Abstract
The multicellular green alga Volvox carteri and its morphologically diverse close relatives (the volvocine algae) are well suited for the investigation of the evolution of multicellularity and development. We sequenced the 138–mega–base pair genome of V. carteri and compared its ~14,500 predicted proteins to those of its unicellular relative Chlamydomonas reinhardtii. Despite fundamental differences in organismal complexity and life history, the two species have similar protein-coding potentials and few species-specific protein-coding gene predictions. Volvox is enriched in volvocine-algal–specific proteins, including those associated with an expanded and highly compartmentalized extracellular matrix. Our analysis shows that increases in organismal complexity can be associated with modifications of lineage-specific proteins rather than large-scale invention of protein-coding capacity.
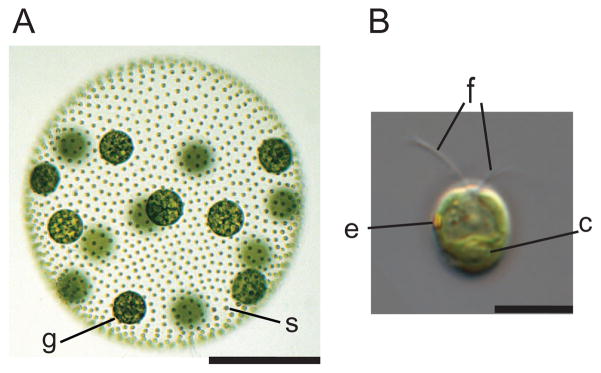
Multicellularity and cellular differentiation evolved independently in diverse lineages, including green and red algae, animals, fungi, plants, Amoebozoa, and Chromalveolates (1) (fig. S1A), yet the genetic changes that underlie these transitions remain poorly understood. The volvocine algae, which include both unicellular and multicellular species with various levels of morphological and developmental complexity, are an appealing model for studying such an evolutionary transition (2) [fig. S2 and supporting online material (SOM) text]. Multicellular Volvox carteri (hereafter Volvox) has two cell types: ~2000 small biflagellate somatic cells that are embedded in the surface of a transparent sphere of glycoprotein-rich extracellular matrix (ECM), and ~16 large reproductive cells (termed gonidia) that lie just below the somatic cell monolayer (2) (Fig. 1A and figs. S1B and S3). The somatic cells resemble those of Chlamydomonas reinhardtii, a model unicellular volvocine alga (3) (Fig. 1B). The changes that are associated with the evolution of Volvox from a Chlamydomonas-like unicellular ancestor have clear parallels in other multicellular lineages but took place more recently than in land plants and animals (4).
Fig. 1.
Volvox and Chlamydomonas. (A) Adult Volvox is composed of ~2000 Chlamydomonas-like somatic cells (s) and ~16 large germline gonidia (g) (scale bar, 200 μm) (fig. S1B). (B) Chlamydomonas cell showing apical flagella (f), chloroplast (c), and eyespot (e) (scale bar, 10 μm). The microscopy used is described in (5).
To begin to characterize the genomic features that are associated with volvocine multicellularity, we sequenced the 138–mega–base pair (Mbp) Volvox genome to ~11.1× redundant coverage (~2.9 million reads) using a whole-genome shotgun strategy (5). The assembly captures over 98% of known mRNA sequences and expressed sequence tags (ESTs) (5). The Volvox nuclear genome is 19.6 Mbp (17%) larger than the Chlamydomonas genome (Table 1), primarily because of increased repeat content in Volvox relative to Chlamydomonas (5) (table S1). Whereas a few repeat families show bursts of expansion in the Volvox and Chlamydomonas lineages, most have changed gradually (fig. S4) (5).
Table 1.
Comparison of the Volvox and Chlamydomonas genomes.
| Species | Genome size (Mbp) | Number of chromosomes | % G and C | Protein-coding loci | % coding | % of genes with introns | Introns per gene | Median intron length (bp) |
|---|---|---|---|---|---|---|---|---|
| V. carteri | 138 | 14* | 56 | 14,520 | 18.0 | 92 | 7.05 | 358 |
| C. reinhardtii | 118 | 17 | 64 | 14,516 | 16.3 | 91 | 7.4 | 174 |
See (15).
