SUMMARY
Until recently, the mechanism of mRNA decay in bacteria was thought to be different from that of eukaryotes. This paradigm changed with the discovery that RppH (ORF176/NudH/YgdP), an Escherichia coli enzyme that belongs to the Nudix superfamily, is an RNA resolution pyrophosphohydrolase that initiates mRNA decay by cleaving pyrophosphate from the 5′-triphosphate. Here we report the 1.9 Å structure of the Nudix hydrolase BdRppH from Bdellovibrio bacteriovorus, a bacterium that feeds on other Gram-negative bacteria. Based on the structure of the enzyme alone and in complex with GTP-Mg2+, we propose a mode of RNA binding similar to that of the nuclear decapping enzyme from Xenopus laevis, X29. In additional experiments, we show that BdRppH can indeed function in vitro and in vivo as an RNA pyrophosphohydrolase. These findings set the basis for the identification of possible decapping enzymes in other bacteria.
INTRODUCTION
The process of mRNA decay in eukaryotes is well characterized. It typically begins with the loss of the 3′-poly(A) tail and removal of the m7Gppp cap by DCP2 (Wang et al., 2002), followed by exonucleolytic degradation of the mRNA body from 5′ to 3′ by Xrn1 or from 3′ to 5′ by the exosome (Garneau et al., 2007). DCP2 belongs to the Nudix superfamily, enzymes that hydrolyze the diphosphate bond of substrates of the form nucleoside diphosphate bound to a moiety x.
Until recently, mRNA degradation was believed to occur by a completely different process in bacteria, in which newly synthesized transcripts bear a 5′-triphosphate rather than a 5′ cap. In Escherichia coli, that process was thought to begin with endonucleolytic cleavage downstream of the 5′ end, most often by RNase E. Such cleavage would generate two fragments, both of which would be rapidly cleared by additional RNase Ecleavage and 3′-to-5′ exonuclease digestion (Schoenberg, 2007).
This paradigm had to be reconsidered when it was discovered that the status of the 5′ end is critical to mRNA decay in bacteria (Bouvet and Belasco, 1992; Celesnik et al., 2007; Emory et al., 1992; Mackie, 1998; Xu and Cohen, 1995). That work showed that the rate constant for RNase E cleavage is significantly higher for RNA substrates bearing a 5′-monophosphate rather than a 5′-triphosphate or 5′-hydroxyl (Celesnik et al., 2007; Mackie, 1998; Tock et al., 2000). The reason for this unexpected specificity became evident with the determination of the crystal structure of E. coli RNase E, whose catalytic domain was found to contain a binding site specific for monophosphorylated RNA 5′ ends. A network of hydrogen bonds between Thr170 and Arg169 and the terminal 5′-phosphate provides the selectivity toward monophosphorylated RNAs (Callaghan et al., 2005). These observations culminated in the discovery that RppH (ORF176/NudH/YgdP), an E. coli Nudix enzyme previously characterized as a diadenosine tetraphosphatase (Bessman et al., 2001), is an RNA pyrophosphohydrolase that initiates mRNA decay in that species by converting the triphosphorylated 5′ ends of primary transcripts to monophosphates (Celesnik et al., 2007; Deana et al., 2008).
Bdellovibrio bacteriovorus is a predatory bacterium that infects and feeds on other Gram-negative bacteria, including some pathogens (Stolp and Petzold, 1962; Stolp and Starr, 1963). B. bacteriovorus has a biphasic lifecycle that alternates between a free-living predatory or “hunt” phase and an intraperiplasmic growth phase (Strauch et al., 2007). In predation, in particular during the period of the intraperiplasmic growth phase, the bacterium is highly dependent on a vast arsenal of hydrolytic enzymes: 204 putative hydrolytic enzymes in a 3.7 Mbp genome. This is the highest density of such enzymes in a bacterial genome reported so far, with the exception of the Buchnera aphidicola genome (Rendulic et al., 2004; van Ham et al., 2003). The highly organized systems employed to secrete these lytic enzymes against only Gram-negative bacteria make B. bacteriovorus an attractive agent as a living antibiotic and a reservoir of potential antimicrobials (Rendulic et al., 2004; Sockett and Lambert, 2004). BdRppH, the product of gene bd0714 (GenBank accession number CAE78673) in B. bacteriovorus, is part of this vast array of hydrolytic enzymes and represents one of six putative Nudix enzymes, which can be recognized by the presence of a sequence encoding the signature motif GX5EX7REUXEEXGU (where U is either Ile, Leu, or Val) (Steyert et al., 2008). In our previous work, we reported that the product of the bd0714 gene is a dGTPase that complements the MutT phenotype in E. coli cells (Steyert et al., 2008). Here we report the structure of BdRppH at 1.9 Å resolution and the characterization of its enzymatic activity, showing that it can function in vitro and in vivo as an RNA pyrophosphohydrolase.
RESULTS
Overall Structure
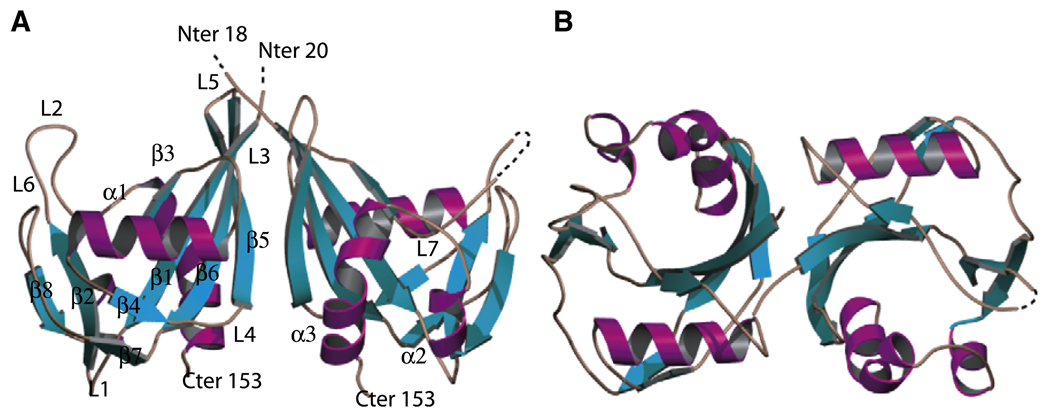
The structure of BdRppH, determined by multiple isomorphous replacement (MIR) to 1.9 Å resolution with a final R/Rfree of 0.22/0.25 (Table 1), contains a dimer in the asymmetric unit of the crystal. The model has excellent geometry, with 91.7% of the residues located in the preferred regions of the Ramachandran plot, and the remaining 8.3% in the allowed region, according to the program PROCHECK (Laskowski et al., 1993; Morris et al., 1992). Each of the two monomers (residues 18–150) is composed of a Nudix domain: a four-stranded mixed-β sheet (β3, β1, β6, β5) and a two-stranded antiparallel β sheet flanked by front (α1) and back (α2, α3) α helices (Figure 1). Residues 1–17 are not observed in either of the two molecules in the asymmetric unit. Furthermore, residues Lys18 and Gly19 are disordered in monomer A, and residues Asn43, Asn44, Ser45, Leu46, and Ala47 are disordered in monomer B (Figure 1).
Table 1.
