini suggest that PHM (the monooxygenase domain of PAM) must be very close to the secretory granule membrane, where cytochrome b561 provides electron equivalents via regeneration of ascorbate. Investigations addressing how PAL interact with PHM are underway.
EXPERIMENTAL PROCEDURES
Production and Purification of His6-PALcc
A CHO-DG44 cell line secreting His6-PALcc-ΔGlyc was selected and grown in CellMax® artificial capillary cartridges as described (De et al., 2006). Spent medium was harvested daily and stored frozen after addition of protease inhibitors. Secreted proteins were concentrated by addition of solid (NH4)2SO4 to 70% saturation (44.2 g/100 ml). Pellets were washed by suspension in 20 mM NaTES, 1.5 M (NH4)2SO4, pH 7.0 (10 ml/pellet from 1 liter) and centrifuged at 16,000 × g for 15 min; supernatants were discarded and pellets were dissolved in 3mM Tris HCl, pH 8.0 (5 ml/pellet from 1 liter). After passage through a 0.22 µm syringe filter, samples (from 0.5 liter of spent medium) were applied to a 5 ml hydrophobic interaction column (HiTrap™ phenyl; Pharmacia) equilibrated with 20 mM NaTES, 1.5 M (NH4)2SO4, pH 7.0 and eluted with a linear gradient to 20 mM NaTES, pH 7.0 over 70 min (flow rate 1.0 ml/min); average recovery to this point was 67%. Fractions enriched in His6-PALcc were pooled based on Coomassie Brilliant Blue staining and Western blot analysis. After concentration using an Amicon stirred flow cell with an YM-30 membrane, samples were loaded onto a Superdex™ 200 gel filtration column (25 × 350 mm) equilibrated with 25 mM NaTES, 100 mM NaCl, pH 7.0. Fractions containing PALcc were identified by SDS-PAGE, pooled and concentrated using an Amicon stirred flow cell. Purity was assessed by Coomassie Brilliant Blue staining after transfer to polyvinylidene difluoride membranes.
Mutagenesis of PALcc
The Stratagene Quick Change Kit (De et al., 2006) was used with pCI.neo.signal.His6. PALcc to mutate single amino acids using the following sense primers (with the corresponding complementary strand) (mutagenic residues underlined with italics):
Pal-R533Q AATAACCTAGTGATTTTCCACCAAGGTGACCATGTTTGGGATGGA
Pal-R533A AATAACCTAGTGATTTTCCACGCAGGTGACCATGTTTGGGATGGA
Pal-R706Q GACCAGTTGTGTGTGGCAGACCAGGAAAATGGCCGAATCCAATGC
Pal-R706A GACCAGTTGTGTGTGGCAGACGCGGAAAATGGCCGAATCCAATGC
Pal-M784Q CCAGTACGCAAGCACTTCGACCAGCCTCATGATATTGTGGCTTCT
Pal-M784A CCAGTACGCAAGCACTTCGACGCGCCTCATGATATTGTGGCTTCT
Pal-D787A AAGCACTTCGACATGCCTCATGCTATTGTGGCTTCTGAAGATGGG
The resulting pCI.neo vectors were sequenced to ensure accurate mutagenesis with no additional changes. The pCI.neo vectors were used for transient expression in pEAK-Rapid cells. Kinetic analyses were performed with 125I-Ac-Tyr-Val-OH-Gly substrate (1–50 µM) (Kolhekar et al., 2002). Western analyses of cell extracts and secreted proteins were performed using PAL antibody JH256 to estimate secretion rates (in all cases, about 2 hours for the cellular content to be secreted); then Westerns were performed with equal PAL activity, to estimate maximal velocity.
PALcc crystallization, Hg-derivatization and preparation of PALcc-substrate crystals
Purified PALcc was concentrated to 11 mg/ml in 25 mM NaTES, 100 mM NaCl, pH 7.0. Crystallization conditions were found by incomplete factorial screening (solution # 37, 0.1 M sodium acetate trihydrate pH = 4.6, 8% w/v PEG4000; Hampton Research -Crystal Screen HR2–110) using hanging-drop vapor diffusion at 293 K. Optimization to 0.1 mM NaAc pH 4.6, 9–12% PEG8000 yielded better crystals; however, crystals were very small and most of them did not diffract. New crystallization conditions were found adding Zn(II) and Fe(III) salts and removing PEG from the original formulation. In the optimized final condition, the crystallization drop was prepared by mixing 1 µl protein solution and 1 µl mother liquor comprising 0.1 mM NaAc pH 4.8, 0.2 mM ZnAc2 and 0.2 mM FeCl3. A small volume of mother liquor was placed at the reservoir (0.2 ml instead of the standard 1 ml) to allow slow equilibration between the drop and mother liquor. Crystals suitable for X-ray diffraction were grown under these conditions.
