Abstract
We show for the first time that it is possible to obtain LNA (Locked Nucleic Acid 1) like binding affinity and biological activity with carbocyclic LNA (cLNA) analogs by replacing the 2’-oxygen atom in LNA with an exocyclic methylene group. Synthesis of the methylene-cLNA nucleoside was accomplished by an intramolecular cyclization reaction between a radical at the 2’-position and a propynyl group at C-4’ position. Only methylene-cLNA modified oligonucleotides showed similar thermal stability and mismatch discrimination properties for complementary nucleic acids as LNA. In contrast, the close structurally related methyl-cLNA analogs showed diminished hybridization properties. Analysis of crystal structures of cLNA modified self-complementary DNA decamer duplexes revealed that the methylene group participates in a tight interaction with a 2’-deoxyribose residue of the 5’-terminal G of a neighboring duplex, resulting in the formation of a CH…O type hydrogen bond. This indicates that the methylene group retains a negative polarization at the edge of the minor groove in the absence of a hydrophilic 2’-substituent and provides a rationale for the superior thermal stability of this modification. In animal experiments, methylene-cLNA ASOs showed similar in vivo activity but reduced toxicity as compared to LNA ASOs. Our work highlights the interchangeable role of oxygen and unsaturated moeities in nucleic acid structure and emphasizes greater use of this bio-isostere to improve the properties of nucleic acids for therapeutic and diagnostic applications.
Introduction
Antisense drug discovery technology represents a powerful method to control gene expression in animals and in cell culture.1 Antisense oligonucleotides (ASOs) bind to their cognate messenger RNA (mRNA) by Watson-Crick base pairing. Upon binding they can modulate splicing, interfere with translation or promote degradation of the mRNA by recrutiment of RNase H or by degradation via the RISC pathway. Chemical modifications have been used extensively in antisense therapeutics to modulate stability, binding affinity, pharmacokinetic and toxicological properties of ASOs.2 First generation ASOs were fully phosphorothioate (PS) modified DNA oligonucleotides, where one of the oxygen atoms in the backbone phosphodiester linkage is replaced with sulfur atom.3 The PS linkage protects the ASO against nuclease degradation and promotes binding to plasma proteins thereby permitting distribution to periphral tissues. However, the PS modification reduces binding affinity for the target RNA and these ASOs are still subject to nuclease mediated degradation.4 Second generation ASOs are chimeric oligonucleotides which have a central gap region of 10 to 16 deoxyribonucleotides flanked on the 3’- and 5’-ends with modified residues such as 2’O-methoxyethyl nucleotides (MOE). The MOE residues improve affinity for target mRNA and further stabilize the olignucleotide against exonuclease digestion.5,6 Currently there are more than 20 second generation antisense oligonucleotides (ASOs) in various stages of clinical trials for a number of indications including hyper-cholesterolemia, diabetes and cancer amongst others. The most advanced second generation ASO, Mipomersen – an inhibitor of Apolipoprotein B 100 (ApoB 100) biosynthesis,7 has shown mean reductions of ∼25% in plasma LDL cholesterol in two phase III clinical trials at a dose of 200 mg/week.8
In addition to MOE, a number of other 2’-modified ribonucleosides such as 2’O-methyl and 2’-fluoro have been investigated for various antisense applications.9 These modifications were found to be tolerated in both the sense and antisense strands of siRNA duplexes10 and have found applications in improving the therapeutic properties of oligonucleotide aptamers.11 2’-fluoro modified ASOs have also found utility as miRNA antagonists where they displayed a remarkable improvement in in vivo activity which has been ascribed to altered sub-cellular pharmacokinetic properties.12 To further improve the binding affinity of oligonucleotides, Wengel and Imanishi independently introduced 2’,4’-methyleneoxy bridged nucleic acids, also commonly known as Locked nucleic acid 1 (LNA)13. LNA modified oligonucleotides show unprecedented improvement in the thermal stability of oligonucleotide duplexes.14 LNA ASOs have also demonstrated promising results for down regulating gene expression,15,16 splice modulation17 and as micro RNA (miRNA) antagonists18 in various animal models. However, some LNA ASOs appear to be associated with an increased risk for causing hepatotoxicity.19
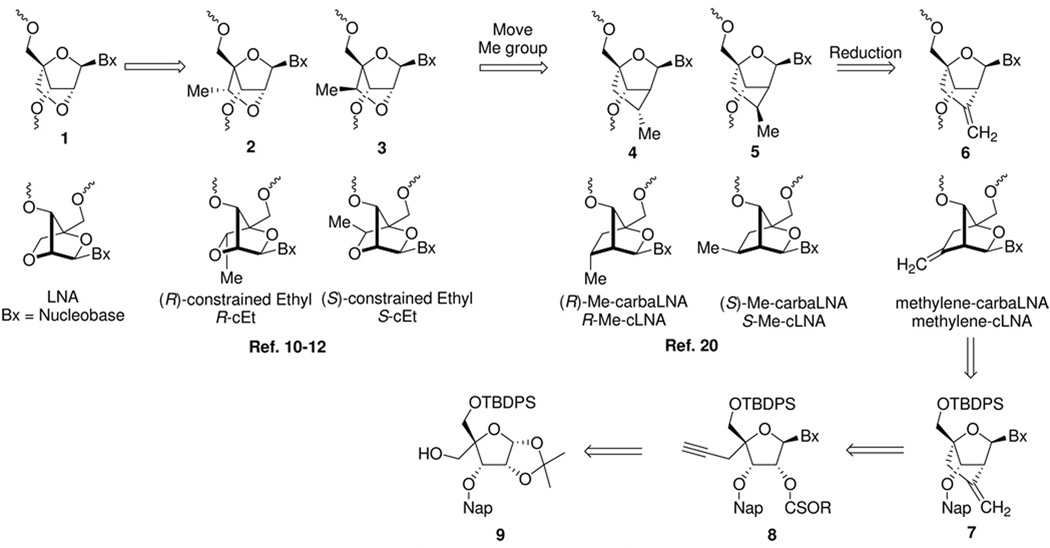
The synthesis of other 2’,4’-conformationally restricted nucleoside analogs has been an active area of interest recently given the successful applications of LNA within the antisense paradigm.20 As part of a comprehensive program aimed at understanding the structure activity relationships (SAR) of ASOs containing LNA and other related conformationally restricted 2’,4’-bridged nucleic acids (BNA), we recently reported the synthesis and biological evaluation of 2’,4’-constrained Ethyl (cEt) modified nucleic acids 2 (R-cEt) and 3 (S-cEt).21–23 We found that replacing LNA with the cEt modifications resulted in an improvement in the therapeutic profile of “gapmer”24 ASOs targeting Mus. musculus phosphatase and tensin homolog (mouse PTEN). We speculated that the improved therapeutic profile of the cEt ASOs was a result of altered interactions of these ASOs with cellular macromolecules. To investigate if changing the position of the alkyl substituent along the 2’,4’-bridging group could provide further improvements in the therapeutic profile, we wished to evaluate substituted carbocyclic LNA (cLNA) analogs 4 (R-methyl-cLNA) and 5 (S-methyl-cLNA) where the 2’ oxygen atom of LNA was replaced with a substituted carbon atom.
