Abstract
Maternal food restriction (FR) during pregnancy results in intrauterine growth-restricted (IUGR) offspring that show rapid catch-up growth and develop metabolic syndrome and adult obesity. However, continued nutrient restriction during nursing delays catch-up growth and prevents development of obesity. Epigenetic regulation of IGF1, which modulates growth and is synthesized and secreted by the liver, may play a role in the development of these morbidities. Control (AdLib) pregnant rats received ad libitum food through gestation and lactation, and FR dams were exposed to 50% food restriction from days 10 to 21. FR pups were nursed by either ad libitum-fed control dams (FR/AdLib) or FR dams (FR/FR). All pups were weaned to ad libitum feed. Maternal FR resulted in IUGR newborns with significantly lower liver weight and, with the use of chromatin immunoprecipitation, decreased dimethylation at H3K4 in the IGF1 region was observed. Obese adult FR/AdLib males had decreased dimethylation and increased trimethylation of H3K4 in the IGF1 region. This corresponded to an increase in mRNA expression of IGF1-A (134 ± 5%), IGF1-B (165 ± 6%), IGF1 exon 1 (149 ± 6%), and IGF1 exon 2 (146 ± 7%) in the FR/AdLib compared with the AdLib/AdLib control group. In contrast, nonobese FR/FR had significantly higher IGF1-B mRNA levels (147 ± 19%) than controls with no difference in IGF1-A, exon 1 or exon 2. Modulation of the rate of IUGR newborn catch-up growth may thus protect against IGF1 epigenetic modifications and, consequently, obesity and associated metabolic abnormalities.
Keywords: intrauterine growth restriction, insulin-like growth factor 1, histone methylation
epidemiological and experimental studies have demonstrated an association between suboptimal intrauterine environment, intrauterine growth restriction (IUGR) and the subsequent development of metabolic and cardiovascular disease in later life. Specifically, maternal undernutrition during pregnancy leads to IUGR with asymmetric reduction in organ growth, such that growth of liver is reduced, whereas that of brain and heart is spared. This altered growth of specific organs is associated with metabolic abnormalities in the adult offspring. Additionally, modulation of postnatal nutrition and, hence, subsequent growth of the IUGR newborn determines the ultimate adult phenotype. For example, IUGR newborns nursed by ad libitum-fed dams exhibit rapid catch-up growth and develop insulin resistance, glucose intolerance, central obesity, type 2 diabetes, hypertension, and cardiovascular disease (38, 41). These adverse health consequences are prevented or diminished in severity when the rapid catch-up growth phase of the newborn is delayed by limiting the availability of nutrients through continued nursing by food-restricted dams (16, 17).
IGF1 plays a key role in mediating perinatal growth and development as part of the growth hormone-IGF-insulin axis (32). IGF1-null mutant rodents have a 40% reduction in growth along with decreased postnatal survival (2, 35). The liver is the predominant source of circulating IGF1 (>80%) although most tissues synthesize IGF1 that has both paracrine and autocrine actions (11, 43, 49). Interestingly, tissue-specific knockouts of hepatic IGF1 result in normal body growth, indicating that extrahepatic IGF1 with autocrine/paracrine action may be sufficient to sustain body growth in these animals. Notably, they have hyperinsulinemia, a reduced age-dependant accumulation of adipose tissue and reduction of plasma IGF, and previous studies conclude that liver-derived IGF1 was associated with metabolic function rather than growth (49, 56). Other studies have found that plasma IGF1 correlates directly with fetal growth, and undernutrition studies have shown reduced IGF1 levels in IUGR (27, 34). Therefore, the findings from genetic and nutrition studies suggest that fetal plasma IGF1, potentially from a nonhepatic source, correlates with fetal growth and that aberrant hepatic IGF1 expression may be programmed during the fetal period but may not result in an abnormal phenotype until later adult life. The underlying mechanisms for the programmed changes in hepatic IGF1 expression may include epigenetic modifications, such as histone methylation.
IGF1 is expressed as two isoforms, which arise from alternative splicing of the six IGF1 exons. Exons 1 and 2 both contain promoter regions and are differentially spliced, as are exons 5 and 6, whereas exons 3 and 4 encode the mature IGF1 peptide and are also present in both isoforms. IGF1-A comprises exon 1 or 2, 3, 4, and 6 and IGF1-B comprises exons 1 or 2, 3, 4, 5, and 6, with exon 2 being the predominate leader exon in the liver (26, 41, 48, 50). It has been shown that development, fasting, and diabetes cause differential regulation of the different transcription start sites (1), possibly as a result of epigenetic modifications to histone structure that result in changes in the ability of the transcriptional machinery to access the DNA and therefore determine gene expression (13, 21, 44). Histone methylation is a key structural alteration that can impact gene expression, either promoting or repressing gene expression, depending on the location (10, 33, 54). Methylation of either DNA or histone proteins requires methyl donors from dietary folate and requires the presence of vitamins B6 and B12, choline, methionine, and a range of methyltransferases (25, 44). Aberrant hypomethylation has been linked with cancer, diabetes, developmental and neurological abnormalities, and cardiovascular and cerebrovascular diseases (9, 30, 31), diseases that are also associated with IUGR offspring (6, 47, 53).
Histone methyltransferase mediates the addition of a methyl group to the lysine in the NH2 terminus tail of histones, and lysine tails can be mono-, di-, or trimethylated. Trimethylation of H3 Lys4 (H3K4) has been well established as a marker on promoter regions of transcriptionally active genes, whereas dimethylation of H3K4 is associated with other areas in the vicinity of active genes and is thought to play an indirect role in promoting expression by recruiting transcriptional machinery (8, 13, 45). Therefore, increased H3K4 di- and trimethylation may be a long-term, heritable modification that is directly linked with active gene expression.