Data Collection and Refinement Statistics
| Native | Hg Derivative | U Derivative | Ho Derivative | GTP Complex | |
|---|---|---|---|---|---|
| Data Collection | |||||
| Space group | P3221 | P3221 | P3221 | P3221 | P212121 |
| Cell dimensions | |||||
| a, b, c (Å) |
a = b = 70.5, c = 100.5 |
a = b = 7 0.2, c = 101.1 |
a = b = 70.1, c = 102.2 |
a = b = 70.2, c = 99.9 |
a = 68.7, b = 68.7, c = 93.0 |
| α, β, γ (°) | α = β = 90, γ = 120 | α = β = 90, γ = 120 | α = β = 90, γ = 120 | α = β = 90, γ = 120 | α = β = γ = 90 |
| Measured reflections | 232,027 | 141,497 | 72,993 | 107,414 | 42,839 |
| Resolution (Å)a | 50–1.9 (1.97–1.90) | 50–2.18 (2.26–2.18) | 50–2.70 (2.80–2.70) | 50–2.00 (2.09–2.00) | 50–2.80 (2.90–2.80) |
| Rsym or Rmerge | 4.3 (39.2) | 4.8 (47.2) | 8.3 (71.6) | 3.7 (77.5) | 5.4 (71.4) |
| I/σI | 65.7 (6.1) | 59.6 (3) | 42.3 (2.3) | 45.8 (2.3) | 31.7 (2.9) |
| Completeness (%) | 99.8 (100) | 95 (86) | 97.7 (90.2) | 99.9 (100) | 98.6 (99.8) |
| Redundancy | 10.0 (9.0) | 8.5 (3.2) | 8.9 (5.6) | 5.4 (5.2) | 3.8 (3.8) |
| Refinement | |||||
| Resolution (Å) | 1.9 | 2 | 2.8 | ||
| Number of reflections | 22,041 | 18,759 | 10,446 | ||
| Rwork/Rfree b | 0.22/0.25 | 0.21/0.25 | 0.23/0.30 | ||
| Number of atoms | 2218 | 2260 | 2253 | ||
| Protein | 2105 | 2127 | 2165 | ||
| Ligand/ion | 15 | 70 | |||
| water | 113 | 118 | 18 | ||
| Metals (per monomer) | 3 | 2 | 2 | 3 | |
| B factors | 40.29 | 45.5 | 69.4 | ||
| Protein | 39.98 | 45.2 | 69.1 | ||
| Ligand/ion | 62.6 | 76.5 | |||
| Water | 46.07 | 49.2 | 68.8 | ||
| Rms deviations | |||||
| Bond lengths (Å) | 0.01 | 0.011 | 0.011 | ||
| Bond angles (°) | 1.29 | 1.36 | 1.42 |
Values in parentheses are for the highest resolution shell.
Rfree is calculated from 5% of the reflections chosen randomly.
Figure 1. Structure of BdRppH.
(A) A “side” representation of BdRppH with β strands shown in cyan, α helices in magenta, and loops in brown. (B) Vertically rotated view (90°) of BdRppH from “above” looking down into the active site.
In BdRppH, the conserved Nudix signature sequence extends from residue Gly54 to Ile77 and folds into the characteristic β-strand-loop-α-helix-loop motif (Gabelli et al., 2001). The hydrogen bonds and salt bridges typically found among residues of the signature sequence are also conserved in this enzyme. For example, the guanidinium of Arg69 is at salt-bridge distances to the carboxylate groups of Glu70 (2.84 Å) and Glu61 (2.55 and 2.79Å). Furthermore, the N terminus of the α helix makes several stabilizing hydrogen bonds such as those from the carboxylate of Glu61 to the main-chain amide of Glu58, from the guanidinium of Arg69 to the main-chain carbonyl of Lys56, and from the mainchain carbonyl of Glu58 to the main-chain amide of Glu61. These interactions help to hold helix α1 of the Nudix fold and the loop preceding it (L3) to β strand b6 of the mixed-b sheet. At the C-terminal end, helix α1 is held via hydrogen bonds between the main-chain carbonyl of Leu75 and the amide of Arg112, located in loop L6 between β7 and β8, and between the carboxylate of Glu74 and the amide of Gly54 (H bonds through water). Finally, the hydrophobic side chains of Leu67, 71, and 75 form a hydrophobic patch on the side of helix α1 that contacts the mixed-sheet β4.
Dimerization
The two monomers in the asymmetric unit form a dimer through a noncrystallographic two-fold axis, in agreement with gel-filtration experiments (data not shown). The monomers are structurally highly similar, with a root-mean-square deviation (rmsd) of 0.7Å over 127 α-carbon atoms. The surface buried upon dimerization of two BdRppH monomers is 1700 Å2, and the surfaces from the two monomers have a shape complementarity coefficient of 0.63 as calculated by the program Sc from the CCP4 software package (CCP4, 1994). This interface involves a combination of hydrophobic interactions (residues 21, 23, 57, 62, 63, 84, 87–89, 98, 100) and hydrogen bonds (residues 20, 59, 61, 62, 64, 83, 85, 89, 91, 102, 142) among 20 residues from each monomer (Figure 2). The monomers are arranged in a head-to-head fashion, similar to that observed in the dimer of GDP-mannose hydrolase from E. coli (GDPMH; Gabelli et al., 2004), in which the rmsd is 2.2Å over 136 α-carbons out of 270, forming the dimer via the three long strands β1, β5, and β6 of the mixed-β sheets. This is especially interesting because the two enzymes have very different catalytic activities: whereas BdRppH hydro-lyzes a diphosphate bond, GDPMH cleaves at a carbon instead of a phosphorus (Gabelli et al., 2004).
Figure 2. The Dimerization Interface.
(A) A frontal view of the BdRppH dimerization interface. Monomer A is colored according to its electrostatic surface potential, and monomer B as a ribbon diagram. (B) View of BdRppH from (A) rotated 90° without the ribbon diagram of monomer B. Instead, key hydrophobic residues of monomer B in the interface are depicted in magenta and important hydrophilic residues are in cyan.
Metal Coordination
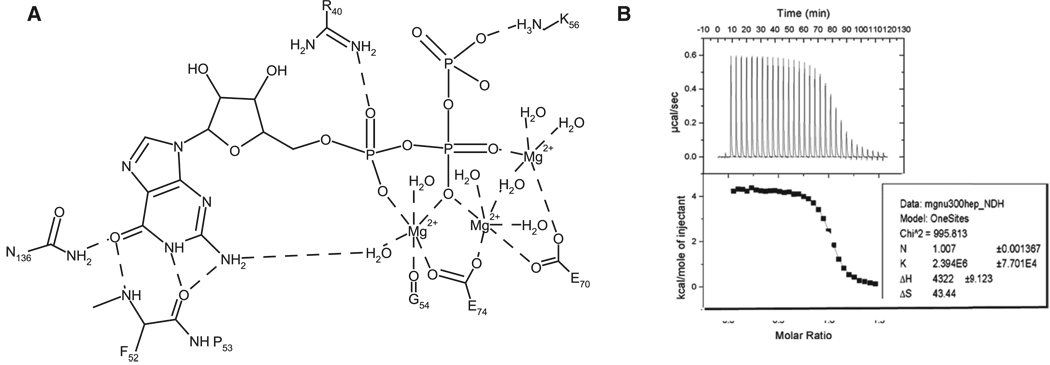
All characterized Nudix enzymes require divalent cations, usually Mg2+ or Mn2+, for catalysis (Frick et al., 1994; Mildvan et al., 2005). BdRppH strongly prefers Mg2+ as the divalent cation, showing little or no activity with Mn2+ (data not shown). BdRppH binds one Mg2+ per monomer with a Kd of 0.36 µM, as measured by isothermal titration calorimetry (ITC) (Figure 3). At 37°C, binding is characterized by unfavorable enthalpy (ΔH = 4.32 kcal/mol), but favorable entropy (−TΔS = −9.12 kcal/mol), probably arising from release of bound water molecules from both Mg2+ and protein.
Figure 3. Mg2+ Coordination in the Binding Site and Thermodynamic Binding Data.