The sequence divergence between Volvox and Chlamydomonas is comparable to that between human and chicken [which diverged ~310 million years ago (Ma)], human and frog (~350 Ma), and Arabidopsis and poplar (~110 Ma), based on the frequency of synonymous transversions at fourfold-degenerate sites (4DTV distance) (5, 6) (table S2). Although conserved synteny between Volvox and Chlamydomonas genomes is evident, these volvocine algae show higher rates of genomic rearrangement than vertebrates and eudicots do (tables S2 to S4 and fig. S5).
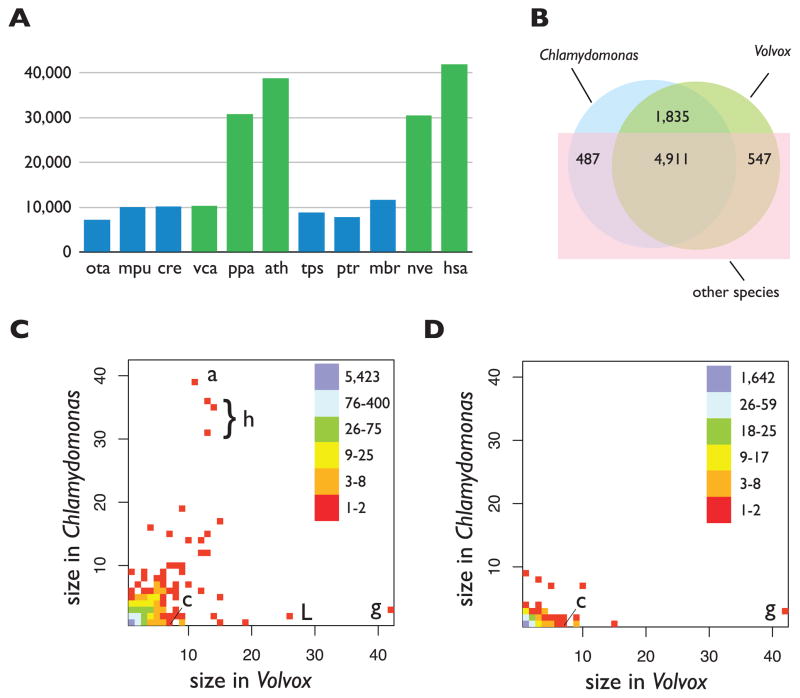
We predicted 14,566 proteins (at 14,520 loci) in Volvox (5) (Table 1 and tables S5 to S7). Volvox and Chlamydomonas have similar numbers of genes (Table 1) (3) and more genes than most unicellular organisms (table S8). Genes in both algae are intron-rich (Table 1), like those of most multicellular organisms (table S8), and introns are longer, on average, in Volvox (fig. S6) (5). Novel protein domains and/or combinations are proposed to have contributed to multicellularity in metazoans (7), and such expansions are evident in both the plant and animal lineages (Fig. 2A and table S9). In contrast, the numbers of domains and combinations in Volvox are very similar to those in Chlamydomonas and other unicellular species (Fig. 2A and table S9) (5). microRNAs (miRNAs) have been identified in Chlamydomonas, most of which have no homologs in Volvox (8, 9). It is likely that Volvox also has miRNAs, but these have yet to be characterized.
Fig. 2.
Comparisons of protein domains and families. (A) Total number of Pfam domains in the multicellular (green) and unicellular (blue) species: Ostreococcus tauri (ota); Micromonas pusilla (mpu); Chlamydomonas reinhardtii (cre); Volvox carteri (vca); Physcomitrella patens (ppa); Arabidopsis thaliana (ath); Thalassiosira pseudonana (tps); Phaeodactylum tricornutum (ptr); Monosiga brevicollis (mbr); Nematostella vectensis (nve); and Homo sapiens (hsa). (B) The numbers of protein families from Volvox, Chlamydomonas, and other species (5) are shown in a Venn diagram. The numbers of Volvox and Chlamydomonas members per protein family are plotted for all families (C) and for the volvocine algae–specific subset (D). In these density plots, the position of each square represents the number of family members in Volvox (x axis) and Chlamydomonas (y axis), with coloring to indicate the total number of families plotted at each position. The Pfam domains for outlier families are abbreviated as follows: a, ankyrin repeat; c, cysteine protease; g, gametolysin; h, histone; L, LRR.