At the time we initiated our synthesis program there were no literature reports describing the synthesis of carbocyclic LNA analogs. We envisaged a strategy where both the R- and S-methyl-cLNA analogs 4 and 5 as well as other cLNA analogs could be prepared from the methylene-cLNA nucleoside 7 by synthetic manipulation of the exocyclic double bond. For example, the double bond can be hydroborated to provide the hydroxymethyl analog or cleaved oxidatively to provide the keto compound, both of which could serve as intermediates for a myriad of chemical transformations (alkylation, fluorination, reduction, oxidation etc.) to provide other cLNA analogs. Retrosynthetically, methylene-cLNA 7 could be prepared from a suitable nucleoside precursor 8 by means of an intramolecular radical cyclization reaction (Figure 1). This strategy was inspired by previous work by Chattopadhyaya,25,26 and Matsuda,27,28 who independently showed that it was possible to form intramolecular carbon-carbon bonds by generating and trapping a radical at the 2’ position of a nucleoside with a suitable acceptor group. We were also motivated to evaluate the antisense properties of the methylene-cLNA 6 itself, as replacing an oxygen atom with a methylene group has been successfully utilized in the anti-viral arena (Entecavir)29 and in the search for novel nucleic acid mimics (cyclohexenyl nucleic acids, CeNA).30
Figure 1.
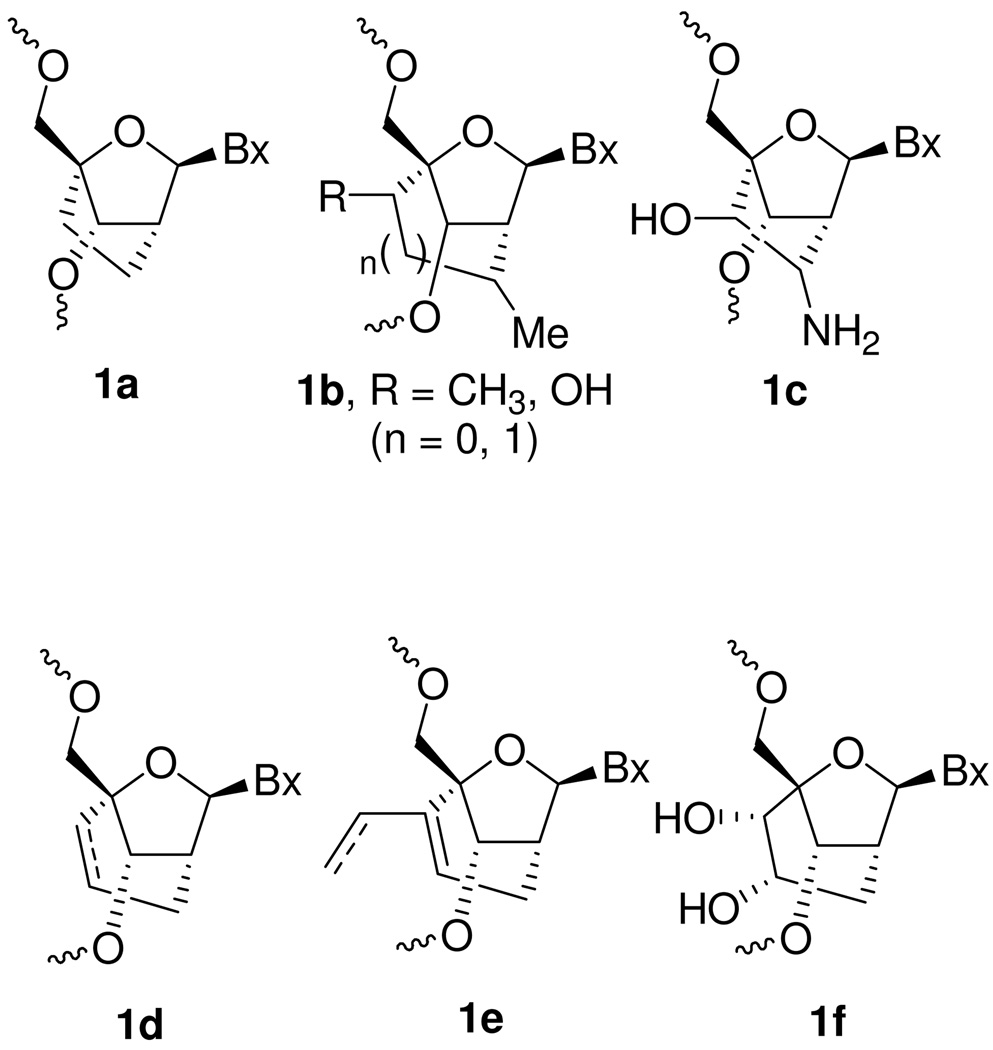
While our work was in progress, Chattopadhyaya and co-workers reported the synthesis and biophysical properties of oligonucleotides containing a mixture of R- and S-methyl-cLNA analogs 4 and 5.31 Since the initial report, Chattopadhyaya has also reported the synthesis and properties of a number of differentially substituted cENA and cLNA analogs (1a–c, Figure 2).32–34 Recently, Nielsen and co-workers reported the synthesis and biophysical properties of a number of carbocyclic ENA analogs using an intramolecular RCM and ene-alkyne metathesis reactions (1d–f, Figure 2).35,36 All the cLNA and cENA analogs reported thus far show moderate to significantly reduced binding affinity for complementary RNA relative to LNA that can be partially restored by introducing hydrophilic groups along the 2’,4’-bridging substituent. Interestingly, both Chattopadhyaya and Nielsen also showed that cLNA analogs appear to recognize complementary DNA less efficiently relative to RNA. They proposed that replacing the 2’oxygen atom with carbon reduces the hydration of the duplex in the minor groove which in turn affects recognition of DNA more severely as compared to RNA.34,35 Recently, Bramsen and co-workers have reported on the utility of cLNA ASOs for positional modification of sense and antisense strands of siRNAduplexes. 37,38 In addition, Corey has shown that cLNA ASOs may have utility in selective targeting of mutant Huntington alleles over wild-type transcripts.39,40 However, there are no reports which conclusively explain why cLNA analogs reported thus far show reduced hybridization properties relative to their “oxa” counterparts. Moreover, there are no reports that describe the in vivo properties of cLNA modified ASOs. In this manuscript we show for the first time that it is possible to obtain LNA like binding affinity with cLNA analogs by replacing the 2’-oxygen with an exocyclic methylene group and rationalize the improved thermodynamic properties using crystal structure data. We also report for the first time, the biological evaluation of cLNA modified ASOs in animal experiments and demonstrate the usefulness of this class of nucleoside modifications for antisense applications.
Figure 2.