(A) Schematic representation of magnesium ion coordination between BdRppH and substrate. (B) The upper panel depicts the raw data collected from the ITC experiments measuring Mg2+ binding to the enzyme; the lower panel shows the plot of the individual integrations of each peak in the raw data and curve fitting to those data, with an inset displaying the results of the fit.
BdRppH-GTP-Mg2+ Complex
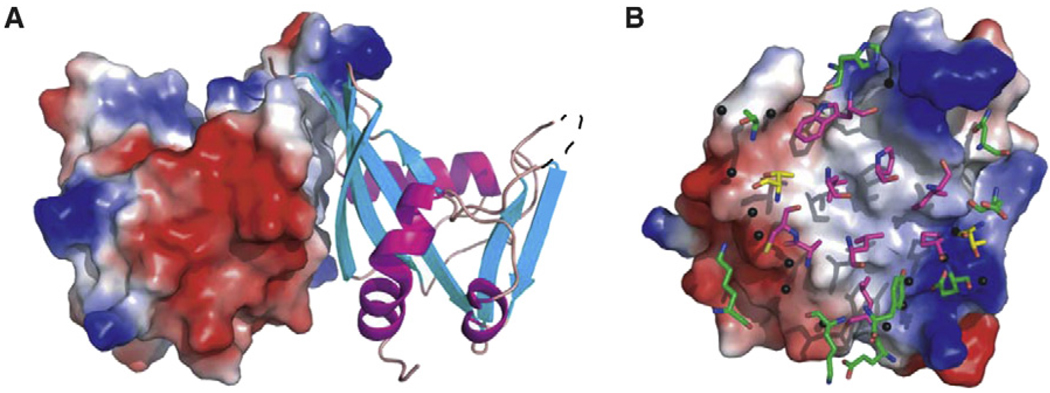
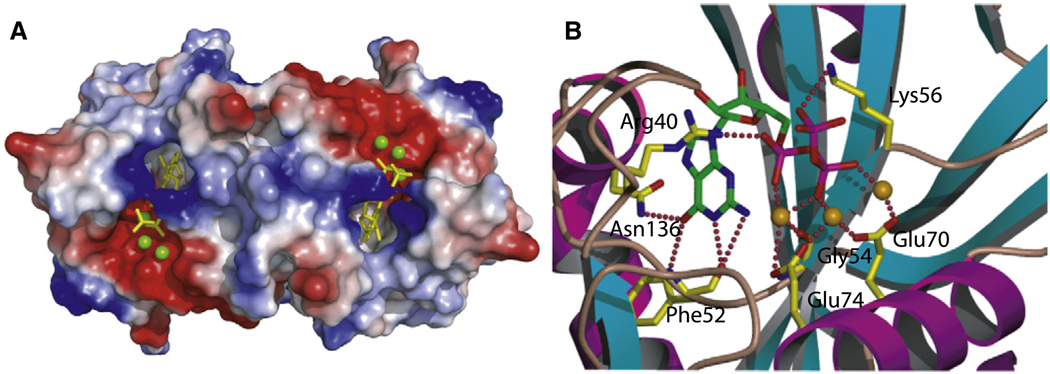
To gain additional insight into the recognition of substrate by BdRppH, the structure of the enzyme in complex with GTP (also referred to as Gppp) and Mg2+ was determined by cocrystallizing the enzyme in the presence of the substrate at pH 7, and then soaking the crystals in the same mother liquor supplemented with 10 mM Mg2+. At pH 7 the enzyme shows little or no activity, thus preventing hydrolysis of GTP, but allowing for residues in the active site to be properly charged (see below). The structure of the protein-substrate-Mg2+ complex showed excellent electron density for one molecule of GTP and three Mg2+ ions bound to each molecule in the asymmetric unit. The two copies of the substrate are bound in the same conformation, with the purine rings in the syn conformation (Figure 4A). The GTP molecules and Mg2+ ions were refined with full occupancy and average temperature factors (76.5) similar to those of the atoms of the protein (69.1).
Figure 4. BdRppH-GTP Binding.
(A) A view from “above” looking down onto the two active sites of BdRppH, with GTP substrate colored in yellow, magnesium ions in green, and the surface of the protein colored according to its electrostatic potential.
(B) Ribbon diagram of the active site of BdRppH showing GTP and key coordinating residues; residues are depicted in yellow, magnesium ions in magenta, and GTP in green.
The purine ring binds in a cleft sandwiched between β sheets β1 and β3 and α helix α3, distal to the Nudix signature sequence residues in helix α1 (Figure 4A). This arrangement and the location of the purine ring is similar to that observed in the nuclear decapping enzyme X29 of Xenopus laevis complexed to m7GpppA (Protein Data Bank [PDB] ID code 2a8t) (Scarsdale et al., 2006), where the purine ring of the adenosine is also bound in the syn conformation in a similar cleft. In the BdRppH-GTP complex, the ring is stabilized by H bonds from the O6 of the purine ring to the nitrogen of Asn136 (2.83 Å) and to the nitrogen of the peptide backbone of Phe52, and between the carbonyl of the peptide backbone of Phe52 and the N2 (3.02 Å) and N1 (2.52 Å) of the purine ring (Figure 4B). The coordination by the amide of Asn136 is equivalent to that provided by Gln184 X29 of X. laevis. Additional H bonds are present between one of the NH groups of the guanidinium of Arg40 to the oxygen O2 of the α-phosphate (2.41 Å), and the amino group of Lys56 to the oxygen O3 of the γ-phosphate (3.21 Å).
Also present are three octahedrally coordinated magnesium ions, liganded by several water molecules and bridging the phosphates of the GTP to the protein. The coordination and arrangement of the metal ions and substrate are similar to those of the structure of the complex of the E. coli ADPRase with magnesium and a nonhydrolyzable substrate analog (PDB ID code 1khz) (Gabelli et al., 2002). Coordination and arrangement of the metal ions are also similar to those of X29 of X. laevis complexed to m7GpppA (PDB ID code 2a8t) (Scarsdale et al., 2006), with the exception that the fourth manganese ion coordinating the m7G cap is missing in the BdRppH structure. This is to be expected, as the mRNA substrate of BdRppH has no m7G cap, but only a triphosphate. The magnesium furthest away from the helix that contains the Nudix signature sequence (α1), is coordinated by the carbonyl of Gly54 (2.17Å), two water molecules (1.89 Å, 1.84 Å), the carboxylate of Glu74 (2.17 Å), an oxygen from the α-phosphate (O1A = 2.17 Å), and an oxygen (O1B = 2.17Å) of the β-phosphate (Figure 3). The central magnesium is coordinated with a water molecule (1.96 Å), the carboxylate of Glu70 (2.18 Å), two water molecules (1.73 Å, 1.94 Å), the carboxylate of Glu74 (2.16 Å), and an oxygen (O2B = 2.17 Å) from the β-phosphate. The third magnesium, closest to helix α1, is coordinated by the carboxylate of Glu70, a molecule of water (2.12Å), and an oxygen (O1B = 2.17Å) from the β-phosphate and these water molecules.
Lanthanides commonly can substitute for Mg2+ in Mg2+ binding sites (Dudev et al., 2005; Gabelli et al., 2001; Glusker, 1991; Kang et al., 2003a, 2003b). Crystals soaked in holmium chloride, one of the heavy-atom derivatives used for the structure determination, revealed holmium bound in a position close to that of the central Mg2+ of the structure of BdRppHGTP-Mg2+. The holmium is coordinated by the carboxylates of glutamate residues Glu70 and Glu73 of the Nudix signature sequence and by three water molecules. This result in combination with the ITC indicates that the middle magnesium is prebound to the active site, and that the other two magnesium ions are recruited with the substrate.