A mercury(II)-derivative crystal was prepared by soaking PALcc crystals in mother liquor supplemented with 1 mM mercury(II) acetate for 12 hours.
To prepare stable crystals of PALcc with the non-peptidic substrate a-hydroxyhippuric acid (HydHipA) (Aldrich), PALcc was crystallized in 0.1 mM NaAc pH 4.8, 0.5 mM HgAc2. PALcc crystals were then soaked in mother liquor supplemented with 5 mM HydHipA for several hours at 293K.
Data collection, structures determination and refinement
Crycooling was achieved for all cases by washing the crystals in mother liquor containing 25 % glycerol and flash freezing in liquid nitrogen. An Hg-mutliwavelength anomalous dispersion (MAD) data to 2.5 Å on a mercury-derivative and a Zn-MAD data to 2.35 Å on a native crystal were collected at beamline X4A at National Synchrotron Light Source (Brookhaven National Laboratory); the statistics of data collection are summarized in Table S1 and S2, respectively (Supplementary information). The HKL2000 software package was used for data reduction and scaling (Otwinowski and Minor, 1997). One molecule was found per asymetric unit. Three Hg atoms were located per molecule and initial phases were calculated and refined with the program autoSHARP (Global Phasing, Cambridge) (Vonrhein et al., 2007), leading to an interpretable electron density map for most of the structure. However, blades 1 and 2 of the propeller could not be built using the phases obtained from the Hg-MAD experiment. Then, an anomalous map was prepared with data taken at 1.28 Å (Zn-edge) using the initial Hg-MAD phases and the Zn site and a metal contact site (Fe) were thus accurately located. With those coordinates, very good phases were calculated and refined using the Zn-MAD data by autoSHARP. Density modification, solvent flattening and automatic building (25–30 % of the structure) were carried out in the SHARP pipeline (Vonrhein et al., 2007). Manual model building was carried out with the program O (Jones et al., 1991) and refinement was performed with Refmac5 (Murshudov et al., 1997). Translation, libration and screw rotation (TLS) anisotropic refinement of rigid bodies was carried out using each blade as a TLS group (Winn et al., 2003). Data collection and model statistics are summarized in Table II.
Diffraction data for the PALcc-HydHipA complex were collected with 1.00 Å radiation at SGX-CAT beamline facilities at Sector 31 of the Advanced Photon Source (Argonne National Laboratory). Data reduction and scaling were carried out as described above. The initial structure was obtained by molecular replacement using the program AMoRe (Navaza, 2001) as implemented in the CCP4 crystallographic software suite (1994) using the coordinates of native PALcc as the search model. The program SKETCHER in CCP4 was used to build the α-hydroxyhippuric acid molecule (HydHipA) and to generate the library file. The HydHipA was manually placed into the “extra density” using the program O (Jones et al., 1991). Structure refinement was performed as described above.
Modeling of a PALcc-[Ala-Ala-Gly(OH)] complex
A model of a (S)-α-hydroxyglycine-extended tripeptide [Ala-Ala-Gly(OH)] bound to PALcc was built by manual insertion of the tripeptide into the Zn active site, using the program O (Jones et al., 1991). The tripeptide was carefully positioned by matching the atoms of the hydroxyglycine moiety with the equivalent atoms of the α-hydroxyhippuric acid, for which experimental data are available.
ACKNOWLEDGMENTS
Mario Bianchet, Sandra Gabelli, and the staff at beamlines X4A and X6A of the National Synchrotron Light Source, Brookhaven National Laboratory, and of Advance Photon Source (APS) at Argonne National Laboratory, are gratefully acknowledged for assistance during synchrotron data collection. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the SGX Collaborative Access Team (SGX-CAT) beamline facilities at Sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company who operates the facility. This work was supported by National Science Foundation grant MCB-0450465 (L.M.A.) and National Institutes of Health grant DK32949 (B.A.E. and R.E.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been submitted to the Protein Data Bank under ID codes 3FVZ and 3FW0 for PALcc and PALcc-substrate, respectively.