We have established a rat model of maternal food restriction during pregnancy that results in IUGR pups. IUGR pups exposed to standard nursing and postweaning diet (food restriction and ad libitum, FR/AdLib) demonstrate rapid catch-up growth and develop adult obesity with increased body fat, insulin resistance, and elevated plasma triglyceride levels. In contrast, IUGR pups that are nursed by FR dams (FR/FR) exhibit delayed catch-up growth and notably do not develop adult obesity and metabolic syndrome (17, 18). We therefore hypothesized that maternal undernutrition-induced IUGR would result in epigenetic modifications of histone structure, specifically H3K4, in the regions of the IGF1 gene of the liver. We further hypothesized that these epigenetic changes, including increased hepatic IGF1 mRNA and plasma IGF1 levels, would be evident in IUGR offspring that develop adult obesity.
MATERIALS AND METHODS
Animals and diet.
A model of maternal FR (50%) during pregnancy was used as previously described (18). Studies were approved by the Animal Research Committee of Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center and were in accordance with the American Association for Accreditation of Laboratory Care and National Institutes of Health guidelines. First-time-pregnant Sprague Dawley rats (Charles River Laboratories, Hollister, CA) were housed in a facility with constant temperature and humidity and a controlled 12-h:12-h light/dark cycle. At 10 days of gestation, pregnant rats were provided either an ad libitum diet of standard laboratory chow (LabDiet 5001; LabDiet, Brentwood, MO; 23% protein, 4.5% fat, 3,030 kcal/kg of metabolizable energy) or a 50% FR diet determined by quantification of normal intake in the ad libitum-fed rats. The respective diets were given from day 10 of pregnancy to term.
Offspring.
Rat dams gave birth normally, and, at day 1 after birth, pups were limited to eight (4 males and 4 females) per litter to normalize rearing although only male offspring were used in this report. Control pups were cross fostered onto ad libitum-fed mothers to continue the control group (AdLib/AdLib). Maternally FR pups (FR) were cross fostered onto either ad libitum-fed mothers (FR/AdLib) or 50% FR mothers (FR/FR). At 3 wk of age, offspring in the three groups were housed individually and weaned to ad libitum feed. Thus two groups of offspring were studied at day 1, control (n = 6) and FR (n = 6), and three groups were studied at 9 mo of age (adult), AdLib/AdLib (n = 6), FR/AdLib (n = 6), and FR/FR (n = 6), to investigate the impact of rapid and delayed catch-up growth, respectively (n = 6 represents 6 offspring from 6 mothers).
Body and liver weights.
Body and liver weights of day 1 newborns and 9-mo adult offspring were measured.
RNA isolation.
Total RNA was extracted from day 1 and adult male liver using the RNA Stat-60 (Tel-Test, Friendswood, TX) according to manufacturer's instructions, treated with DNase-I (Qiagen, Valencia, CA), and then quantified by absorption at A260nm measured by UV spectrophotometry. Gel electrophoresis confirmed the integrity of the samples.
Quantitative real-time PCR.
Liver mRNA levels of IGF1-A, IGF1-B, IGF1 exon 1 (IGF1-E1), IGF1 exon 2 (IGF1-E2), and GAPDH were measured using real-time RT-PCR. cDNA was synthesized from 5 μg of DNase-treated total RNA using a Superscript III First-Strand kit (Invitrogen, Carlsbad, CA), with the inclusion of no superscript and no RNA samples to confirm both RNA and reagent purity. Target primers and probes were designed using Primer Express software (Applied Biosystems, Foster City, CA) as previously described (26); target probes were labeled with carboxyfluorescein fluorescent reporter dye. cDNA, probe, and primers were added to TaqMan Universal PCR Master Mix (Applied Biosystems), and samples were run on an ABI Prism 7000. GAPDH CT values were assessed for stable expression across all groups to ensure suitability for use as a reference gene, and relative quantification of PCR products was calculated on the basis of value differences between the target and GAPDH control using the comparative CT method. This method is predicated on an amplification efficiency of 100 (46). Cycle parameters were 50°C for 2 min, 95°C for 10 min, and then 40 cycles at 95°C for 15 s and 60°C for 60 s.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) with antidimethyl-H3K4 and antitrimethyl-H3K4 (Millipore, Billerica, MA) was performed as described previously (25). A quantity of 100 mg of liver tissue from day 1 and adult male rats was ground in liquid nitrogen and cross linked with formaldehyde, with a final concentration of 1%, for 10 min. The chromatin was sonicated (Sonic Dismembrator, model 100; Fisher Scientific, Pittsburgh, PA) 12 times for 10 s on ice at the highest level to generate chromatin fragments of 500–2,000 bp. The sonicated chromatin was quantified on the basis of DNA content at A260, and on the basis of this estimate the chromatin equivalent of 100 μg of DNA was used in each immunoprecipitation (IP). Fifty microliters of antidimethyl-H3K4 and antitrimethyl-H3K4 (Millipore) were used for each IP. DNA samples without the addition of an antibody and a sample without the addition of DNA were included to verify the purity of the DNA and the reagents, respectively. Following IP, cross linking was reversed, and a DNeasy Tissue Kit (Qiagen) was used to purify the DNA from the IP samples. Real-time PCR was used to quantitate the amount of IGF1-promoter 1 (IGF1-P1), promoter 2 (IGF1-P2), exon 5 (IGF1-E5), exon 6 (IGF1-E6), 3′UTR-3 (IGF1-U3), and 3′UTR-4 (IGF1-U4) sequence (26) that was immunoprecipitated.