Results and Discussion
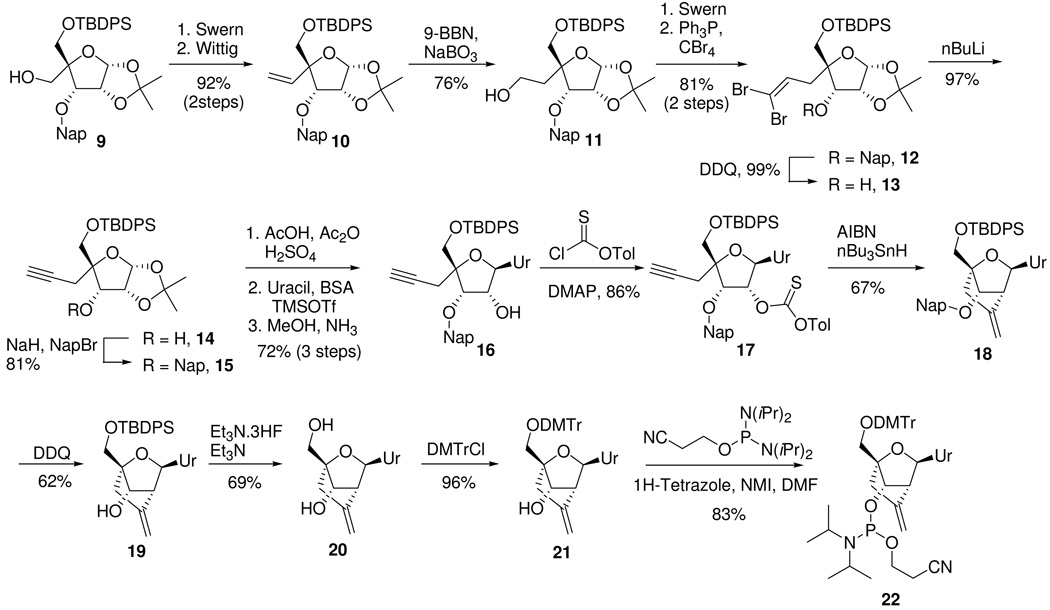
Synthesis of the methylene-cLNA nucleoside commenced from the known 5’-O-TBDPS-3’O-(2-methylnapthalene)-allofuranose derivative 9 (Scheme 1).22 Swern oxidation41 of the primary alcohol followed by a Wittig reaction provided olefin 10 in excellent yield (92%, 2 steps). Hydroboration of the double bond using 9-BBN/sodium perborate42 provided the primary alcohol 11 in good yield (76%). A Swern oxidation of the primary alcohol followed by a Corey-Fuchs43 reaction provided the dibromo olefin 12 (81%, 2 steps). Attempts to convert 12 directly to the alkyne 15 using nBuLi were unsuccessful. Treatment of 12 with nBuLi resulted in the formation of a dark green colored solution indicative of single electron transfer into the neighboring naphthlene ring.44 To avoid this problem, the naphthyl group (Nap) was deprotected with DDQ45 to provide 13 (99%), which was further treated with nBuLi to provide the alkyne 14 efficiently (97%). Reprotection of the 3’-hydroxyl group as the naphthyl ether provided the desired alkyne 15 (81%). Acetolysis of the 1,2-isopropylidene group, followed by synthesis of the nucleoside by means of a Vorbruggen reaction46 with persilylated uracil and deprotection of the 2’-O-acetyl group provided nucleoside 16 in good yield (72%, 3 steps). The 2’-hydoxyl group in 16 was converted to the thiocarbonate 17 by treatment with tolyl-chlorothionoformate (86%). Treatment of the thiocarbonate 17 with tributyltin hydride and AIBN provided the cyclized nucleoside 18 by means of intramolecular radical cyclization reaction. Interestingly, Chattopadyaya had previously reported that unsubstituted alkynes do not participate efficiently in intramolecular radical cyclization reactions due to decomposition of the alkyne moiety under the reaction conditions employed (AIBN/nBu3SnH).25 However, in case of nucleoside 17, no further protection of the alkyne was required for efficient cyclization to occur. It is conceivable that in case of 17, the radical cyclization occurs faster than the competing decomposition reactions. The naphthyl protecting group was next cleanly removed using DDQ to provide 19 (62%). Deprotection of the 5’O-TBDPS ether provided nucleoside 20 (69%). Protection of the 5’-hydroxyl group as the 4,4’-dimethoxytrityl (DMTr) ether (96%) followed by a phosphitylation reaction provided the desired amidite 22 in good yield (83%).
Scheme 1.
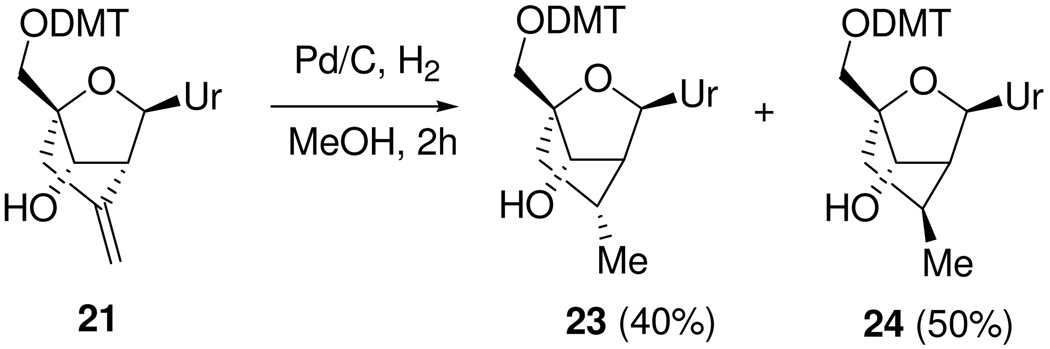
The structure of the methylene-cLNA nucleoside was established by NMR spectroscopy. For the 2’,4’ cyclized nucleoside 18, the H1’, H2’ and H3’ proton resonances appear as singlets, which is consistent with the furanose ring being locked in the Northern conformation. In addition, the identity of the methylene-cLNA nucleoside was further confirmed by reducing the exocyclic double bond in the 5’O-DMTr protected nucleoside 21 using catalytic hydrogenation to provide a separable mixture of the known nucleosides 23 and 24 (supporting information).31
Thermal stability and mis-match discrimination properties of cLNA analogs
All the modified nucleoside phosphoramidites were incorporated into oligonucleotides using standard phosphoramidite chemistry described previously.22 We first evaluated the cLNA analogs in an oligonucleotide sequence used by Imanishi13 to evaluate the hybridization properties of LNA and compared them to R-cEt, S-cEt and LNA modified oligonucleotides (Table 1). In this sequence, methylene-cLNA (A2, ΔTm +4.8 °C/mod. vs. RNA, −1.6 °C/mod. vs. DNA) showed similar thermal stability versus RNA as LNA (A7, ΔTm +4.5 °C/mod. vs. RNA, +0.1 °C/mod. vs. DNA), S-cEt (A6, ΔTm +4.5 °C/mod. vs. RNA, −0.1 °C/mod. vs. DNA) and R-cEt (A5, ΔTm +4.7 °C/mod. vs. RNA, −1.1 °C/mod. vs. DNA) but reduced stability versus complementary DNA as compared to LNA and S-cEt modified oligonucleotides.
Table 1. Evaluation of methylene-cLNA, R-Me-cLNA and S-Me-cLNA in thermal denaturation experiments.
Thermal stability measurements of methylene-cLNA, R-Me-cLNA, S-Me-cLNA, R-cEt, S-cEt and LNA versus complementary RNA and DNA in the Imanishi sequence
 | |||||||
|---|---|---|---|---|---|---|---|
| Oligomer | Modification | Sequencea (5’ – 3’) |
Tmb (°C) vs RNA |
ΔTm (°C)/mod. |
Tm (°C) vs DNA |
ΔTm (°C)/mod. |
(Tm RNA-Tm DNA)/ # mod. (°C) |
| A1 | DNA | d(GCGTTTTTTGCT) | 45.6 | 0 | 50.9 | 0 | 0 |
| A2 | Methylene-cLNA | d(GCGTTUTTTGCT) | 50.4 | +4.8 | 49.3 | −1.6 | +1.1 |
| A3 | R-Me-cLNA | d(GCGTTUTTTGCT) | 48.5 | +2.9 | 47.8 | −3.1 | +0.7 |
| A4 | S-Me-cLNA | d(GCGTTUTTTGCT) | 47.6 | +2.0 | 47.0 | −3.9 | +0.6 |
| A5 | R-cEtc | d(GCGTTUTTTGCT) | 50.3 | +4.7 | 49.8 | −1.1 | +0.5 |
| A6 | S-cEtc | d(GCGTTUTTTGCT) | 50.1 | +4.5 | 50.8 | −0.1 | −0.7 |
| A7 | LNAc | d(GCGTTUTTTGCT) | 50.1 | +4.5 | 51.0 | +0.1 | −0.9 |
Bold letter indicates modified residue;
Tm values were measured in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA, sequence of RNA complement 5’-r(AGCAAAAAACGC)-3’ and of DNA complement 5’-d(AGCAAAAAACGC)-3’;
Ref. 11
Chattopadhyaya had previously reported the hybridization properties of a mixture of the R-methyl and S-methyl thymine nucleobase modified cLNA analogs in a different Tm sequence.31 We were able to separate these nucleosides at the 5’O-DMTr stage and evaluate the individual isomers in thermal denaturation studies. Evaluation of the R-Me-cLNA (A3, ΔTm +2.9 °C/mod. vs. RNA, −3.1 °C/mod. vs. DNA) in the Imanishi sequence showed that this isomer did not hybridize complementary RNA or DNA as efficiently as LNA or methylene-cLNA oligonucleotides. The S-Me-cLNA (A4, ΔTm +2.0 °C/mod. vs. RNA, −3.9 °C/mod. vs. DNA) showed the poorest hybridization properties compared to all the analogs evaluated. In general the cLNA analogs exhibited slightly improved selectivity (as measured by Tm RNA – Tm DNA/ number of modifications) for RNA versus DNA as compared to their “oxa” counterparts but the increase was very modest.