Catalytic Base
Based on the existing body of work, at least four mechanisms of hydrolysis have been postulated for Nudix enzymes. In those, the catalytic base is either the second glutamate of the signature sequence, or a glutamate or histidine in the loop equivalent to L6 of BdRppH (Mildvan et al., 2005). In GDPMH (Gabelli et al., 2004) and ADPRase (Gabelli et al., 2001), the catalytic base that deprotonates a water molecule for nucleophilic attack (GDPMH His124, ADPRase Glu162) and one of the ligands to the catalytic metal (GDPMH Gln123, ADPRase Glu164) are contained in the loop equivalent to L6. In E. coli MutT, a monomer, and in Ap4A pyrophosphatase (Conyers et al., 2000), the second glutamic acid of the signature sequence acts not only as one of the ligands of the catalytic metal but also as the catalytic base (MutT Glu53) (Mildvan et al., 2005). BdRppH, as a canonical Nudix enzyme, also has a glutamic acid (Glu70) in the position equivalent to this MutT residue, and can complement the MutT phenotype in E. coli (Steyert et al., 2008). However, in the BdRppH complex structure, Glu70 is directly coordinating two magnesium ions, an unlikely arrangement for a catalytic base. BdRppH contains two histidines (His115 and His116) in loop L6 (Figure 1 and Figure 5) that occupy positions structurally equivalent to the catalytic bases of GDPMH and ADPRase. However, the positions of these histidines in the substrate complex structure are such that they could not act to activate a molecule of water for hydrolysis.
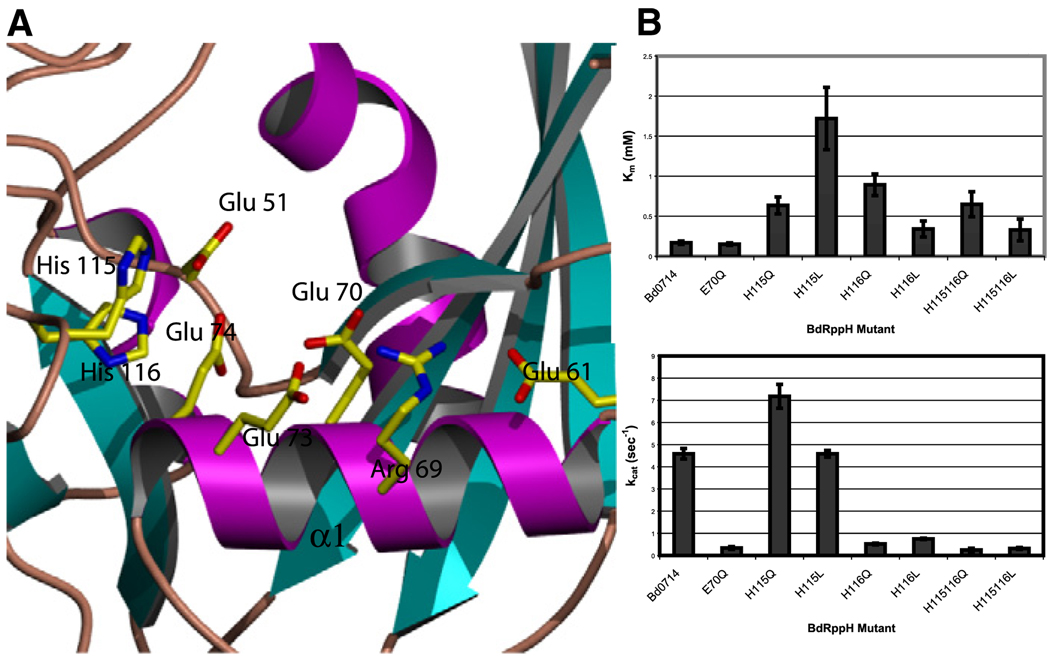
Figure 5. BdRppH Active Site and Kinetic Effects of Mutation of the Possible Catalytic Bases.
(A) View of the active site showing the residues in the Nudix signature sequence. Yellow is used for carbon, blue for nitrogen, and red for oxygen; β strands are shown in cyan, α helices in magenta, and loops in brown.
(B) Values ofKm and kcat for the wild-type enzyme and the mutations of residues Glu70, His116, and His115. All experiments were done in triplicate.
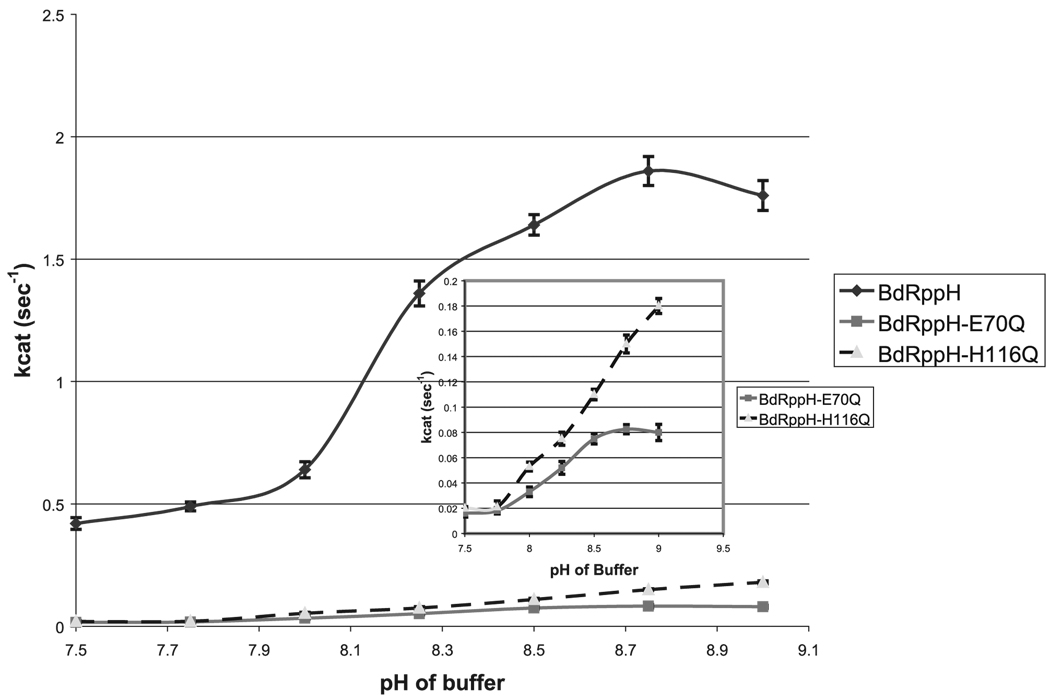
As the substrate-protein complex structure did not suggest a directly identifiable mechanism or identify a clear residue as the catalytic base, potential catalytic bases such as Glu70, His115, and His116 were each mutated to glutamine. Kinetic analysis of the E7OQ-to-Gln mutant, using dGTP as substrate, revealed only a 13.5-fold drop in kcat, with no effect on Km (Figure 5). Also, this mutation, although resulting in a lower kcat, produced no shift or alteration in the pH profile. These results give a strong indication that Glu70 is probably not the catalytic base, and strengthens the argument that it is only one of the metal ligands. Mutation of His116 to Gln caused an 8.8-fold drop in kcat and 5.3-fold increase in Km. However, this mutation resulted in a significant change in the pH profile of the enzyme, indicating that His116 may be indirectly involved in catalysis (Figure 6). Mutation of the same residue to Leu caused a 6-fold drop in kcat, and only a 2-fold increase in Km (Figure 5). Mutating His115 to Gln resulted in a 1.5-fold increase in kcat and 3.7-fold increase of Km, whereas mutation to Leu increased Km by 10-fold with no change in kcat (Figure 5). These results suggest that mutation of His115 affects substrate binding, but has little or no effect on the catalytic steps of the enzyme. Double mutation of His115 and 116 to Gln or Leu resulted in 18-fold and 14-fold drops in kcat, respectively, and 3.8- and 2-fold increases in Km. The weak effect of these mutations may be a result of some flexibility in loop L6 that allows alternative residues or a solvent hydroxyl to function as the catalytic base.