Western blot.
Protein was extracted from day 1 and adult liver using a radioimmuno precipitation assay buffer that contained a protease inhibitor cocktail (Roche, Manheim, Germany). Supernatant protein concentration was determined by BCA assay (Pierce, Rockford, IL). An IGF1 antibody (Upstate Biotechnology, Lake Placid, NY) diluted 1:1,000 and a GAPDH (Chemicon, Temecula, CA) diluted 1:5,000 were used. One hundred micrograms of protein were mixed with a loading buffer and reducing agent, boiled for 10 min, and separated on a 12% Bis-Tris gel (BioRad, Richmond, CA). The protein was transferred to a PVDF membrane for 1 h in an ice-water bath. Nonspecific antibody binding was blocked by incubation for 1 h at room temperature with 5% nonfat dry milk in 0.1% Tris-buffered saline with Tween (TBST). The membrane was then incubated with the appropriate antibody in 5% milk in TBST for 1 h at room temperature and then washed three times for 5 min in TBST. The membrane was washed with the secondary antibody goat anti-rabbit horseradish peroxidase conjugate (Upstate Biotechnology) diluted 1:5,000 for 1 h at room temperature and then washed three times for 5 min in TBST. Supersignal West Pico Chemiluminescent Substrate (Pierce) was used to detect the targeted protein. The band intensity was analyzed using a BioRad GS-800 densitometer, and IGF1 intensity was normalized to GAPDH.
Plasma.
Excess pups at 1 day of age during culling (3–5 pups per litter from 6 litters per group) were decapitated, and blood was collected in heparinized capillary tubes. At 9 mo of age, one male from each adult litter (n = 6 per group) was fasted overnight, and blood was collected via cardiac puncture in heparinized and EDTA-aprotinin (780 KIU/ml of blood) tubes for determination of plasma IGF1 levels. IGF1 concentrations were determined by a radioimmunoassay rat-specific IGF1 kit (Diagnostic Systems Laboratories, Webster, TX).
Statistics.
Data from newborn control and FR groups were compared using a Student's unpaired t-test. Data from the AdLib/AdLib, FR/AdLib, and FR/FR groups in adult life were compared using one-way ANOVA. All data are the means ± SE with weights in grams and plasma in nanograms/milliliter and are presented as a percentage of the age-matched control group for mRNA, ChIP DNA, and protein.
RESULTS
Body and liver weights.
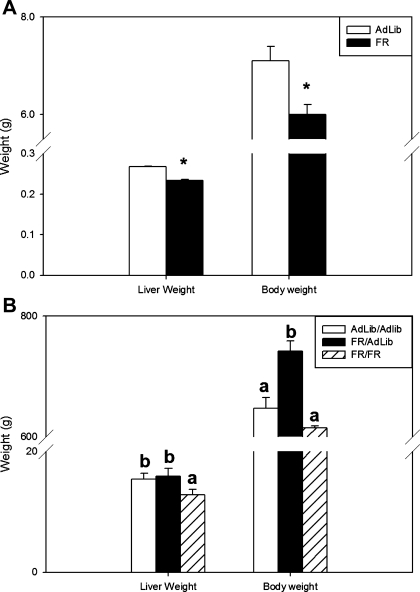
At day 1, FR newborns had significantly decreased body weights compared with control newborns (6.0 ± 0.2 vs. 7.1 ± 0.3 g, P < 0.01). The liver weight in FR newborns was also decreased compared with controls (233 ± 3 vs. 277 ± 2 mg, P < 0.01) (Fig. 1A).
Fig. 1.
A: newborn liver weight and body weight (g). Open bars represent ad libitum-fed animals (AdLib), and solid bars represent food-restricted animals (FR); asterisk denotes differences between AdLib and FR newborns, P < 0.01. B: adult liver weight and body weight (g). Open bars represent AdLib/AdLib (control), solid bars represent FR/AdLib (FR during pregnancy), and hatched bars represent FR/FR (FR during pregnancy and lactation); a and b denote a significant difference between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR, P < 0.05.
At 9 mo, FR/AdLib males were heavier than AdLib/AdLib controls (742 ± 17 vs. 647 ± 18 g, P < 0.01), whereas liver weight in the animals was not different (15.9 ± 1.3 vs. 15.4 ± 1.0 g). In contrast, the FR/FR males had body weights (615 ± 150 g) similar to that of AdLib/Adlib, whereas the liver weight in this group remained significantly lower than controls (12.8 ± 0.9 g, P < 0.01) (Fig. 1B).
Hepatic GAPDH and IGF1 mRNA expression.
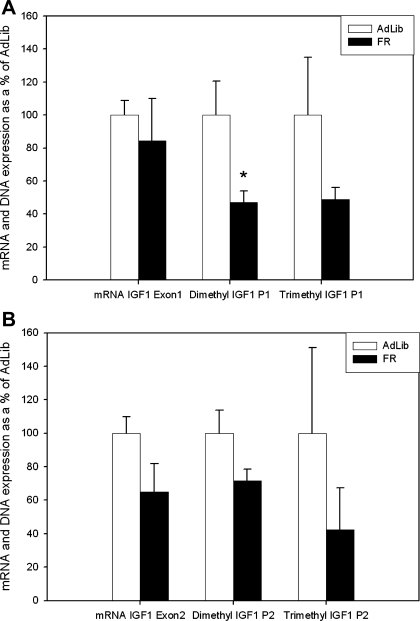
No significant difference was found between GAPDH CT values in newborn AdLib [25.5 ± 0.1 Cycle number (CN)] or FR (25.6 ± 0.2 CN) groups. Similarly there was no significant difference between GAPDH CT values in the AdLib/AdLib (23.8 ± 0.3 CN), FR/AdLib (24.4 ± 0.2 CN), or FR/FR (23.6 ± 0.4 CN) groups. At 1 day of age there was no significant difference in hepatic IGF1 mRNA expression between FR and AdLib groups (Fig. 2 and Table 1).