We next evaluated the mismatch selectivity of the cLNA analogs versus mismatched RNA complements as reported by Imanishi47 and compared them to LNA (Table 2). Methylene-cLNA A2 and R-Me-cLNA A3 showed comparable (−6 °C) mismatch selectivity while the S-Me-cLNA A4 showed slightly improved (−8 °C) mismatch selectivity for the G-U mismatched pair as compared to LNA (−5 °C). All the cLNA analogs showed excellent selectivity for the C-U (−14 to −16 °C) and the U-U (−12 to −13 °C) mismatched pair which were essentially identical to that observed for LNA.
Table 2.
Mismatch discrimination of DNA, methylene-cLNA, R-Me-cLNA, S-Me-cLNA and LNA versus mismatched RNA complements
| Tm (ΔTm = Tm(mismatch) –Tm (match)) (°C) | |||||
|---|---|---|---|---|---|
| Oligomer | Sequence (5’ – 3’)a | X = A (match) |
G | C | U |
| A1 (DNA) | d(GCGTTTTTTGCT) | 45.6 | 41.5 (−4.1) | 32.3 (−13.3) | 31.8 (−13.8) |
|
A2 (methylene- cLNA) |
d(GCGTTUTTTGCT) | 50.4 | 44.6 (−5.8) | 34.9 (−15.5) | 37.2 (−13.2) |
| A3 (R-Me-cLNA) | d(GCGTTUTTTGCT) | 48.5 | 42.3 (−6.2) | 34.8 (−13.7) | 36.3 (−12.2) |
| A4 (S-Me-cLNA) | d(GCGTTUTTTGCT) | 47.6 | 39.8 (−7.8) | 33.4 (−14.2) | 35.3 (−12.3) |
| A8 (LNA)c | d(GCGTTUTTTGCT) | 50.1 | 45.0 (−5.1) | 35.2 (−14.9) | 37.2 (−12.9) |
Bold letter indicates modified residue;
Tm values were measured in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA, sequence of RNA complement 5’-r(AGCAAAXAACGC)-3’;
Ref. 11
We also evaluated the hybridization properties of methylene-cLNA and the R-Me-cLNA in a different oligonucleotide sequence first described by Herdewijn for the evaluation of HNA and related analogs (Table 3).30 This sequence is a three base extension of a 9-mer sequence used by Wengel to evaluate the hybridization properties of LNA and allows for three incorporations of U/T monomers at different positions of the oligonucleotide.48 In this sequence, the methylene-cLNA (B2, ΔTm +7.1 °C/mod. vs. RNA, +4.0 °C/mod. vs. DNA) showed excellent hybridization properties which were superior to that of R-Me-cLNA (B3, ΔTm +5.4 °C/mod. vs. RNA, +1.8 °C/mod. vs. DNA) as well as S-cEt (B4, ΔTm +6.4 °C/mod. vs. RNA, +3.5 °C/mod. vs. DNA) and LNA (B5, ΔTm +6.1 °C/mod. vs. RNA, +3.6 °C/mod. vs. DNA) modified oligonucleotides. As seen with the Imanishi sequence, the methylene-cLNA (+1.1 °C) and R-Me-cLNA (+1.6 °C) oligonucleotides showed only slightly improved selectivity for RNA versus DNA as compared to LNA (+0.5 °C) and S-cEt (+0.8 °C).
Table 3.
Thermal stability measurements of methylene-cLNA, R-Me-cLNA, S-cEt LNA versus complementary RNA and DNA in the Herdewijn sequence
| Oligomer | Modification | Sequence (5’ – 3’) |
Tma (°C) vs RNA |
ΔTm (°C)/mod. |
Tm (°C) vs DNA |
ΔTm (°C)/mod. |
(Tm RNA-Tm DNA)/ # mod. (°C) |
|---|---|---|---|---|---|---|---|
| B1 | DNA | d(CCAGTGATATGC) | 42.6 | 0 | 48.7 | 0 | 0 |
| B2 | Methylene- cLNA |
d(CCAGUGAUAUGC) | 64.0 | +7.1 | 60.7 | +4.0 | +1.1 |
| B3 | R-Me-cLNA | d(CCAGUGAUAUGC) | 58.9 | +5.4 | 54.2 | +1.8 | +1.6 |
| B4 | S-cEt | d(CCAGUGAUAUGC) | 61.8 | +6.4 | 59.3 | +3.5 | +0.8 |
| B5 | LNA | d(CCAGUGAUAUGC) | 61.0 | +6.1 | 59.4 | +3.6 | +0.5 |
Bold letter indicates modified residue;
Tm values were measured in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA, sequence of RNA complement 5’-r(GCAUAUCACUGG)-3’ and of DNA complement 5’-d(GCATATCACTGG)-3’.
Thus it appears that replacing the 2’-oxygen atom in LNA with a sp3 hybridized carbon atom (analogs 4 and 5) but not a sp2 hybridized carbon atom (Methylene-cLNA 6), results in loss of binding affinity for complementary nucleic acids. Moreover, changing the orientation of the methyl group at the sp3 carbon from the (S) (edge of minor groove) to the (R) (in the minor groove) configuration appears to diminish binding affinity for complementary DNA and RNA. Interestingly, Chattopadhayaya recently reported the thermal stability of oligonucleotides containing a cLNA analog where the 2’-oxygen atom is replaced with an unsubstituted methylene group.34 Even in that case, no improvement in binding affinity relative to the mixture of methyl-substituted analogs 4/5 was observed. Consistent with the reports of Nielsen and Chattopadhyaya, we observed an enhancement in recognition of complementary DNA versus complementary RNA for the cLNA analogs, although the magnitude of stabilization was small and somewhat sequence dependent. Nonetheless, the methylene-cLNA analog 6 represents the first example of a cLNA that shows LNA like binding affinity without the added need for hydrophilic substituents along the 2’-4’-bridging group to restore duplex hydration.
It is conceivable that the methylene group in 6 does not disrupt the water of hydration around the backbone phosphodiester linkage and in the minor groove and this results in improved binding affinity for complementary nucleic acids relative to analogs 4 and 5 which have a saturated carbon atom at the same position. To test this hypothesis, we examined the crystal structure of methylene-cLNA, R- and S-Me-cLNA modified oligonucleotides incorporated into a self-complementary DNA decamer duplex. 49
The crystal structures of decamer duplexes with sequence d(GCGTAU*ACGC), whereby U* is methylene-cLNA-U, R-Me-cLNA-U, or S-Me-cLNA-U (referred to as Vindec E1, Rmedec E2 and Smedec E3, respectively), were determined at resolutions of between 1.35 and 1.57 Å (Table 4). The three duplexes adopt an A-form geometry with average values for helical rise and twist of ca. 2.8 Å and 32°, respectively. All of them exhibit a kink of ca. 12° into the major groove that is localized between the T4 and A5 residues (nucleotides of one strand are numbered 1 to 10 and those in the complementary strand are numbered 11 to 20; the modified cLNA residues are U*6 and U*16). The local compression of that groove goes along with an extended variant of the backbone at residue A5, that is, the α and γ torsion angles are in the ap range instead of in the standard sc− and sc+ ranges, respectively. As expected, the conformations of sugars are virtually without exception of the C3’-endo type. In particular, the sugars of all six cLNA nucleotides fall into this pucker range.