To investigate protein evolution in Chlamydomonas and Volvox, we constructed families containing both orthologs and paralogs from 20 diverse species, including animals, plants, fungi, protists, and bacteria (5) (table S10). We assigned 9311 (64%) Volvox and 9189 (63%) Chlamydomonas protein sequences to 7780 families (Fig. 2B), of which 80% (5423) contain one ortholog from each alga (table S11). 1835 families (26%) contain orthologs only from Volvox and Chlamydomonas (that is, they are volvocine-specific) (Fig. 2B). Only 32 EST-supported Volvox gene models lack detectable homologs in Chlamydomonas or other species (5) (tables S12 and S13), suggesting that limited protein-coding innovation occurred in the Volvox lineage.
Gene-family expansion or contraction is an important source of adaptive variation (10, 11). In a density plot of proteins per family in Volvox versus Chlamydomonas (Fig. 2C), most points lie on or near the diagonal, showing that the majority of families have approximately equal membership from each alga. Exceptions include the gametolysin/VMP (Volvox matrix metalloprotease) family, whose substrates are cell-wall/ECM proteins (12) (Fig. 2C, g), and a family containing leucine-rich repeat proteins (LRRs), whose functions in green algae have not been well defined (Fig. 2C, L). Conversely, families containing core histones and ankyrin repeats have more members in Chlamydomonas (Fig. 2C, a). In contrast, the subset of 1835 volvocine algae–specific families (5) shows a strikingly different distribution (Fig. 2D), with a significant bias toward more members in Volvox (P = 2 × 10−120, heterogeneity chi-squared test). These families include ECM proteins such as VMPs and pherophorins that both participate in ECM biogenesis (12), and an algal subgroup of cysteine proteases (Fig. 2D, c).
Although some of the genomic differences between Volvox and Chlamydomonas may reflect environmental adaptations that have not been extensively investigated (5), we expected many of the changes to be in protein families that are associated with the large differences in organismal complexity. Therefore, we investigated in detail pathways related to key developmental processes that are either novel or qualitatively different in Volvox relative to Chlamydomonas (2). These include the following: protein secretion and membrane trafficking [potentially involved in cytoplasmic bridge formation via incomplete cytokinesis (13, 14)]; the cytoskeleton [potentially involved in Volvox-specific basal body rotation, inversion, and asymmetric cell division (15)]; ECM and cell-wall proteins [involved in ECM expansion, sexual differentiation, and morphogenesis (12) (fig. S2)]; and cell-cycle regulation (potentially involved in cell division patterning or asymmetric cell division). The components of these pathways are nearly identical in Volvox and Chlamydomonas (table S14). Transcription-related proteins also have highly similar repertoires in the two species (fig. S7 and table S15) (5). Thus, with three exceptions (see below), we found little difference in the complements of proteins that might underlie developmental complexity in Volvox.