Fig. 2.
A: newborn IGF1 exon 1 mRNA expression and H3K4 dimethylation and trimethylation of promoter 1 (P1) on exon 1. Open bars represent AdLib, and solid bars represent FR; asterisk denotes differences between AdLib and FR newborns, P < 0.05. B: newborn IGF1 exon 2 mRNA expression and H3K4 dimethylation and trimethylation of H3K4 promoter 2 (P2) on exon 2. Open bars represent AdLib, and solid bars represent FR.
Table 1.
Newborn and adult mRNA expression of regions of IGF1
| Treatment | Age | IGF1 Exon 1 | IGF1A | IGF1 Exon 2 | IGF1B |
|---|---|---|---|---|---|
| AdLib | Newborn | 100 ± 8.8 | 100 ± 22.8 | 100 ± 9.9 | 100 ± 9.0 |
| FR | Newborn | 84.3 ± 25.8 | 211.9 ± 98.5 | 64.8 ± 17.1 | 104 ± 30.7 |
| AdLib/AdLib | Adult | 100 ± 9.6 | 100 ± 6.9 | 100 ± 8.8 | 100 ± 5.0 |
| FR/AdLib | Adult | 149.3 ± 6.6 | 133.7 ± 5.3 | 146 ± 7.7 | 164.9 ± 6.3 |
| FR/FR | Adult | 74.6 ± 8.3 | 99.3 ± 6.8 | 86.3 ± 8.3 | 146.8 ± 21.0 |
Values are means ± SE. mRNA expression of regions of IGF1 relative to GADPH is expressed as a percentage of expression levels of controls for the age group. The results are from newborns at 1 day of age of ad libitum-fed animals (AdLib, controls) vs. food-restricted pregnancy animals (FR), adults at 9 mo of age of AdLib/AdLib (control) vs. FR/AdLib (food-restricted pregnancy and AdLib lactation), and FR/FR (food-restricted pregnancy and lactation). * and † Differences between adult treatment groups, P < 0.05.
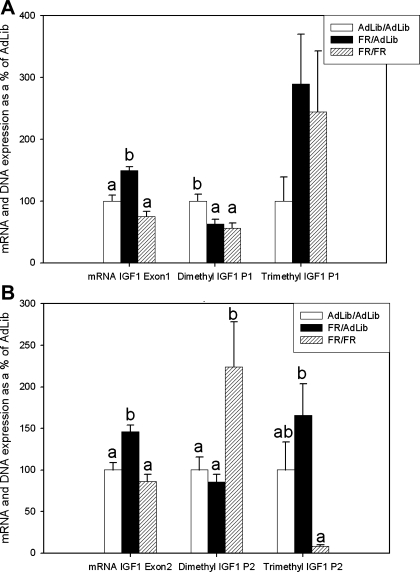
At 9 mo of age there was significantly increased mRNA expression of IGF1-A (134 ± 5%), IGF1-B (165 ± 6%), IGF1-E1 (149 ± 6%, Fig. 3A), and IGF1-E2 (146 ± 7%, Fig. 3B) in FR/AdLib compared with AdLib/AdLib. In contrast, in the FR/FR group there was only a significant difference in IGF1-B (147 ± 19%) (Table 1).
Fig. 3.
A: adult IGF1 exon 1 mRNA expression and H3K4 dimethylation and trimethylation of P1 on exon 1. Open bars represent AdLib/AdLib (control), solid bars represent FR/AdLib (FR during pregnancy), and hatched bars represent FR/FR (FR during pregnancy and lactation); a and b denote a significant difference between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR, P < 0.05. B: adult IGF1 exon 2 mRNA expression and H3K4 dimethylation and trimethylation of P2 on exon 2. Open bars represent AdLib/AdLib (control), solid bars represent FR/AdLib (FR during pregnancy), and hatched bars represent FR/FR (FR during pregnancy and lactation); a and b denote a significant difference between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR, P < 0.05.
ChIP.
At 1 day of age H3K4 dimethylation was lower in the FR compared with the AdLib newborn at IGF1-P1 (47.2 ± 6.7%), IGF1-E5 (67.2 ± 7.4%), and IGF1-U3 (61.1 ± 6.7%). There were, however, no significant differences in the level of trimethylation at any IGF1 site in the FR and AdLib groups (Fig. 2, Table 2).
Table 2.