Table 4.
Selected crystal and refinement data.
| Oligonucleotide | Vindec | Rmedec | Smedec |
|---|---|---|---|
| Sequence | d(GCGTAU*ACGC) | ||
| Modified nucleotide (U*) | Methylene-cLNA-U E1 | R-Me-cLNA-U E2 | S-Me-cLNA-U E3 |
| Space group | Orthorhombic P212121 | ||
| Unit cell constants [Å] α=β=γ=90° | a=24.42 b=44.11 c=45.84 | a=26.10 b=43.49 c=45.97 | a=24.47 b=44.11 c=45.95 |
| Resolution [Å] | 1.57 | 1.51 | 1.35 |
| Outer shell [Å] | 1.63–1.57 | 1.56–1.51 | 1.40–1.35 |
| No. of unique reflections | 7,592 | 7,451 | 11,119 |
| Completeness (outer shell) [%] | 99.4 (99.7) | 99.4 (99.7) | 98.7 (91.7) |
| R-merge (outer shell) [%] | 6.7 (24.8) | 5.7 (52.7) | 5.9 (43.6) |
| R-work [%] | 16.3 | 17.2 | 14.6 |
| R-free [%] | 20.8 | 23.7 | 18.6 |
| No. of DNA atoms | 408 | 408 | 408 |
| No. of waters | 65 | 48 | 78 |
| R.m.s. deviations bonds [Å] | 0.022 | 0.019 | 0.022 |
| R.m.s. deviations angles [°] | 2.5 | 2.1 | 2.4 |
| Avg. B-factor, DNA atoms [Å2] | 15.0 | 22.0 | 11.9 |
| Avg. B-factor, solvent [Å2] | 24.1 | 32.4 | 22.2 |
| PDB entry code (to be assigned) | XXXX | YYYY | ZZZZ |
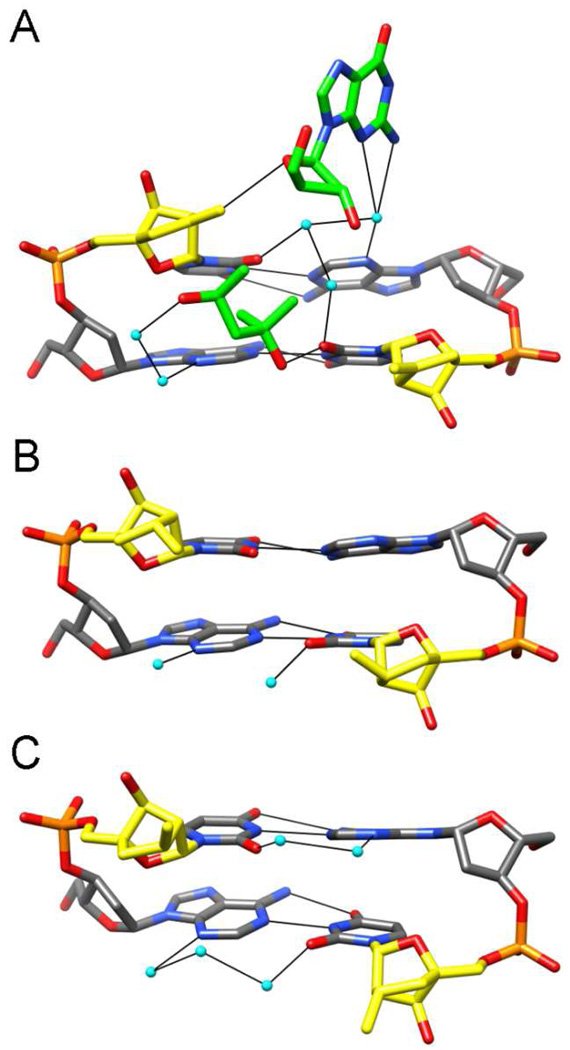
The overall packing motifs in the three orthorhombic crystal lattices of the Vindec, Rmedec and Smedec structures are quite similar. Terminal base pairs of duplexes stack against the minor grooves of neighboring DNAs such that each duplex acts as a receptor of two base pairs from nearest neighbors. However, there are subtle differences in the closeness of the inter-duplex contacts that allow insights into the individual electrostatic potentials of cLNA sugars. Thus, in the case of the Vindec duplex, the sugar of residue U*6 engages in a tight interaction with the 2′-deoxyribose moiety of the 5′-terminal G from a neighboring strand (Figure 3A). This leads to formation of a C-H…O-type hydrogen bond between the outer methylene carbon from U*6 and O4′ of G1# (# marks a symmetry mate; C…O distance 3.50 Å). By comparison, the packing of duplexes in the Rmedec and Smedec lattices is altered such that the neighboring terminal guanidine does not approach the floor of the minor groove and therefore the R- or S-methyl substituents of cLNA sugars (Figure 3 B and C). These observations provide an indication that the methylene-cLNA moiety retains a negative polarization at the edge of the minor groove in the absence of a hydrophilic 2′-substituent. Conversely, the R- and S-Me-cLNA sugars in the Rmedec and Smedec structures are poorly hydrated and unable to extend the water structure around the edges of nucleobases toward the phosphate backbone. The interaction between the methylene moiety of U*6 in the Vindec structure is in some ways reminiscent of the environment of the 2′-O-allyl substituent in the crystal structure of a decamer duplex of the same sequence containing 2′-O-modified uridines at positions 6 and 16.50 The outer methyleneic carbon of the 2′-allyl substituent at U6 displays contacts to four water molecules with distances <3.3 Å. Among these waters, two bridge the outer methylene carbon atoms of the substituents at U6 and U16, whereby the =C(H)2…OW distances are 2.88 Å and 3.28 Å for allyl(U6) and 3.45 Å and 3.51 Å for allyl(U16). The allyl moieties themselves are spaced at > 5 Å across the minor groove and are thus too far apart to allow for effective π-π stacking. Overall, the structural data are in line with the superior RNA affinity of methylene-cLNA modified oligonucleotides and demonstrate that methylene-cLNA analogs offer benefits over other cLNA analogs without the need of introducing hydrophilic chemistry.
Figure 3. X-ray crystallographic studies of DNAs containing carbocyclic LNA analogs.
The central A5pU*6:A15pU*16 base-pair steps viewed into the minor grooves of the (A) Vindec E1, (B) Rmedec E2, and (C) Smedec E3 crystal structures. The strand with residues A5 and U*6 is on the left, carbon, oxygen, nitrogen and phosphorus atoms are colored in gray, red, blue and orange, respectively, and carbon atoms of cLNA sugars are highlighted in yellow. Carbon atoms of a 5′-terminal G from a neighboring duplex and an MPD (2-methyl-2,4-pentanediol) molecule in the Vindec crystal are highlighted in green, water molecules are cyan, and hydrogen bonds are thin solid lines.