Figure 6. pH Profile of Wild-Type BdRppH and Mutants of Potential Catalytic Bases.
Values of kcat versus pH of BdRppH, BdRppH-E70Q, and H116Q over a wide pH range are shown. y axis units are kcat values in s−1 and the×axis is in units of pH. All experiments were done in triplicate.
RNA Pyrophosphohydrolase Activity In Vitro
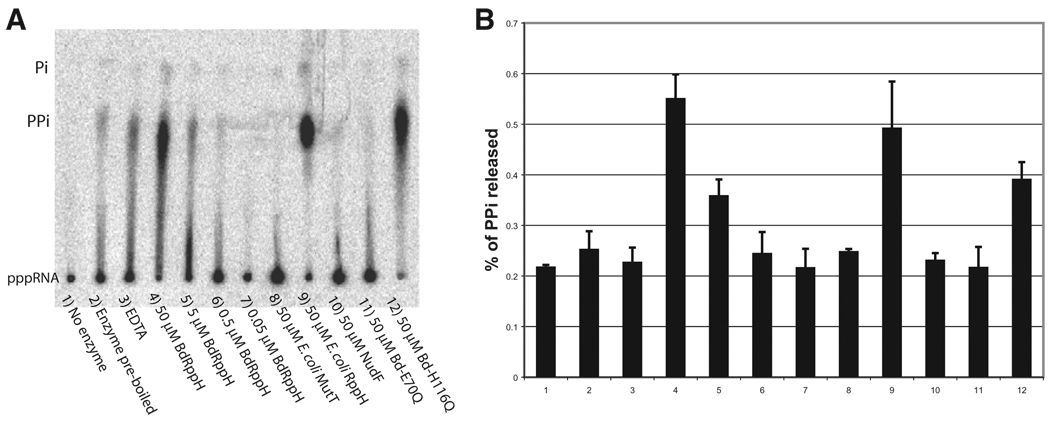
Based on an unusually high sequence similarity of 53% to the Nudix fold region of eukaryotic enzymes involved in nuclear decapping(seeFigureS1available online), suchas the U8 snoRNA decapping enzyme from X. laevis, BdRppH was tested in vitro for pyrophosphohydrolase activity against a 5′-triphosphate RNA. When added to an RNA 43-mer of sequence GGAA[UC]7UAUG [CU]10Cwith a γ-32P-labeled triphosphate at the 5′ end, BdRppH showed concentration-dependent RNA pyrophosphohydrolase activity (Figure 7) resembling that of E. coli RppH (Deana et al., 2008). The pyrophosphohydrolase activity of BdRppH dropped 5′0-fold when Glu70, one of the metal ligands in the Arg-Glu portion of the signature sequence, was mutated to Gln (Figure 7), as is the case for E. coli RppH (Deana et al., 2008). However, mutation of His116 to Gln caused a 2-fold drop in RNA pyrophosphohydrolase activity, arguing against a role for that residue as the catalytic base used in the RNA pyrophosphate activity.
Figure 7. In Vitro Analysis of the RNA Pyrophosphohydrolase Activity of BdRppH.
(A) TLC assay demonstrating RNA pyrophosphohydrolase activity.
(B) Bar graphs show the activity of each lane relative to E. coli RppH (lane 9); values represent background-corrected spot density measured by ImageQuant. All experiments were done in triplicate.
Complementation In Vivo
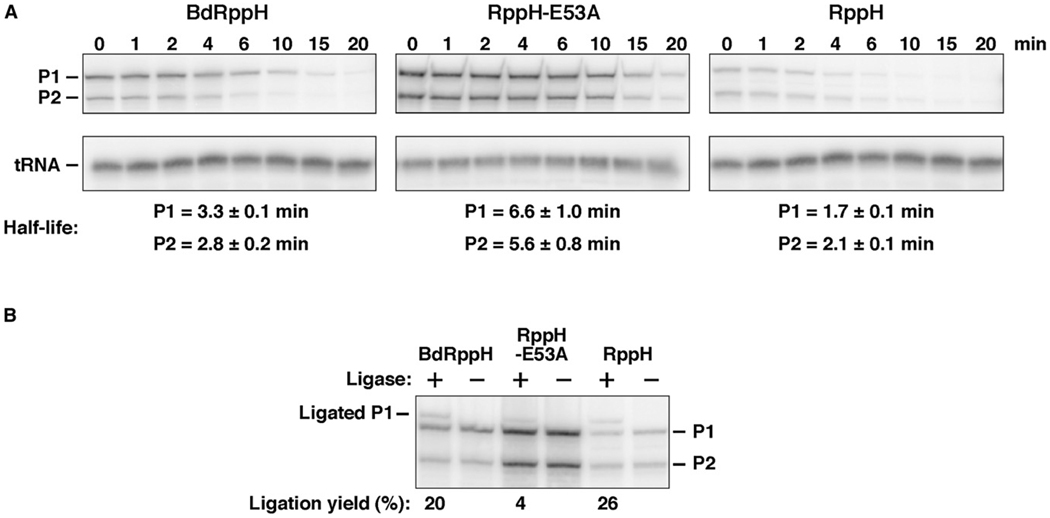
The ability of BdRppH to function as an RNA pyrophosphohydrolase in vivo was investigated by testing its ability to complement RppH deficiency in E. coli. An E. coli strain lacking the gene that encodes RppH was transformed with plasmids encoding BdRppH, E. coli RppH, or a catalytically inactive RppH mutant (RppH-E53A) (Deana et al., 2008), and the effect of each protein on the phosphorylation state and decay of rpsT mRNA was examined (Figure 8). Normally, RppH triggers rapid degradation of the rpsT P1 and P2 transcripts in E. coli; however, the absence of that enzyme nearly abolishes pyrophosphate removal from rpsT mRNA, thereby impeding its decay (Deana et al., 2008). Plasmid-encoded BdRppH can complement the defects caused by RppH deficiency in E. coli (Figure 8), accelerating the decay of rpsT mRNA and markedly increasing the percentage of rpsT P1 messages that are monophosphorylated at the 5′ end, as judged from the ligation yield in a phosphorylation assay by ligation of oligonucleotides (PABLO) assay (Celesnik et al., 2007). Plasmid-encoded RppH has a similar effect, whereas RppH-E53A fails to complement these cells. These results suggest that BdRppH has the requisite in vivo activity to function as an RNA pyrophosphohydrolase in B. bacteriovorus.
Figure 8. BdRppH Complementation of RppH Deficiency in E. coli.
(A) An E. coli strain from which the chromosomal rppH gene had been deleted was transformed with any of three plasmids encoding BdRppH, RppH, or RppHE53A, and northern blotting was used to monitor the decay of the E. coli rpsT P1 and P2 transcripts after transcription inhibition with rifampicin.
(B) The 5′-phosphorylation state of the rpsT P1 transcript was examined in the same three strains by PABLO analysis (Deana et al., 2008), in which monophosphorylated (but not triphosphorylated) mRNAs underwent splinted ligation to a DNA oligonucleotide, thereby reducing their electrophoretic mobility (Celesnik et al., 2007).