Newborn and adult levels of IGF1 regions of DNA associated with H3K4 dimethylation and trimethylation
| Group | Methyl | Promoter 1 | Promoter 2 | Exon 5 | Exon 6 | UTR 3 | UTR 4 |
|---|---|---|---|---|---|---|---|
| AdLib | Di | 100 ± 20.4 | 100 ± 13.7 | 100 ± 13.0 | 100 ± 21.3 | 100 ± 13.2 | 100 ± 27.8 |
| FR | Di | 47.2 ± 6.7a | 71.4 ± 7.1 | 67.2 ± 7.4a | 70.9 ± 7.6 | 61.1 ± 6.7a | 74.7 ± 15.6 |
| AdLib | Tri | 100 ± 34.9 | 100 ± 51.0 | 100 ± 32.2 | 100 ± 32.6 | 100 ± 41.6 | 100 ± 37.4 |
| FR | Tri | 48.7 ± 7.3 | 42.3 ± 25.0 | 65.7 ± 11.0 | 60.7 ± 6.6 | 25.7 ± 4.3 | 31.25 ± 6.6 |
| AdLib/ AdLib | Di | 100 ± 11.0c | 100 ± 15.6b | 100 ± 14.9c | 100 ± 37.1 | 100 ± 28.6b | 100 ± 20.0 |
| FR/AdLib | Di | 62.6 ± 7.7b | 85.3 ± 9.5b | 49.6 ± 3.8b | 50.0 ± 11.3 | 127.8 ± 28.6b | 70.3 ± 9.5 |
| FR/FR | Di | 55.7 ± 8.3 | 223.7 ± 54.5c | 95.7 ± 11.6c | 36.9 ± 4.7 | 233.3 ± 28.6c | 64.9 ± 12.0 |
| AdLib/AdLib | Tri | 100 ± 39.3 | 100 ± 33.6d,e | 100 ± 40.7 | 100 ± 48.8 | 100 ± 33.3 | 100 ± 20.1d |
| FR/AdLib | Tri | 289.5 ± 81.0 | 165.7 ± 37.9e | 181.4 ± 42.5 | 98.8 ± 58.7 | 204.8 ± 90.5 | 419.1 ± 134e |
| FR/FR | Tri | 244.5 ± 98.2 | 8.1 ± 2.0d | 82.3 ± 20.9 | 130.6 ± 75.2 | 54.8 ± 30.0 | 152.3 ± 36.1d |
Values are means ± SE. DNA is normalized to input level and expressed as a percent of control. Results are shown from 1 day of age of AdLib (controls) vs. FR (food-restricted pregnancy), from 9 mo of age of AdLib/AdLib (control) vs. FR/AdLib (food-restricted pregnancy and AdLib lactation), and FR/FR (food-restricted pregnancy and lactation). aDifferences between AdLib and FR newborns. b and cDifferences between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR in dimethylation. d and eDifferences between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR in trimethylation, P < 0.05.
At 9 mo of age there was significantly decreased H3K4 dimethylation at IGF1-P1 (62.6 ± 7.7%) and IGF1-E5 (49.6 ± 3.8%) and increased H3K4 trimethylation at IGF1-U4 (419.1 ± 134.0%) in the FR/AdLib group compared with AdLib/AdLib. Adult FR/FR offspring also had decreased H3K4 dimethylation at IGF1-P1 (55.7 ± 8.3%) and additionally had increased dimethylation at IGF1-P2 (223.7 ± 54.5%) and IGF1-U3 (233.3 ± 28.6%), and, whereas there were no differences in trimethylation compared with AdLib/AdLib, IGF-P2 (8.1 ± 2.0%) was significantly decreased compared with FR/AdLib (165.7 ± 37.9%) (Fig. 3, Table 2).
IGF1 protein.
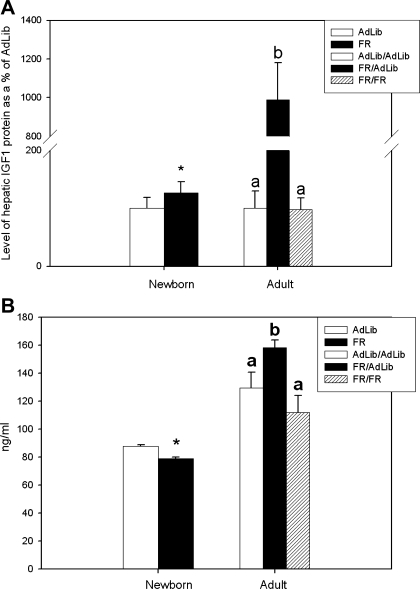
At 1 day of age, FR newborns had increased hepatic IGF1 protein levels compared with AdLib offspring (126.4 ± 18.9 vs. 100 ± 18.7%, P < 0.05) (Fig. 4A). At 9 mo of age, hepatic IGF1 protein was significantly increased in FR/AdLib compared with FR/FR and AdLib/AdLib (990 ± 190 vs. 98 ± 20 and 100 ± 30%, P < 0.001) (Fig. 4A).
Fig. 4.
A: quantification of newborn and adult liver IGF1 peptide relative to GAPDH and expressed as a percentage of AdLib (newborn) and AdLib/Adlib (adult). Newborn open bar represents AdLib, and solid bars represent FR; adult open bars represent AdLib/AdLib (control), solid bars represent FR/AdLib (FR during pregnancy), and hatched bar represents FR/FR (FR during pregnancy and lactation); asterisk denotes differences between AdLib and FR newborns; a and b denote a significant difference between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR, P < 0.05. B: newborn and adult plasma IGF1 concentrations (ng/ml). Newborn open bar represents AdLib, and solid bars represent FR; adult open bar represents AdLib/AdLib (control), solid bar represents FR/AdLib (FR during pregnancy), and hatched bar represents FR/FR (FR during pregnancy and lactation); asterisk denotes differences between AdLib and FR newborns, P < 0.01; a and b denote a significant difference between the adult groups AdLib/AdLib, FR/AdLib, and FR/FR, P < 0.05.
Plasma IGF1.