Biological evaluation of antisense oligonucleotides containing carbocyclic LNA analogs
Biological evaluation of the methylene-cLNA 6 and the R-Me-cLNA 4 modifications was carried out using two different oligonucleotide sequences (14- and 18-mer) targeting mouse PTEN (Table 5). The 14-mer ASO sequence has a 10-base deoxynucleotide “gap” flanked on the 5’ and the 3’ end with two modified residues (2–10-2 design) and was described by us previously to characterize the in vivo properties of the R- and S-cEt and LNA “gapmer” ASOs.23,51 We first evaluated the ASOs in thermal stability experiments. As seen in the Tm sequences, methylene cLNA ASO C1 showed similar thermal stability as LNA ASO C3 while the R-Me-cLNA ASO C2 showed lower stability. The slightly improved thermal stability of LNA ASO C3 could be attributed to the presence of 5-methyl substituents on the pyrimidine nucleobases, which are known to improve the thermal (approx. +0.5 °C/mod.)52 and nuclease stability of oligonucleotides.53 The thermal stability observations were essentially recapitulated in the 18-mer sequence where the methylene-cLNA D1 ASO was similar to LNA ASO D3 and both were slightly better than R-Me-cLNA D2. However, the influence of the 5-methyl groups was not as prominent for 18-mer sequence as compared to the 14-mer sequence. It is conceivable that the 18-mer ASO is capable of forming a more stable duplex (∼1.7 helical turns) as compared to the 14-mer ASO (∼1.3 helical turns) and is less influenced by the added stabilization provided by 5-methyl groups on the pyrimidine nucleobases.
Table 5.
Thermal stability measurements and in vitro acitivity of gapmer cLNA ASOs targeting mouse PTEN
| ASO | Modification | Sequence (5’ – 3’)a | Tmb (°C) | IC50 (µM)c, | ED50 (mg/kg) |
|---|---|---|---|---|---|
| C1 | Methylene-cLNA (PS) | CUTAGCACTGGCCU | 61.9 | 3.9 | 4.9 |
| C2 | R-Me-cLNA (PS) | CUTAGCACTGGCCU | 57.3 | 7.9 | 8.7 |
| C3 | LNAd (PS) | mCTTAGCACTGGCmCT | 65.4 | 2.8 | 3.5 |
| D1 | Methylene-cLNA (PS) | CUGCTAGCCTCTGGATUU | 60.5 | 4.9 | 4 |
| D2 | R-Me-cLNA (PS) | CUGCTAGCCTCTGGATUU | 58.8 | 8.9 | 17 |
| D3 | LNA (PS) | mCTGCTAGCCTCTGGATTT | 61.6 | 7.7 | 4 |
Bold letters indicate modified nucleosides in ‘wings’ flanking a central DNA ‘gap’ region, all inter-nucleosidic linkages are phosphorothioate, T = Thymine, U = Uracil, A = Adenine, G = Guanine, C = cytosine, mC = 5-methyl cytosine;
Tm values were measured in 10 mM sodium phosphate buffer (pH 7.2) containing 100 mM NaCl and 0.1 mM EDTA; Sequence of RNA complement for ASOs C1, C2 and C3: 5’-r(UCAAGGCCAGUGCUAAGAGU)-3’ and for ASOs D1, D2 and D3: 5’-r(UCAAAUCCAGAGGCUAGCAG)-3’
IC50 values for ASOs C1–3 and D1–3 were determined in mouse bEND cells using electroporation;
LNA ASO C3 with U/C nucleobases in the wings had a Tm of 61.3 °C.
The ASOs were next evaluated in cell culture experiments in brain endothelial cells (bEND) using electroporation to assist ASO delivery. For the 14-mer ASOs, the activity in cell culture essentially parallels the Tm data. LNA ASO C3 (2.8 µM) showed the best activity while the cLNA ASOs C1 (3.9 µM) and C2 (7.9 µM) were slighly less potent. For the 18-mer ASOs, the methylene-cLNA ASO D1 was the most potent (4.9 µM) while the R-Me-cLNA ASO D2 (8.9 µM) and LNA ASO D3 (7.7 µM) were slightly less active.
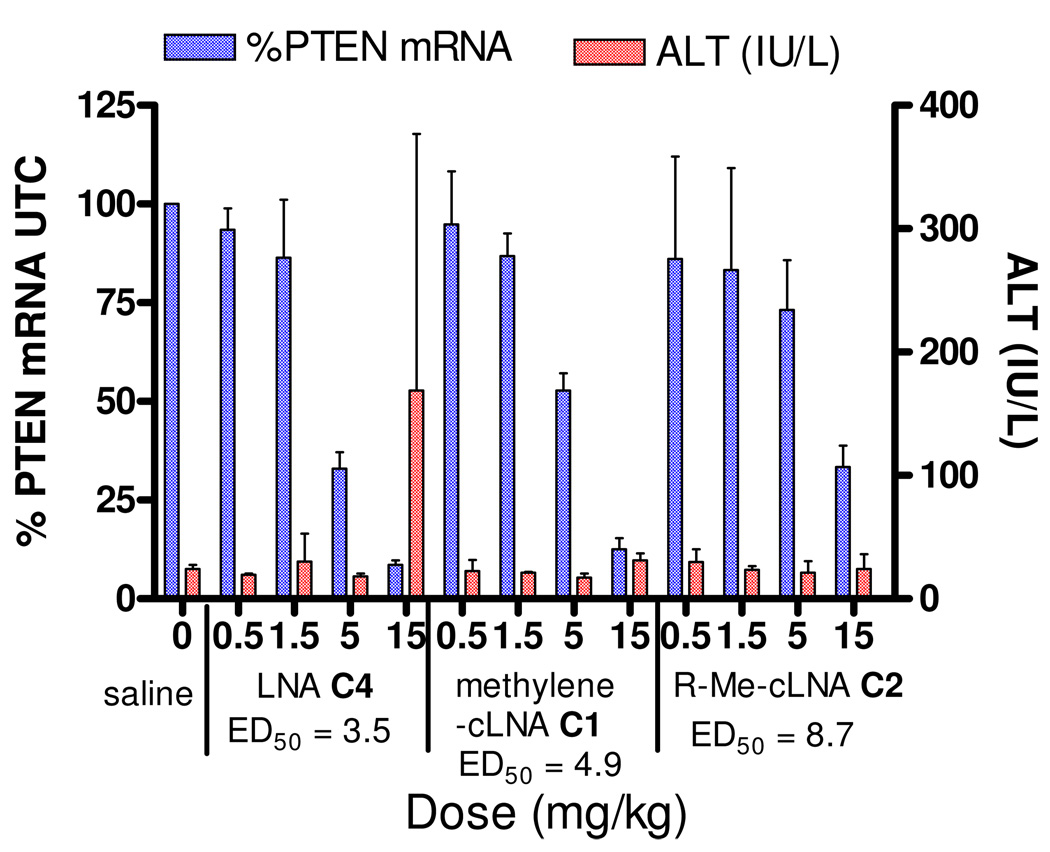
ASOs C1, C2 and C3 were evaluated in animal experiments using a sub-chronic dosing schedule. Mice (n = 4 per dose group) were injected i.p with 0.5, 1.5, 5 and 15 mg/kg twice a week for three weeks, of ASOs C1, C2 and C3 (Figure 4). PTEN mRNA levels in liver were measured using quantitative RT-PCR 72 hours after injection of the last ASO dose and normalized to saline treated animals using protocols described previously.23 Plasma alanine amino transferase (ALT) levels, a well established marker for liver injury, were measured at sacrifice. The LNA ASO C3 served as a positive control for the experiment. Dose dependent down regulation of PTEN mRNA in liver tissue was observed for all the ASOs tested. The LNA ASO C3 was the most potent with an ED50 of 3.5 mg/kg. The methylene-cLNA ASO C1 was slightly less potent (ED50 = 4.9 mg/kg) while the R-Me-cLNA ASO C2 (ED50 = 8.7 mg/kg) was the least potent ASO. In this experiment, slight elevations in plasma ALT levels were observed for the LNA ASO C4 while no plasma ALT elevations were observed for ASOs C1 and C2 at the highest dose (15 mg/kg) tested. 54 For ASOs C1 and C4, maximal knockdown (>90%) of liver PTEN mRNA was observed for mice in the high dose group.