DISCUSSION
The structure of BdRppH presented here is, to our knowledge, the first structure of a bacterial RNA pyrophosphohydrolase. Overall, the structure has the general characteristics of other Nudix enzymes: a four-stranded mixed-β sheet and a two-stranded antiparallel β sheet flanked by front and back α helices. Interestingly BdRppH appears to be the prototype of a new subfamily of Nudix hydrolases. Although it is a pyrophosphohydrolase, it forms a head-to-head dimer via its three long mixed-β sheets β1, β5, and β6, similar to E. coli GDPMH, a hydrolase that cleaves at carbon instead of phosphorus (Gabelli et al., 2004). BdRppH is similar in structure to the prototypical Nudix enzyme MutT from E. coli (rmsd of 2.1 Å over 98 carbons out of 134), but our kinetic experiments show that unlike MutT, BdRppH may not use a glutamate in helix α1 as the catalytic base. Although two histidine residues from loop L6 are in a position from which they could act as the catalytic base, when GTP is the substrate, mutation of the candidate residues identified from the structure shows only minor effects on the kinetic parameters. Most probably, the flexibility of loop L6 allows different residues or a solvent hydroxyl to deprotonate the attacking water molecule. In addition, in in vitro tests of BdRppH RNA pyrophosphohydrolase activity, only mutation of Glu70, a Mg2+ ligand, negatively affects activity, whereas BdRppH-H116Q shows some activity, suggesting that the two activities may operate with slightly different mechanisms.
BdRppH shows significant similarities to eukaryotic enzymes involved in decapping. Although the similarity is not very extensive, it does have a feature similar to Box B of the human Dcp2 (Piccirillo et al., 2003). This is defined by a small patch of positively charged residues (Lys128, Arg137, Lys138, Lys142, Lys145) that runs along the α helices α2 and α3. These may supply an RNA binding surface similar to that proposed for the decapping enzyme from X. laevis, X29 (Scarsdale et al., 2006), and recently shown by biochemical and NMR studies of Dcp2 to actually bind RNA (Deshmukh et al., 2008; She et al., 2008). This mode of binding is supported by the structure of the BdRppH-GTP complex that shows that GTP is bound in a position and conformation similar to those of the nucleotide in the m7GpppA-X29 complex. Another possibility is that the first 19 amino acids of BdRppH, which are disordered and not seen in the electron density map, become ordered in the presence of RNA and form a binding surface for the substrate.
Recently, it was shown that BdRppH hydrolyzes dGTP and that this activity can complement MutT deficiency in E. coli (Steyert et al., 2008). Whether this activity is another primary function of BdRppH or an adventitious activity only important in a MutT deficient mutant remains an interesting question. Interestingly, E. coli RppH does not have this secondary activity (M.J. Bessman, personal communication), but hydrolyzes Ap4A instead. In an attempt to identify which sequence differences might be responsible for this activity difference, we aligned, using structural information, the sequences of BdRppH, E. coli MutT and RppH, and X. laevis X29 (Figure S2). As expected, all four proteins contain an arginine at the position equivalent to BdRppH Arg40 and a phenylalanine at BdRppH position 52. However, although BdRppH and EcMutT have a lysine residue at BdRppH position 56, but RppH does not. In BdRppH, Lys56 participates in the stabilization of the γ-phosphate of the Gppp substrate. Its absence in RppH may explain the lack of reactivity of RppH toward Gppp. It is noteworthy that RppH has a long insertion starting at the position equivalent to Gly93 of BdRppH. This insertion, present in other Nudix enzymes of known structure that hydrolyze Ap4A, forms a helix that is involved in substrate binding, including the phosphate portion (Fletcher et al., 2002; Swarbrick et al., 2000). These differences in the way BdRppH and E. coli RppH appear to recognize the phosphate portion of the substrate may be responsible for the difference in their Gppp hydrolyzing activity.
Nudix hydrolases, enzymes found throughout the three kingdoms of life, are good candidates to fill the role of mRNA pyrophosphohydrolases/decapping enzymes, as they catalyze the cation-dependent hydrolysis of a wide range of nucleoside diphosphates linked to another moiety × to yield NMP plus P-X. In eukaryotes, the degradation of deadenylated mRNAs begins with the removal of the m7Gppp cap by a Nudix enzyme (Dcp2) (Wang et al., 2002). In E. coli, recent studies have shown that the Nudix hydrolase RppH can initiate mRNA decay by removing pyrophosphate from the 5′ ends of triphosphorylated transcripts (Deana et al., 2008), resulting in monophosphorylated intermediates that are significantly more susceptible to RNase E cleavage (Celesnik et al., 2007; Deana et al., 2008; Mackie, 1998). Thus, although differing in detail, the mechanism of mRNA degradation in bacteria is conceptually similar to that in eukaryotes: a triphosphate at the 5′ end confers stability on the mRNA, much like the m7Gppp cap of eukaryotic mRNA. In both cases, a Nudix enzyme is important for removing that protection and triggering mRNA degradation.
Our in vitro experiments show that the B. bacteriovorus Nudix enzyme BdRppH can hydrolyze the 5′ triphosphate of RNA, liberating monophosphorylated RNA and pyrophosphate. In E. coli, BdRppH can complement RppH deficiency, restoring normal mRNA degradation. Hence, BdRppH has the in vitro and in vivo activities required for it to function as an RNA pyrophosphohydrolase in B. bacteriovorus.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
BdRppH was cloned, expressed, and purified as described before (Steyert et al., 2008). Briefly, BL21 (DE3) cells (Novagen), transformed with a pET 24a containing the BdRppH coding sequence, were grown aerobically at 37°C in LB medium to an OD A600nm of ~.6. The culture was induced with 1 mM isopropyl-β-D-thiogalactopyranoside, and grown for an additional 3 hr. The culture was centrifuged, the supernatant was discarded, and the cell pellet was frozen at −80°C. The BL21 (DE3) cell pellet (12 g) was resuspended in 120 ml of 50 mM Tris (pH 7.5), 1 mM EDTA, 1 mM DTT buffer (TED buffer) and lysed. The lysed cells were centrifuged and the soluble fraction was brought to 30% ammonium sulfate via the slow addition of saturated ammonium sulfate, and centrifuged. The supernatant of the ammonium sulfate precipitation was subjected to hydrophobic-interaction chromatography, followed by a 15S cation-exchange column. Fractions containing BdRppH were loaded onto a gel-filtration column, and then eluted with TED buffer. The gel-filtration eluate was concentrated to 10 mg/ml and stored at −80°C.
Site-Directed Mutagenesis
The sequences of coding and complementary noncoding mismatch primers used for the mutations are as follows: H116Q, 5′-CCT CGG GCC AAG CAT CAA ATG ATG CTG GAA TGG ATC-3′ and 5′-GAT CCA TTC CAG CAT CAT TTG ATG CTT GGC CCG AGG-3′; H115Q, 5′-GAG CCT CGG GCC AAG CAA CAC ATG ATG CTG GAA TGG ATC-3′ and 5′-CAT TCC AGC ATC ATG TGT TGC TTG GCC CGA GGC TC-3′; H115L, 5′-GCC TCG GGC CAA GCT TCA AAT GAT GCT GG-3′ and 5′-CCA GCA TCA TGT GAA GCT TGG CCC GAG GC-3′; H116L, 5′-CCT CGG GCC AAG CAT CTC ATG ATG CTG GAA TG-3′ and 5′-CAT TCC AGC ATC ATG AGA TGC TTG GCC CGA GG-3′; H115116L, 5′-GGC GAG CCT CGG GCC AAG CTT CTC ATG ATG CTG GAA TGG ATC-3′ and 5′-GAT CCA TTC CAG CAT CAT GAG AAG CTT GGC CCG AGG CTC GCC-3′; H115116Q, 5′-GGC GAG CCT CGG GCC AAG CAA CAA ATG ATG CTG GAA TGG ATC C-3′ and 5′-GGA TTC ATT CCA GCA TCA TTT GTT GCT TGG CCC GAG GCT CGC C-3′; E70Q was made as described previously in Steyert et al. (2008).