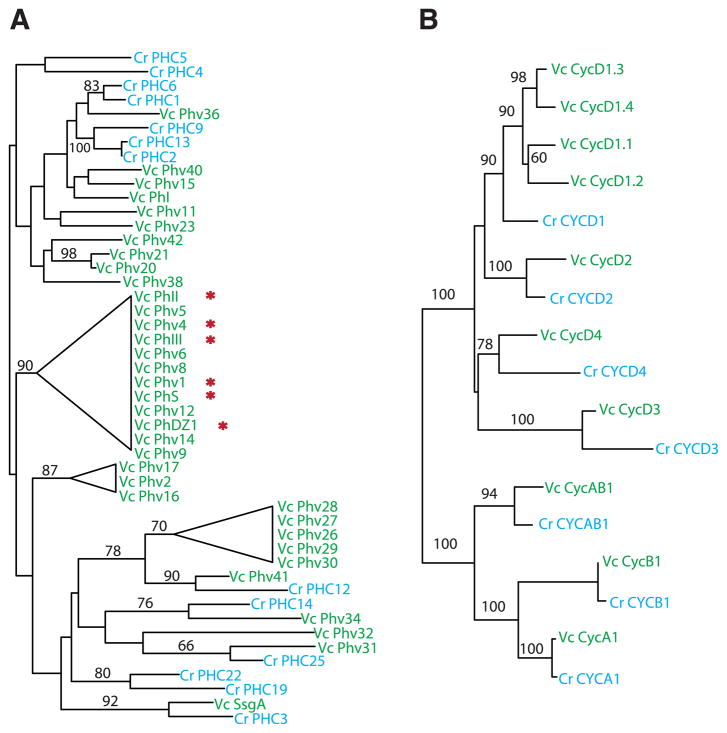
The ECM composes up to 99% of an adult Volvox spheroid and is larger and more structurally complex than the ancestral Chlamydomonas-like cell wall from which it was derived (12) (fig. S3 and SOM text). These changes are mirrored by at least two dramatic changes in ECM protein family size in Volvox as compared with Chlamydomonas: pherophorins (49 versus 27 members) and VMPs (42 versus 8 members) (Fig. 3A, fig. S8, and table S14). We found expanded Volvox-specific clades of pherophorins and VMPs as well as species-specific duplications in both algae (Fig. 3A and fig. S8). Besides their role in ECM structure, Volvox pherophorins have evolved into a diffusible sex-inducer glycoprotein that has replaced nitrogen deprivation (used in Chlamydomonas and other volvocine algae) as the trigger for sexual differentiation (16). The co-option of an ECM protein for sexual signaling shows parallels in the sexual agglutinins of Chlamydomonas that are themselves related to cell-wall/ECM proteins (17). The Volvox ECM proteins, pherophorins, and VMPs diversified and then presumably were recruited to novel developmental roles in Volvox, thus representing a source of adaptive plasticity that is specific to the volvocine algae.
Fig. 3.
Diversification of key protein families with known or predicted roles in Volvox development. Unrooted maximum likelihood trees (5) are shown for pherophorins (A) and cyclins (B). Protein sequences are from Volvox (Vc, green) and Chlamydomonas (Cr, blue). Incomplete gene models were not included; Volvox-specific clades with poorly resolved branches are collapsed into triangles; bootstrap support ≥50% is indicated on branches. Red asterisks indicate pherophorins whose mRNA levels are up-regulated by a sex inducer (16).
The Volvox and Chlamydomonas cell cycles are fundamentally similar, but Volvox has evolved additional regulation of timing, number, and types of cell divisions (symmetric and asymmetric) among different subsets of embryonic cells (2). The division program of males and females is further modified during sexual development to produce sperm and eggs (18). Whereas most of the core cell-cycle proteins of Volvox and Chlamydomonas have a 1:1 orthology relationship, the cyclin D family is notably larger in Volvox. In addition to three D cyclins that have Chlamydomonas orthologs (Cycd2, Cycd3, and Cycd4), Volvox has four D1-related cyclins (Cycd1.1 to Cycd1.4), whereas Chlamydomonas has only one (Fig. 3B). D cyclins bind cyclin-dependent kinases and target them to phosphorylate retinoblastoma (RB)–related proteins (19). In Chlamydomonas, the RB-related protein MAT3 controls the timing and extent of cell division (20), so it is plausible that the expanded D-type cyclin family in Volvox plays a role in regulating its cell division program during development.
The genetic changes that brought about the evolution of multicellular life from unicellular progenitors remain obscure (2, 21, 22). For example, many proteins associated with animal multicellularity, such as cadherins and receptor tyrosine kinases (23), evolved in the unicellular ancestor of animals and are specific to its descendants. Other critical components of metazoan multicellularity, including key transcription factors and signaling molecules, are absent from the closest unicellular relatives of animals (22), suggesting that animal multicellularity also involved protein-coding innovation. Our comparisons of Volvox and Chlamydomonas indicate that, with the interesting exceptions of pherophorins, VMPs, and D cyclins, the developmental innovations in the Volvox lineage did not involve major changes in the ancestral protein repertoire. This is consistent with previous observations indicating co-option of ancestral genes into new developmental processes without changes in copy number or function (24–26). However, our analyses do suggest that the expansion of lineage-specific proteins occurred preferentially in Volvox and provided a key source of developmental innovation and adaptation. Further studies of gene regulation (27) and the role of noncoding RNAs (28) will be enabled by the Volvox genome sequence, allowing a more complete understanding of the transformation from a cellularly complex Chlamydomonas-like ancestor to a morphologically and developmentally complex “fierce roller.”