At 1 day of age, FR newborns had decreased plasma IGF1 compared with control offspring (79 ± 1 vs. 88 ± 1 ng/ml, P < 0.01) (Fig. 4B). Conversely at 9 mo of age, FR/AdLib males had significantly increased plasma IGF1 levels compared with FR/FR and control (158.5 ± 5.3 vs. 111.9 ± 12.2 and 129.4 ± 11.4 ng/ml, P < 0.01) (Fig. 4B).
DISCUSSION
We have previously shown that maternal FR during pregnancy results in IUGR offspring that when normally nursed show rapid catch-up growth and develop adult obesity and insulin resistance. However, continued nutrient restriction during nursing delays catch-up growth and prevents development of adult obesity. This study presents novel findings of epigenetic modifications to the histone structure associated with the IGF1 gene in IUGR offspring with differing adult phenotypes. Notably, IUGR newborns that undergo rapid catch-up growth and develop adult obesity (FR/AdLib) have increased postnatal hepatic IGF1 mRNA levels, likely a result of IGF1 histone and chromatin structure modifications to H3K4 trimethylation. Conversely, IUGR animals with delayed catch-up growth and absence of adult obesity (FR/FR) have levels similar to that of controls. Thus modulation of the rate of IUGR newborn catch-up growth may partially protect against IGF1 epigenetic modifications.
Consistent with previous studies (17), the IUGR newborns have reduced body and liver weights, including reduced plasma IGF1 levels. Notably, IUGR newborns clearly demonstrate disrupted hepatic histone methylation status in the IGF1 region. Specifically, H3K4 dimethylation was significantly decreased at the IGF1-P1, IGF1-E5, and IGF1-U3, whereas dimethylation of IGF1-P2, IGF1-E6, and IGF1-U4 as well as trimethylation of all IGF1 regions showed a decreased trend. Of particular interest from the methylation data is the large variation seen within the AdLib animals compared with the relatively small variation among the FR animals. This may demonstrate a critical window in which AdLib control animals are beginning to have increased dimethylation and trimethylation of IGF1, yet the developmentally delayed FR animals have yet to reach this period of growth. Despite the decreased H3K4 methylation in IUGR newborns, hepatic IGF1 mRNA expression was unaltered although protein expression was increased, suggesting a disruption in translation efficiency and exocytosis of the IGF1 protein. This concept is supported by the decreased plasma IGF1 level seen in IUGR, which is predominantly of liver origin.
IUGR newborns suckled by a normally fed control dam (FR/AdLib) exhibit rapid catch-up growth and as adults develop obesity and insulin resistance (17, 18). Consistent with increased growth, FR/AdLib offspring have markedly elevated plasma IGF1 levels, upregulation of hepatic IGF1 protein, and increased mRNA expression of multiple IGF1 regions. Furthermore, there was a persistent decrease in dimethylation of H3K4 at the IGF1-P1 and IGF1-E5. In contrast, trimethylation of H3K4 at IGF1-U4 is increased. Across all regions of IGF1, FR/AdLib offspring showed a trend toward decreased dimethylation, as these previously dimethylated lysine tails become further methylated as evidenced by the corresponding increased trimethylation. This overall increase in methylation can result in a structural change to the histone complex that has been shown to correspond with an increase in expression as we have seen also.
IUGR newborns continued to be suckled by a FR dam (FR/FR) exhibited delayed catch-up growth and absence of adult obesity (17, 18). FR/FR adult offspring have body weights comparable to the control although liver weight is lower. Nonetheless, plasma IGF1 levels and hepatic IGF1 protein expression are similar to the control offspring. Expression of hepatic mRNA for IGF1-E1, IGF1-E2, and IGF1A is the same as controls; however, mRNA from IGF1B shows an increase similar to the rapid catch-up group. Consistent with FR/AdLib, the FR/FR offspring too showed decreased dimethylation at IGF1-P1. However, in contrast to FR/AdLib, FR/FR had increased dimethylation at IGF1-P2 and IGF1-U3 and decreased trimethylation at IGF1-P2. Whereas the FR/AdLib offspring demonstrated decreasing dimethylation and increasing trimethylation across all regions of IGF1, the FR/FR offspring show trends of increasing dimethylation and decreasing trimethylation at IGF1-P2 and U3 and decreasing dimethylation and increased trimethylation at IGF1-P1, E6, and U4, possible evidence of a chromatin structure that is not as transcriptionally active as the more open FR/AdLib group, yet with increased activity compared with controls.
The link between IUGR and the development of adult morbidities such as metabolic syndrome has been extensively demonstrated in humans and various animal models (5, 20, 38, 55) although the etiology of these diseases remains unknown. Epigenetic changes may be one of the underlying mechanisms (8, 10, 21, 24, 26, 36, 53), as numerous studies have linked epigenetic modifications of DNA and histones with the development of cancer and aging (7, 19, 22, 23, 39, 44). Epigenetic instability, such as loss of imprinting and relaxation of the inactive X chromosome (3, 7, 37) as well as loss of methylation in mice at a rate of 0.01% per month (4, 22), has been associated with aging and specifically linked to environmental differences, such as nutrition and toxic substances during postnatal life (21, 23). In addition, mutations in the IGF1 pathway leading to reduced IGF1 levels have been shown to double the lifespan of Caenorhabditis elegans (29, 34) and extend the lifespan of drosophilia by 48% (14, 52). In humans, high levels of IGF1 are related to the development of cancer although some studies have shown that increased IGF1 has a beneficial impact on numerous diseases associated with aging. These studies have focused on the association of IGF1 with age-related disease, rather than actual lifespan (12, 51). It is apparent in humans that, although IGF1 is involved in aging and the development of age-associated morbidities, it is a dynamic system that requires a balance for the maximum benefit. Accordingly it could be postulated that overly increased IGF1 levels could thereby decrease lifespan, and aberrant epigenetic modifications that persist through life may further confound the problem.