Each coding strand primer and noncoding strand primer was complementary to the noncoding strand, except at the appropriate mismatch (underlined) in the codon designating the amino acid to be mutated. The wild-type BdRppH gene in pET 24a vector was used as the template for the PCR to generate single or double mutants of BdRppH, following the instruction manual of the QuikChange site-directed mutagenesis kit (Stratagene). DH-5α E. coli cells were transformed with the resulting plasmids. Plasmid DNA was purified using the QIAGEN miniprep kit, and sequenced to verify mutations. BL21 (DE3) E. coli cells were transformed with plasmids containing mutations for protein expression.
Calorimetry
The enthalpy of Mg2+ binding to BdRppH was measured in a VP-ITC microcalorimeter (Microcal) with a reaction cell volume of 1.4 ml. Purified protein was extensively dialyzed overnight against buffer (50 mM HEPES [pH 8], 200 mM NaCl, 1mMTCEP) at 4°C. All glassware and buffer were thoroughly pretreated using Chelex 100 resin throughout. All solutions were degassed immediately before use. In a typical binding experiment, the calorimeter cell contained a protein solution in buffer or buffer alone at 3′3K (30°C). The sample was titrated with 25, 10 µl injections of a concentrated Mg2+ solution (3′0 µl) added at 3.5 min intervals stirring at 300 rpm. A correction factor for the increase in reaction volume was applied to both reactant concentrations. The integrated heat effects were analyzed by nonlinear regression methods using the Microcal Origin 5 software package. The resulting data were fitted to two identical binding sites per dimer and reported per mol of Mg2+ bound. The enthalpy of binding, ΔH (kcal/mol), was directly determined from the heat released, which is independent of the binding model. The association binding constant, K (M−1), was estimated by fitting the binding curve, and the entropy, ΔS (cal/ K mol), was calculated from the standard expression ΔG = −RTlnK = ΔH − TDS. Values and uncertainties for n, K, and ΔH are weighted averages and their errors, and weighted standard deviations from three or four titration runs, using the x2 values and errors recovered from the fits. Values and uncertainties for the rest of the parameters are determined from these weighted averages and the propagation of their errors.
Colorimetric Assay
The standard reaction mix of 50 µl consisted of 50 mM Tris (pH 8.5), 5 mM MgCl2, 1 mM DTT, 0.25 units of inorganic pyrophosphatase, 30 nM BdRppH, and 1 mM nucleotide substrate. The reactions were run at 37°C for 10 min, and quenched by addition of 10 µl of 100 mM EDTA. Quenched reactions were analyzed for inorganic orthophosphate by diluting the 50 µl reaction with 250 µl of H2O, followed by the addition of 700 µl of 6:1 (0.42% ammonium molybdate in 1 N H2SO4):(10% ascorbic acid). This mixture was incubated for 20 min at 42°C and the absorbance was measured at 820 nm (Fiske and Subbarow, 1925).
Kinetics
Steady-state initial rates were measured by the colorimetric assay described above. The reaction mixture (50 µl) contained 30 nM wild-type or mutant BdRppH and varying concentrations of dGTP (0.0625–1 mM). Km and kcat were calculated by fitting the Michaelis-Menten equation, using nonlinear least squares, to initial rate values as a function of substrate concentration. The dependence of kcat on pH was determined at six different pH values as previously described (Harris et al., 2000). For these measurements the following buffers were used: HEPES for pH value 7.5, and Tris-HCl for pH values 7.75, 8, 8.25, 8.5, 8.75, and 9. The kcat for wild-type enzyme and BdRppH-E70Q were measured at saturating substrate concentrations (2 and 4 mM dGTP).
Crystallization
BdRppH crystals grew by vapor diffusion in hanging drops containing a mixture of 2 µl of protein at 10 mg/ml with an equal volume of a reservoir solution of 12%–16% PEG 4000, 0.05 M Na acetate (pH 4.6), and 0.5–1 M ammonium acetate. Single crystals grew in 1–5 days. Crystals had P3221 symmetry and contained two monomers in the asymmetric unit. Crystals with holmium were cocrystallized in 12%–16% PEG 4000, 0.05 M Na acetate (pH 4.6), and 0.5–1Mammoniumacetate with 1mMHoCl2. Crystals with GTP were cocrystallized in 12%–16% PEG 4000, 0.05 M Na acetate (pH 7), and 0.5–0.6 M ammonium acetate with 10 mM GTP and soaked in 10 mM MgCl2 in the crystallization buffer. These crystals had P212121 symmetry and also contained two molecules in the asymmetric unit. Data were collected at beamline X6A of the National Synchrotron Light Source (Brookhaven National Laboratory) with a wavelength of 1.0 Å. The images were collected using a Quantum 210 charge-coupled device (CCD) detector (ADSC) and processed with the HKL suite (Otwinowski and Minor, 1997). The SGX mail-in program of the Advanced Photon Source (Argonne National Laboratory) was also used to collect data under similar conditions.
Structure Determination
The structure of BdRppH was determined by multiple isomorphous replacement. Diffraction data from crystals derivatized with mercury acetate, uranyl acetate, and holmium chloride were used with the program SOLVE (Terwilliger, 2000, 2003; Terwilliger and Berendzen, 1999) to calculate the initial phases (Z score of 31.21, and figure of merit of 0.39–2.8 Å). The density calculated with the initial phases, modified with the program RESOLVE, gave an interpretable electron density map (Terwilliger, 2000, 2003; Terwilliger and Berendzen, 1999). RESOLVE was only able to trace 26 residues via its automatic chaintracing feature. The model was built interactively with the program O (Jones and Kjeldgaard, 1997; Jones et al., 1991) and refined using REFMAC 5.0 (CCP4, 1994) with individual restrained B factors. Anisotropic refinement using translation, libration, and screw rotation (TLS) of rigid bodies was carried out using each monomer as a TLS group (CCP4, 1994). Water molecules were placed automatically by the program ARP in peaks greater than 3.0σ in Fo – Fc maps. Refinement was monitored by calculating Rfree using 5% of the data set aside for crossvalidation. The final model was refined to an R of 22% and an Rfree of 25%.
Structures of the other crystals were determined by molecular replacement with the program AMoRe (Navaza, 2001) using the coordinates of the final native structure as the search model. The models were built and refined as described above. Drawings were prepared with PyMOL, ESPript, and MOLSCRIPT (DeLano, 2002; Gouet et al., 1999; Kraulis, 1991).
Detection of BdRppH RNA Pyrophosphohydrolase Activity
A 43-mer sequence of GGAA[UC]7UAUG[CU]10C RNA was transcribed and purified as previously described (Acker et al., 2007). The DNA template sequence used for transcription was 5′-GAG AGA GAG AGA GAG AGA GAG CAT AGA GAG AGA GAG AGA TTC C-3′. Briefly, the RNA was synthesized by in vitro transcription in a reaction mixture containing transcription buffer, DTT, NTPs, clamp oligo, oligo template, and T7 polymerase. The product of the reaction was purified via phenol and chloroform washes and further purified by elution from an acrylamide gel, and 5′ labeling of the synthesized RNA was done with γ-labeled [32P]ATP using PNK (New England Biolabs), following the manufacturer’s protocol.