Supplementary Material
Acknowledgments
The work conducted by the Joint Genome Institute of the U.S. Department of Energy is supported by the Office of Science of the U.S. Department of Energy under contract number DE-AC02-05CH11231 and by NIH grant R01 GM078376 and a Coypu Foundation grant to J.U.; a grant from the Natural Sciences and Engineering Research Council–Canada to A.M.N.; NSF grants IBN-0444896 and IBN-0744719 to S.M.M.; Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research numbers 20247032 and 22570203 to I.N.; and NIH grant 5 P41 LM006252 to J.J. We thank M. Cipriano for Pfam annotations; E. Hom, E. Harris, and M. Stanke for Augustus u9 gene models; and R. Howson for artwork. Sequence data from this study are deposited at the DNA Databank of Japan/European Molecular Biology Laboratory/GenBank under the project accession no. ACJH00000000.
Footnotes
References and Notes
- 1.Baldauf SL. Science. 2003;300:1703. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 2.Kirk DL. Bioessays. 2005;27:299. doi: 10.1002/bies.20197. [DOI] [PubMed] [Google Scholar]
- 3.Merchant SS, et al. Science. 2007;318:245. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herron MD, Hackett JD, Aylward FO, Michod RE. Proc Natl Acad Sci USA. 2009;106:3254. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Materials and methods are available as supporting material on Science Online.
- 6.Reisz RR, Müller J. Trends Genet. 2004;20:237. doi: 10.1016/j.tig.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Putnam NH, et al. Science. 2007;317:86. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 8.Zhao T, et al. Genes Dev. 2007;21:1190. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. Nature. 2007;447:1126. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 10.Lespinet O, Wolf YI, Koonin EV, Aravind L. Genome Res. 2002;12:1048. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J. Trends Ecol Evol. 2003;18:292. [Google Scholar]
- 12.Hallmann A. Int Rev Cytol. 2003;227:131. doi: 10.1016/s0074-7696(03)01009-x. [DOI] [PubMed] [Google Scholar]
- 13.Green KJ, Kirk DL. J Cell Biol. 1981;91:743. doi: 10.1083/jcb.91.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green KJ, Viamontes GI, Kirk DL. J Cell Biol. 1981;91:756. doi: 10.1083/jcb.91.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirk D. Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge Univ. Press; Cambridge: 1998. [Google Scholar]
- 16.Hallmann A. Plant J. 2006;45:292. doi: 10.1111/j.1365-313X.2005.02627.x. [DOI] [PubMed] [Google Scholar]
- 17.Ferris PJ, et al. Plant Cell. 2005;17:597. doi: 10.1105/tpc.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferris P, et al. Science. 2010;328:351. doi: 10.1126/science.1186222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Veylder L, Beeckman T, Inzé D. Nat Rev Mol Cell Biol. 2007;8:655. doi: 10.1038/nrm2227. [DOI] [PubMed] [Google Scholar]
- 20.Fang S-C, de los Reyes C, Umen JG. PLoS Genet. 2006;2:e167. doi: 10.1371/journal.pgen.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buss LW. The Evolution of Individuality. Princeton Univ. Press; Princeton, NJ: 2006. [Google Scholar]
- 22.King N, et al. Nature. 2008;451:783. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King N, Hittinger CT, Carroll SB. Science. 2003;301:361. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 24.Miller SM, Kirk DL. Development. 1999;126:649. doi: 10.1242/dev.126.4.649. [DOI] [PubMed] [Google Scholar]
- 25.Nishii I, Ogihara S, Kirk DL. Cell. 2003;113:743. doi: 10.1016/s0092-8674(03)00431-8. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Q, Fowler R, Tam LW, Edwards L, Miller SM. Dev Genes Evol. 2003;213:328. doi: 10.1007/s00427-003-0332-x. [DOI] [PubMed] [Google Scholar]
- 27.Carroll SB. Cell. 2008;134:25. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 28.Peterson KJ, Dietrich MR, McPeek MA. Bioessays. 2009;31:736. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.