Although it is clear that epigenetics plays a role in the alteration of transcription of the IGF1 gene into mRNA, other additional mechanisms may also be altered that would lead to the observed discordance between mRNA and peptide levels within the liver of IUGR newborns. Altered mRNA stability arising from different splice variants of IGF1 (40) and disruption in transcription or peptide exocytosis may account for the build up of the IGF1 peptide in IUGR livers, where no explanatory increase in mRNA is present. Goya et al. (28) have shown that the treatment of underfed diabetic rats with insulin leads to the partial recovery of IGF1 mRNA expression, and insulin combined with refeeding returned mRNA levels to normal. They also demonstrated, using fetal rat hepatocytes, that insulin treatment leads to increased stability of IGF1 mRNA as opposed to an increase in gene transcription (28). We have previously shown that plasma insulin levels in FR/AdLib offspring are decreased at 3 wk and increased at 9 mo of age (18). Although no data for plasma insulin have been collected in our animals, on the basis of data from 3 wk and 9 mo, it is possible that a reduced insulin level in the IUGR newborn may correspond with a reduction in IGF1 mRNA stability at day 1; similarly, an increase in IGF1 stability in the adult may occur in accordance with the increased insulin. IGF1 is not normally considered to be stored in vesicles within the cell but is rather secreted as it is produced (15, 42). A decreased rate of exocytosis from the hepatocyte may account for a build up of peptide even in the absence of increased mRNA and plasma IGF1.
In conclusion, this study demonstrates that IUGR leads to epigenetic modifications in the liver and alterations in hepatic IGF1 mRNA and peptide and plasma IGF1 levels. Most importantly, rapid catch-up growth during the postnatal period in IUGR offspring leads to increased H3K4 trimethylation with increased hepatic mRNA and plasma IGF1, which may partly contribute to an increase in liver and body weights. Thus modulation of the rate of IUGR newborn catch-up growth may protect against IGF1 epigenetic modifications.
GRANTS
This work was supported by National Institute of Child Health and Human Development Grant R03HD060241 and American Heart Association Grant 0865125.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
The authors thank Linda Day and Stacy Behare for assistance with animals.
REFERENCES
- 1.Adamo ML, Ben-Hur H, Roberts CT, Jr, LeRoith D. Regulation of start site usage in the leader exons of the rat insulin-like growth factor-I gene by development, fasting, and diabetes. Mol Endocrinol 5: 1677–1686, 1991 [DOI] [PubMed] [Google Scholar]
- 2.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75: 73–82, 1993 [PubMed] [Google Scholar]
- 3.Bandyopadhyay D, Medrano EE. The emerging role of epigenetics in cellular and organismal aging. Exp Gerontol 38: 1299–1307, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res 30: 2365–2373, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker DJ. The developmental origins of adult disease. Eur J Epidemiol 18: 733–736, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Barker DJ, Clark PM. Fetal undernutrition and disease in later life. Rev Reprod 2: 105–112, 1997 [DOI] [PubMed] [Google Scholar]
- 7.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics 165: 2055–2062, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Boushey CJ, Beresford SA, Omenn GS, Motulsky AG. A quantitative assessment of plasma homocysteine as a risk factor for vascular disease. Probable benefits of increasing folic acid intakes. JAMA 274: 1049–1057, 1995 [DOI] [PubMed] [Google Scholar]
- 10.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet 1: R95–R101, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Carro E, Torres-Aleman I. Serum insulin-like growth factor I in brain function. Keio J Med 55: 59–63, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Ceda GP, Dall'Aglio E, Maggio M, Lauretani F, Bandinelli S, Falzoi C, Grimaldi W, Ceresini G, Corradi F, Ferrucci L, Valenti G, Hoffman AR. Clinical implications of the reduced activity of the GH-IGF-I axis in older men. J Endocrinol Invest 28: 96–100, 2005 [PubMed] [Google Scholar]
- 13.Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Mol Endocrinol 19: 563–573, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292: 104–106, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Daughaday WH, Rotwein P. Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr Rev 10: 68–91, 1989 [DOI] [PubMed] [Google Scholar]
- 16.Desai M, Gayle D, Babu J, Ross MG. Permanent reduction in heart and kidney organ growth in offspring of undernourished rat dams. Am J Obstet Gynecol 193: 1224–1232, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Desai M, Gayle D, Babu J, Ross MG. Programmed obesity in intrauterine growth-restricted newborns: modulation by newborn nutrition. Am J Physiol Regul Integr Comp Physiol 288: R91–R96, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Desai M, Gayle D, Babu J, Ross MG. The timing of nutrient restriction during rat pregnancy/lactation alters metabolic syndrome phenotype. Am J Obstet Gynecol 196: 555 e551–e557, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Eriksson JG, Forsen T, Tuomilehto J, Jaddoe VW, Osmond C, Barker DJ. Effects of size at birth and childhood growth on the insulin resistance syndrome in elderly individuals. Diabetologia 45: 342–348, 2002 [DOI] [PubMed] [Google Scholar]