SUPPLEMENTAL DATA
Supplemental Data include two figures and two tables and can be found with this article online at …
REFERENCES
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chang YH, Chuang LY, Hwang CC. Mechanism of proton transfer in the 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. J Biol Chem. 2007;282:34306–34314. doi: 10.1074/jbc.M706336200. [DOI] [PubMed] [Google Scholar]
- Chen P, Solomon EI. O2 activation by binuclear Cu sites: noncoupled versus exchange coupled reaction mechanisms. Proc Natl Acad Sci U S A. 2004a;101:13105–13110. doi: 10.1073/pnas.0402114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Solomon EI. Oxygen activation by the noncoupled binuclear copper site in peptidylglycine alpha-hydroxylating monooxygenase. Reaction mechanism and role of the noncoupled nature of the active site. J Am Chem Soc. 2004b;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]
- Crespo A, Marti MA, Roitberg AE, Amzel LM, Estrin DA. The catalytic mechanism of peptidylglycine alpha-hydroxylating monooxygenase investigated by computer simulation. J Am Chem Soc. 2006;128:12817–12828. doi: 10.1021/ja062876x. [DOI] [PubMed] [Google Scholar]
- De M, Bell J, Blackburn NJ, Mains RE, Eipper BA. Role for an essential tyrosine in peptide amidation. J Biol Chem. 2006;281:20873–20882. doi: 10.1074/jbc.M513886200. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- Francisco WA, Merkler DJ, Blackburn NJ, Klinman JP. Kinetic mechanism and intrinsic isotope effects for the peptidylglycine alpha-amidating enzyme reaction. Biochemistry. 1998;37:8244–8252. doi: 10.1021/bi973004y. [DOI] [PubMed] [Google Scholar]
- Hall DR, Bond CS, Leonard GA, Watt CI, Berry A, Hunter WN. Structure of tagatose-1,6-bisphosphate aldolase. Insight into chiral discrimination, mechanism, and specificity of class II aldolases. J Biol Chem. 2002;277:22018–22024. doi: 10.1074/jbc.M202464200. [DOI] [PubMed] [Google Scholar]
- Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- Hege T, Baumann U. Protease C of Erwinia chrysanthemi: the crystal structure and role of amino acids Y228 and E189. J Mol Biol. 2001;314:187–193. doi: 10.1006/jmbi.2001.5124. [DOI] [PubMed] [Google Scholar]
- Iliadi KG, Avivi A, Iliadi NN, Knight D, Korol AB, Nevo E, Taylor P, Moran MF, Kamyshev NG, Boulianne GL. nemy encodes a cytochrome b561 that is required for Drosophila learning and memory. Proc Natl Acad Sci U S A. 2008;105:19986–19991. doi: 10.1073/pnas.0810698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Karpusas M, Nolte M, Benton CB, Meier W, Lipscomb WN, Goelz S. The crystal structure of human interferon beta at 2.2-A resolution. Proc Natl Acad Sci U S A. 1997;94:11813–11818. doi: 10.1073/pnas.94.22.11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katopodis AG, May SW. Novel Substrates and Inhibitors of Peptidylglycine α-Amidating Monooxygenase. Biochemistry. 1990;29:4541–4548. doi: 10.1021/bi00471a006. [DOI] [PubMed] [Google Scholar]
- Katopodis AG, Ping DS, Smith CE, May SW. Functional and structural characterization of peptidylamidoglycolate lyase, the enzyme catalyzing the second step in peptide amidation. Biochemistry. 1991;30:6189–6194. doi: 10.1021/bi00239a016. [DOI] [PubMed] [Google Scholar]
- Kent UM, Fleming PJ. Purified cytochrome b561 catalyzes transmembrane electron transfer for dopamine beta-hydroxylase and peptidyl glycine alpha-amidating monooxygenase activities in reconstituted systems. J Biol Chem. 1987;262:8174–8178. [PubMed] [Google Scholar]
- Klinman JP. The copper-enzyme family of dopamine beta-monooxygenase and peptidylglycine alpha-hydroxylating monooxygenase: resolving the chemical pathway for substrate hydroxylation. J Biol Chem. 2006;281:3013–3016. doi: 10.1074/jbc.R500011200. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Bell J, Shiozaki EN, Jin L, Keutmann HT, Hand TA, Mains RE, Eipper BA. Essential Features of the Catalytic Core of Peptidyl-a-hydroxyglycine a-Amidating Lyase. Biochemistry. 2002;41:12384–12394. doi: 10.1021/bi0260280. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Roberts MS, Jiang N, Johnson RC, Mains RE, Eipper BA, Taghert PH. Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J Neurosci. 1997;17:1363–1376. doi: 10.1523/JNEUROSCI.17-04-01363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding. Chem Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Kocourek A, Braun M, Tschesche H, Huber R, Bode W, Maskos K. Substrate specificity determinants of human macrophage elastase (MMP-12) based on the 1.1 A crystal structure. J Mol Biol. 2001;312:731–742. doi: 10.1006/jmbi.2001.4954. [DOI] [PubMed] [Google Scholar]