For the decapping assay, a 20 µl solution containing 50 mM Tris (pH 8.5), 5 mM MgCl2, 1 mM DTT, 1 unit/µl RNasin (Promega), and 15 nM 5′ end-labeled RNA (35,000 cpm) was incubated with either purified BdRppH (50 µM, 0.5 µM, 0.05 µM), MutT (10 µM), RppH (50 µM), orf209 (50 µM), BdRppH-E70Q (50 µM), or BdRppH-H116Q (50 µM) for 60 min at room temperature. Reaction samples were quenched with 5 µl of EDTA (100 mM, pH 8.0) and analyzed by TLC on PEI-cellulose (J.T. Baker) developed with potassium phosphate buffer (0.3 M, pH 7.5). Band or spot intensities were compared by using a Molecular Dynamics Storm 820 phosphorimager and ImageQuant software.
Complementation of RppH deficiency in E. coli was tested by transforming E. coli strain JW2798Δkan (Deana et al., 2008) with any of three otherwise identical plasmids encoding BdRppH, RppH, or RppH-E53A, each bearing an amino-terminal hexahistidine tag. Measurements of the decay rate of rpsT mRNA and the phosphorylation state of the rpsT P1 transcript were performed as previously described (Celesnik et al., 2007; Deana et al., 2008).
Supplementary Material
Supplemental Data include two figures and can be found with this article online at http://www.cell.com/structure/supplemental/S0969-2126(09)00075-6.
ACKNOWLEDGMENTS
We thank M.J. Bessman (Department of Biology, Johns Hopkins University) for helpful discussions, Sarah Mitchell for her expertise in making radiolabeled RNA, and Yuko Oku for advice and expertise in performing the in vitro RNA pyrophosphohydrolase assay. Beamline X6A of the National Synchrotron Light Source (Brookhaven National Laboratory) and SGX mail-in program of the Advanced Photon Source (Argonne National Laboratory) are gratefully acknowledged. These studies were supported by grants to L.M.A. (GM066895) and J.G.B. (GM35769) from the National Institutes of Health.
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein Data Bank for RNA pyrophosphohydrolase BdRppH, RNA pyrophosphohydrolase BdRppH in complex with holmium, RNA pyrophosphohydrolase in complex with dGTP, and RNA pyrophosphohydrolase in complex with GTP and Mg2+ under ID codes 3EES, 3EEU, 3EF5, and 3FFU, respectively.
REFERENCES
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Walsh JD, Dunn CA, Swaminathan J, Weldon JE, Shen J. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A) J. Biol. Chem. 2001;276:37834–37838. doi: 10.1074/jbc.M107032200. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed]
- Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conyers GB, Wu G, Bessman MJ, Mildvan AS. Metal requirements of a diadenosine pyrophosphatase from Bartonella bacilliformis: magnetic resonance and kinetic studies of the role of Mn2+ Biochemistry. 2000;39:2347–2354. doi: 10.1021/bi992458n. [DOI] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Deshmukh MV, Jones BN, Quang-Dang DU, Flinders J, Floor SN, Kim C, Jemielity J, Kalek M, Darzynkiewicz E, Gross JD. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- Dudev T, Chang LY, Lim C. Factors governing the substitution of La3+ for Ca2+ and Mg2+ in metalloproteins: a DFT/CDM study. J. Am. Chem. Soc. 2005;127:4091–4103. doi: 10.1021/ja044404t. [DOI] [PubMed] [Google Scholar]
- Emory SA, Bouvet P, Belasco JG. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- Fletcher JI, Swarbrick JD, Maksel D, Gayler KR, Gooley PR. The structure of Ap(4)A hydrolase complexed with ATP-MgF(x) reveals the basis of substrate binding. Structure. 2002;10:205–213. doi: 10.1016/s0969-2126(02)00696-2. [DOI] [PubMed] [Google Scholar]
- Frick DN, Weber DJ, Gillespie JR, Bessman MJ, Mildvan AS. Dual divalent cation requirement of the MutT dGTPase. Kinetic and magnetic resonance studies of the metal and substrate complexes. J. Biol. Chem. 1994;269:1794–1803. [PubMed] [Google Scholar]
- Gabelli SB, Bianchet MA, Bessman MJ, Amzel LM. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 2001;8:467–472. doi: 10.1038/87647. [DOI] [PubMed] [Google Scholar]
- Gabelli SB, Bianchet MA, Ohnishi Y, Ichikawa Y, Bessman MJ, Amzel LM. Mechanism of the Escherichia coli ADP-ribose pyrophosphatase, a Nudix hydrolase. Biochemistry. 2002;41:9279–9285. doi: 10.1021/bi0259296. [DOI] [PubMed] [Google Scholar]
- Gabelli SB, Bianchet MA, Azurmendi HF, Xia Z, Sarawat V, Mildvan AS, Amzel LM. Structure and mechanism of GDP-mannose glycosyl hydrolase, a Nudix enzyme that cleaves at carbon instead of phosphorus. Structure. 2004;12:927–935. doi: 10.1016/j.str.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Glusker JP. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Harris TK, Wu G, Massiah MA, Mildvan AS. Mutational, kinetic, and NMR studies of the roles of conserved glutamate residues and of lysine-39 in the mechanism of the MutT pyrophosphohydrolase. Biochemistry. 2000;39:1655–1674. doi: 10.1021/bi9918745. [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A 47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kang LW, Gabelli SB, Bianchet MA, Xu WL, Bessman MJ, Amzel LM. Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family. J. Bacteriol. 2003a;185:4110–4118. doi: 10.1128/JB.185.14.4110-4118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang LW, Gabelli SB, Cunningham JE, O’Handley SF, Amzel LM. Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis. Structure. 2003b;11:1015–1023. doi: 10.1016/s0969-2126(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Kraulis J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;277:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA. 2003;9:1138–1147. doi: 10.1261/rna.5690503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Scarsdale JN, Peculis BA, Wright HT. Crystal structures of U8 snoRNA decapping Nudix hydrolase, X29, and its metal and cap complexes. Structure. 2006;14:331–343. doi: 10.1016/j.str.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Schoenberg DR. The end defines the means in bacterial mRNA decay. Nat. Chem. Biol. 2007;3:535–536. doi: 10.1038/nchembio0907-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockett RE, Lambert C. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat. Rev. Microbiol. 2004;2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- Steyert SR, Messing SA, Amzel LM, Gabelli SB, Pineiro SA. Identification of Bdellovibrio bacteriovorus HD100 Bd0714 as a Nudix dGTPase. J. Bacteriol. 2008;190:8215–8219. doi: 10.1128/JB.01009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolp H, Petzold H. Untersuchungen uber einen obligat parasitischen mikroorganismus mit lytischer aktivat fur Pseudomonas-bakterien. J. Phytopathol. 1962;45:364–390. [Google Scholar]
- Stolp H, Starr MP. Bdellovibrio bacteriovorus Gen. Et Sp. N., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- Strauch E, Schwudke D, Linscheid M. Predatory mechanisms of Bdellovibrio and like organisms. Future Microbiol. 2007;2:63–73. doi: 10.2217/17460913.2.1.63. [DOI] [PubMed] [Google Scholar]
- Swarbrick JD, Bashtannyk T, Maksel D, Zhang XR, Blackburn GM, Gayler KR, Gooley PR. The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L. J. Mol. Biol. 2000;302:1165–1177. doi: 10.1006/jmbi.2000.4085. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- van Ham RC, Kamerbeek J, Palacios C, Rausell C, Abascal F, Bastolla U, Fernandez JM, Jimenez L, Postigo M, Silva FJ, et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Cohen SN. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include two figures and can be found with this article online at http://www.cell.com/structure/supplemental/S0969-2126(09)00075-6.