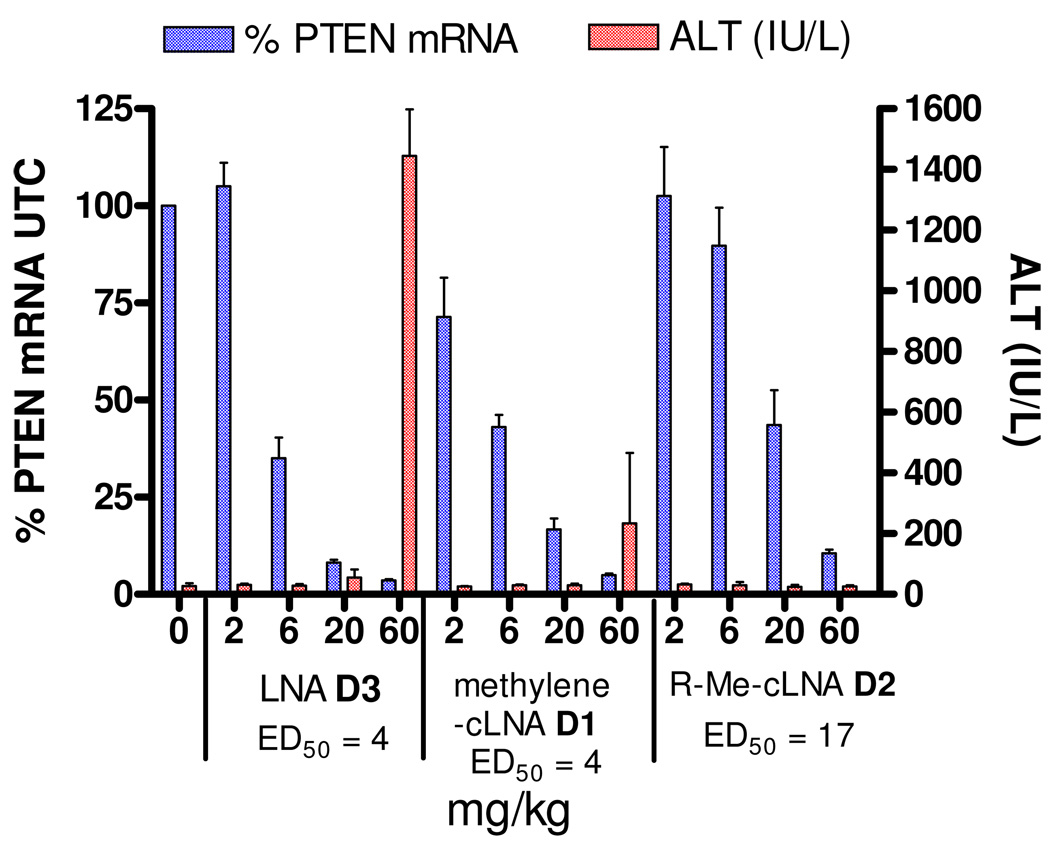
ASOs D1, D2 and D3 were also evaluated in animal experiments. Mice (n = 4 per dose group) were injected i.p. with 2, 6, 20 and 60 mg/kg of ASOs D1, D2 and D3 and the plasma ALT levels and PTEN mRNA levels in liver were measured 72 hours after the last injection (Figure 5). Dose dependent down regulation of PTEN mRNA in liver were observed for all ASOs tested. The methylene-cLNA ASO D1 and LNA ASO D4 exhibited very similar potencies (ED50 = 4 mg/kg) while the R-Me LNA ASO was significantly less potent (ED50 = 17 mg/kg). The big shift in the potency curve for ASO D2 was especially surprising since all these ASOs exhibited very similar IC50 values for down regulating PTEN mRNA in cell culture experiments (Table 5). The Methylene-cLNA ASO D1 and the R-Me-cLNA ASO D2 showed modest to no elevations in ALT levels at the highest does evaluated while the LNA ASO was toxic at the high dose.
Conclusion
We show for the first time that it is possible to obtain LNA like binding affinity using cLNA analogs by replacing the 2’-oxygen atom with an exocyclic methylene group. Methylene-cLNA 6 modified ASOs showed improved hybridization for complementary nucleic acids relative to the close structurally related methyl substituted cLNA analogs 4 and 5. We also show that in the case of cLNA analogs 4 and 5, the relative orientation of the methyl group (inside the minor groove for R-Me-cLNA 4 and edge of the minor groove for S-Me-cLNA 5) affects the thermal stability of duplexes containing these modifications. The cLNA modifications appear to improve the recognition of cognate RNA over DNA, but the magnitude of stabilization is small and somewhat sequence dependent. Analysis of crystal structures of the cLNA analogs incorporated into a self complementary DNA decamer duplex revealed that the methylene group participates in a tight interaction with a 2’-deoxyribose residue of the 5’-terminal G of a neighbouring duplex resulting in the formation of a CH…O type hydrogen bond. This interaction shows that the methylene-cLNA retains a negative polarization at the edge of the minor groove in the absence of a hydrophilic 2′-substituent and provides a rationale for the enhanced thermal stability of methylene-cLNA modified oligonucleotides. Conversely, the sugar residues in the R- and S-Me-cLNA analogs are poorly hydrated and unable to extend the water structure around the edges of nucleobases toward the phosphate backbone.
Evaluation of gapmer cLNA ASOs in cell culture indicated that the methylene-cLNA ASOs were generally as active as LNA while the R-Me-cLNA ASOs were slightly less potent. However, it appears that the potency differences are magnified when the ASOs were tested in animal experiments. The in vivo data for the 18-mer ASOs D1, D2 and D3 was especially remarkable since all of them showed very similar thermal stability and activity in cell culture but the R-Me-cLNA ASO D2 was 4-fold less active in animals. This suggests that subtle differences in structure between closely related chemically modified ASOs can have a significant impact of on their in vivo properties. In this regard, our work helps to further understand the intricate nature of structure activity relationships of chemically modified ASOs and lays the groundwork for identifying optimal designs of cLNA analogs for future in vivo applications. Moreover, this work also highlights the interchangeable role of oxygen and unsaturated moeities in nucleic acid structure and emphasizes greater use of this bio-isostere to improve the properties of nucleic acids for therapeutic and diagnostic applications.
Supplementary Material
Figure 4. PTEN mRNA down regulation in liver and plasma ALT levels for mice treated for three weeks with methylene-cLNA ASO C1, R-Me-cLNA ASO C2 and LNA ASO C3.
Figure 5. PTEN mRNA down regulation in liver and plasma ALT levels for mice treated with single dose of Methylene-cLNA ASO D1, R-Me-cLNA D2 and LNA ASO D3.
Scheme 2.
Acknowledgments
This work was supported in part by NIH grant R01 GM55237 (to M.E.). We would like to thank Merry Daniel for help with the crystallization experiments and Dr. Karsten Schmidt for HRMS measurements. Vanderbilt University is a member institution of LS-CAT at the Advanced Photon Source (Argonne, IL). Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38.
Footnotes
Supporting Information Available: General experimental procedures for synthesis of 10–28; 1H and 13C NMR spectra and analytical data for all new compounds; 31P NMR spectra for all phosphoramidites; analytical data for oligonucleotides and dose response curves for cell culture experiments are provided. Atomic coordinates and structure factor data for the three crystal structures have been deposited in the Protein Data Bank (http://www.rcsb.org; awaiting entry code assignments). The material is available free of charge at http://pubs.acs.org.