- Luo Y, Samuel J, Mosimann SC, Lee JE, Tanner ME, Strynadka NC. The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization. Biochemistry. 2001;40:14763–14771. doi: 10.1021/bi0112513. [DOI] [PubMed] [Google Scholar]
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, et al. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc Natl Acad Sci U S A. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIninch JK, McIninch JD, May SW. Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase. J Biol Chem. 2003;278:50091–50100. doi: 10.1074/jbc.M303828200. [DOI] [PubMed] [Google Scholar]
- Merkler DJ. C-terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extended peptides and the importance of the amide to bioactivity. Enzyme Microb Technol. 1994;16:450–456. doi: 10.1016/0141-0229(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots. Proc Natl Acad Sci U S A. 2002;99:14752–14757. doi: 10.1073/pnas.232361199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Gerratana B, Stapon A, Townsend CA, Rosenzweig AC. Crystal structure of carbapenam synthetase (CarA) J Biol Chem. 2003;278:40996–41002. doi: 10.1074/jbc.M307901200. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Penning TM, Bennett MJ, Smith-Hoog S, Schlegel BP, Jez JM, Lewis M. Structure and function of 3α-hydroxysteroid dehydrogenase. Steroids. 1997;62:101–111. doi: 10.1016/s0039-128x(96)00167-5. [DOI] [PubMed] [Google Scholar]
- Ping D, Katopodis AG, May SW. Tandem Stereochemistry of Peptidylglycine a-Monooxygenase and Peptidylamidoglycolate Lyase, the Two Enzymes Involved in Peptide Amidation. J Am Chem Soc. 1992;114:3998–4000. [Google Scholar]
- Prigge ST, Eipper BA, Mains RE, Amzel LM. Dioxygen Binds End-On to Mononuclear Copper in a Precatalytic Enzyme Complex. Science. 2004;304:864–867. doi: 10.1126/science.1094583. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of peptidylglycine alpha-hydroxylating monooxygenase. Science. 1997;278:1300–1305. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Substrate-mediated electron transfer in peptidylglycine alpha-hydroxylating monooxygenase. Nat Struct Biol. 1999;6:976–983. doi: 10.1038/13351. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Mains RE, Eipper BA, Amzel LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagenhauf E, Etges R, Metcalf P. The crystal structure of the Leishmania major surface proteinase leishmanolysin (gp63) Structure. 1998;6:1035–1046. doi: 10.1016/s0969-2126(98)00104-x. [DOI] [PubMed] [Google Scholar]
- Sharma SD, Raghuraman G, Lee MS, Prabhakar NR, Kumar GK. Intermittent hypoxia activates peptidylglycine alpha-amidating monooxygenase in rat brain stem via reactive oxygen species-mediated proteolytic processing. J Appl Physiol. 2009;106:12–19. doi: 10.1152/japplphysiol.90702.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- Toney JH, Hammond GG, Fitzgerald PM, Sharma N, Balkovec JM, Rouen GP, Olson SH, Hammond ML, Greenlee ML, Gao YD. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-beta-lactamase. J Biol Chem. 2001;276:31913–31918. doi: 10.1074/jbc.M104742200. [DOI] [PubMed] [Google Scholar]
- Trijbels F, Vogels GD. Degradation of allantoin by Pseudomonas acidovorans. Biochim Biophys Acta. 1966;113:292–301. doi: 10.1016/s0926-6593(66)80068-1. [DOI] [PubMed] [Google Scholar]
- Tsubaki M, Takeuchi F, Nakanishi N. Cytochrome b561 protein family: expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta. 2005;1753:174–190. doi: 10.1016/j.bbapap.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]