- 21.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res 600: 46–57, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 102: 10604–10609, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Q, McKnight RA, Yu X, Callaway CW, Lane RH. Growth retardation alters the epigenetic characteristics of hepatic dual specificity phosphatase 5. FASEB J 20: 2127–2129, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Fu Q, McKnight RA, Yu X, Wang L, Callaway CW, Lane RH. Uteroplacental insufficiency induces site-specific changes in histone H3 covalent modifications and affects DNA-histone H3 positioning in day 0 IUGR rat liver. Physiol Genomics 20: 108–116, 2004 [DOI] [PubMed] [Google Scholar]
- 26.Fu Q, Yu X, Callaway CW, Lane RH, McKnight RA. Epigenetics: intrauterine growth retardation (IUGR) modifies the histone code along the rat hepatic IGF-1 gene. FASEB J 23: 2438–2449, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gicquel C, Le Bouc Y. Hormonal regulation of fetal growth. Horm Res 65, Suppl 3: 28–33, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Goya L, de la Puente A, Ramos S, Martin MA, Escriva F, Alvarez C, Pascual-Leone AM. Regulation of IGF-I and -II by insulin in primary cultures of fetal rat hepatocytes. Endocrinology 142: 5089–5096, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature 408: 255–262, 2000 [DOI] [PubMed] [Google Scholar]
- 30.Hankey GJ, Eikelboom JW. Homocysteine and stroke. Curr Opin Neurol 14: 95–102, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Herran A, Garcia-Unzueta MT, Amado JA, Lopez-Cordovilla JJ, Diez-Manrique JF, Vazquez-Barquero JL. Folate levels in psychiatric outpatients. Psychiatry Clin Neurosci 53: 531–533, 1999 [DOI] [PubMed] [Google Scholar]
- 32.Holt RI. Fetal programming of the growth hormone-insulin-like growth factor axis. Trends Endocrinol Metab 13: 392–397, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet Suppl 33: 245–254, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Kenyon C. A conserved regulatory system for aging. Cell 105: 165–168, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75: 59–72, 1993 [PubMed] [Google Scholar]
- 36.MacLennan NK, James SJ, Melnyk S, Piroozi A, Jernigan S, Hsu JL, Janke SM, Pham TD, Lane RH. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics 18: 43–50, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Martin GM. Epigenetic drift in aging identical twins. Proc Natl Acad Sci USA 102: 10413–10414, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev 85: 571–633, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Novak K. Epigenetics changes in cancer cells. MedGenMed 6: 17, 2004 [PMC free article] [PubMed] [Google Scholar]
- 40.Ohlsen SM, Dean DM, Wong EA. Characterization of multiple transcription initiation sites of the ovine insulin-like growth factor-I gene and expression profiles of three alternatively spliced transcripts. DNA Cell Biol 12: 243–251, 1993 [DOI] [PubMed] [Google Scholar]
- 41.Pell JM, Saunders JC, Gilmour RS. Differential regulation of transcription initiation from insulin-like growth factor-I (IGF-I) leader exons and of tissue IGF-I expression in response to changed growth hormone and nutritional status in sheep. Endocrinology 132: 1797–1807, 1993 [DOI] [PubMed] [Google Scholar]
- 42.Plisetskaya EM. Some of my not so favorite things about insulin and insulin-like growth factors in fish. Comp Biochem Physiol B Biochem Mol Biol 121: 3–11, 1998 [DOI] [PubMed] [Google Scholar]
- 43.Randhawa R, Cohen P. The role of the insulin-like growth factor system in prenatal growth. Mol Genet Metab 86: 84–90, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Santos-Reboucas CB, Pimentel MM. Implication of abnormal epigenetic patterns for human diseases. Eur J Hum Genet 15: 10–17, 2007 [DOI] [PubMed] [Google Scholar]
- 45.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108, 2008 [DOI] [PubMed] [Google Scholar]
- 47.Schwartz J, Morrison JL. Impact and mechanisms of fetal physiological programming. Am J Physiol Regul Integr Comp Physiol 288: R11–R15, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Shemer J, Adamo ML, Roberts CT, Jr, LeRoith D. Tissue-specific transcription start site usage in the leader exons of the rat insulin-like growth factor-I gene: evidence for differential regulation in the developing kidney. Endocrinology 131: 2793–2799, 1992 [DOI] [PubMed] [Google Scholar]
- 49.Sjogren K, Jansson JO, Isaksson OG, Ohlsson C. A transgenic model to determine the physiological role of liver-derived insulin-like growth factor I. Minerva Endocrinol 27: 299–311, 2002 [PubMed] [Google Scholar]
- 50.Smith PJ, Spurrell EL, Coakley J, Hinds CJ, Ross RJ, Krainer AR, Chew SL. An exonic splicing enhancer in human IGF-I pre-mRNA mediates recognition of alternative exon 5 by the serine-arginine protein splicing factor-2/alternative splicing factor. Endocrinology 143: 146–154, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Tang BL. SIRT1, neuronal cell survival and the insulin/IGF-1 aging paradox. Neurobiol Aging 27: 501–505, 2006 [DOI] [PubMed] [Google Scholar]
- 52.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110, 2001 [DOI] [PubMed] [Google Scholar]
- 53.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20: 63–68, 2004 [DOI] [PubMed] [Google Scholar]
- 54.Wolffe AP. Packaging principle: how DNA methylation and histone acetylation control the transcriptional activity of chromatin. J Exp Zool 282: 239–244, 1998 [PubMed] [Google Scholar]
- 55.Wu G, Bazer FW, Cudd TA, Meininger CJ, Spencer TE. Maternal nutrition and fetal development. J Nutr 134: 2169–2172, 2004 [DOI] [PubMed] [Google Scholar]
- 56.Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA 96: 7324–7329, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]