References
- 1.Bennett CF, Swayze EE. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 2.Swayze EE, Bhat BIn. In: Antisense Drug Technology: Principles, Strategies, and Applications. 2nd ed. Crooke ST, editor. Boca Raton: CRC Press; 2008. pp. 143–182. [Google Scholar]
- 3.Eckstein F. Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 4.Geary RS, Yu RZ, Levin AA. Curr. Opin. Investig. Drugs. 2001;2:562–573. [PubMed] [Google Scholar]
- 5.Martin P. Helv. Chim. Acta. 1995;78:486–504. [Google Scholar]
- 6.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, Egli M. Nat. Struct. Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 7.Crooke RM, Graham MJ, Lemonidis KM, Whipple CP, Koo S, Perera RJ. J. Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, Tribble DL, Flaim JD, Crooke ST. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 9.Prakash TP, Bhat B. Curr. Top. Med. Chem. 2007;7:641–649. doi: 10.2174/156802607780487713. [DOI] [PubMed] [Google Scholar]
- 10.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 11.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 12.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obika S, Nanbu D, Hari Y, Andoh J-I, Morio K-I, Doi T, Imanishi T. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 14.Wengel J. Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 15.Sapra P, Wang M, Bandaru R, Zhao H, Greenberger LM, Horak ID. Nucleosides, Nucleotides and Nucleic Acids. 2010;29:97–112. doi: 10.1080/15257771003597733. [DOI] [PubMed] [Google Scholar]
- 16.Fluiter K, Frieden M, Vreijling J, Rosenbohm C, De Wissel MB, Christensen SM, Koch T, Orum H, Baas F. Chembiochem. 2005;6:1104–1109. doi: 10.1002/cbic.200400419. [DOI] [PubMed] [Google Scholar]
- 17.Graziewicz MA, Tarrant TK, Buckley B, Roberts J, Fulton L, Hansen H, Orum H, Kole R, Sazani P. Mol. Ther. 2008;16:1316–1322. doi: 10.1038/mt.2008.85. [DOI] [PubMed] [Google Scholar]
- 18.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP, Bennett CF. Nucl. Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou C, Chattopadhyaya J. Curr. Opin. Drug Discov. Devel. 2009;12:876–898. [PubMed] [Google Scholar]
- 21.Swayze EE, Seth PP. PCT Int. Appl. 2007 WO, 2007090071. [Google Scholar]
- 22.Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, Kinberger GA, Prakash TP, Migawa MT, Bhat B, Swayze EE. J. Org. Chem. 2010;75:1569–1581. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- 23.Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, Witchell D, Swayze EE. J. Med. Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 24.Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 25.Xi Z, Rong J, Chattopadhyaya J. Tetrahedron. 1994;50:5255–5272. [Google Scholar]
- 26.Xi Z, Agback P, Plavec J, Sandström A, Chattopadhyaya J. Tetrahedron. 1992;48:349–370. [Google Scholar]
- 27.Sukeda M, Shuto S, Sugimoto I, Ichikawa S, Matsuda A. J. Org. Chem. 2000;65:8988–8996. doi: 10.1021/jo000967l. [DOI] [PubMed] [Google Scholar]
- 28.Sukeda M, Ichikawa S, Matsuda A, Shuto S. J. Org. Chem. 2003;68:3465–3475. doi: 10.1021/jo0206667. [DOI] [PubMed] [Google Scholar]
- 29.Bisacchi GS, Chao ST, Bachard C, Daris JP, Innaimo S, Jacobs GA, Kocy O, Lapointe P, Martel A, Merchant Z, Slusarchyk WA, Sundeen JE, Young MG, Colonno R, Zahler R. Bioorg. Med. Chem. Lett. 1997;7:127–132. [Google Scholar]
- 30.Wang J, Verbeure B, Luyten I, Lescrinier E, Froeyen M, Hendrix C, Rosemeyer H, Seela F, Van Aerschot A, Herdewijn P. J. Am. Chem. Soc. 2000;122:8595–8602. [Google Scholar]
- 31.Srivastava P, Barman J, Pathmasiri W, Plashkevych O, Wenska M, Chattopadhyaya J. J. Am. Chem. Soc. 2007;129:8362–8379. doi: 10.1021/ja071106y. [DOI] [PubMed] [Google Scholar]
- 32.Zhou C, Plashkevych O, Chattopadhyaya J. J. Org. Chem. 2009;74:3248–3265. doi: 10.1021/jo900391n. [DOI] [PubMed] [Google Scholar]
- 33.Zhou C, Liu Y, Andaloussi M, Badgujar N, Plashkevych O, Chattopadhyaya J. J. Org. Chem. 2009;74:118–134. doi: 10.1021/jo8016742. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Liu Y, Dupouy C, Chattopadhyaya J. J. Org. Chem. 2009;74:6534–6554. doi: 10.1021/jo901009w. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Hansen MH, Albæk N, Steffansen SI, Petersen M, Nielsen P. J. Org. Chem. 2009;74:6756–6769. doi: 10.1021/jo9013657. [DOI] [PubMed] [Google Scholar]
- 36.Albæk N, Petersen M, Nielsen P. J. Org. Chem. 2006;71:7731–7740. doi: 10.1021/jo061225g. [DOI] [PubMed] [Google Scholar]
- 37.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, Babu BR, Hojland T, Abramov M, Van Aerschot A, Odadzic D, Smicius R, Haas J, Andree C, Barman J, Wenska M, Srivastava P, Zhou C, Honcharenko D, Hess S, Muller E, Bobkov GV, Mikhailov SN, Fava E, Meyer TF, Chattopadhyaya J, Zerial M, Engels JW, Herdewijn P, Wengel J, Kjems J. Nucl. Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjaer N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, Herdewijn P, Wengel J, Kjems J. Nucl. Acids Res. 2010 doi: 10.1093/nar/gkq341. doi:10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagnon KT, Hu J, Swayze EE, Bennett CF, Potier P, Montaillier C, Roesch E, Lemaitre M, Randolph J, Deleavey G, Damha M, Corey D. CHDI's 5th Annual Huntington's Disease Therapeutics Conference. 2010 [Google Scholar]
- 41.Mancuso AJ, Huang S-L, Swern D. J. Org. Chem. 1978;43:2480–2482. [Google Scholar]
- 42.Peng CG, Damha MJ. Nucl. Acids Res. 2005;33:7019–7028. doi: 10.1093/nar/gki1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corey EJ, Fuchs PL. Tetrahedron Lett. 1972;13:3769–3772. [Google Scholar]
- 44.Holy NL. Chem. Rev. 1974;74:243–277. [Google Scholar]
- 45.Xia J, Abbas SA, Locke RD, Piskorz CF, Alderfer JL, Matta KL. Tetrahedron Lett. 2000;41:169–173. [Google Scholar]
- 46.Vorbrüggen H, Bennua B. Chem. Ber. 1981;114:1279–1286. [Google Scholar]
- 47.AbdurRahman SM, Seki S, Obika S, Yoshikawa H, Miyashita K, Imanishi T. J. Am. Chem. Soc. 2008;130:4886–4896. doi: 10.1021/ja710342q. [DOI] [PubMed] [Google Scholar]
- 48.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 49.Egli M, Usman N, Rich A. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 50.Egli M, Minasov G, Tereshko V, Pallan PS, Teplova M, Inamati GB, Lesnik EA, Owens SR, Ross BS, Prakash TP, Manoharan M. Biochemistry. 2005;44:9045–9057. doi: 10.1021/bi050574m. [DOI] [PubMed] [Google Scholar]
- 51.Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ, Black C, Seth PP, Swayze EE, Bhat B. J. Med. Chem. 2010;53:1636–1650. doi: 10.1021/jm9013295. [DOI] [PubMed] [Google Scholar]
- 52.Freier SM, Altmann KH. Nucl. Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terrazas M, Kool ET. Nucl. Acids Res. 2009;37:346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elevations in ALT levels using LNA ASO C3 have been reproduced in several animal experiments using the described dosing regimen.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.